RNA and Protein Determinants Mediate Differential Binding of miRNAs by a Viral Suppressor of RNA Silencing Thus Modulating Antiviral Immune Responses in Plants
Abstract
:1. Introduction
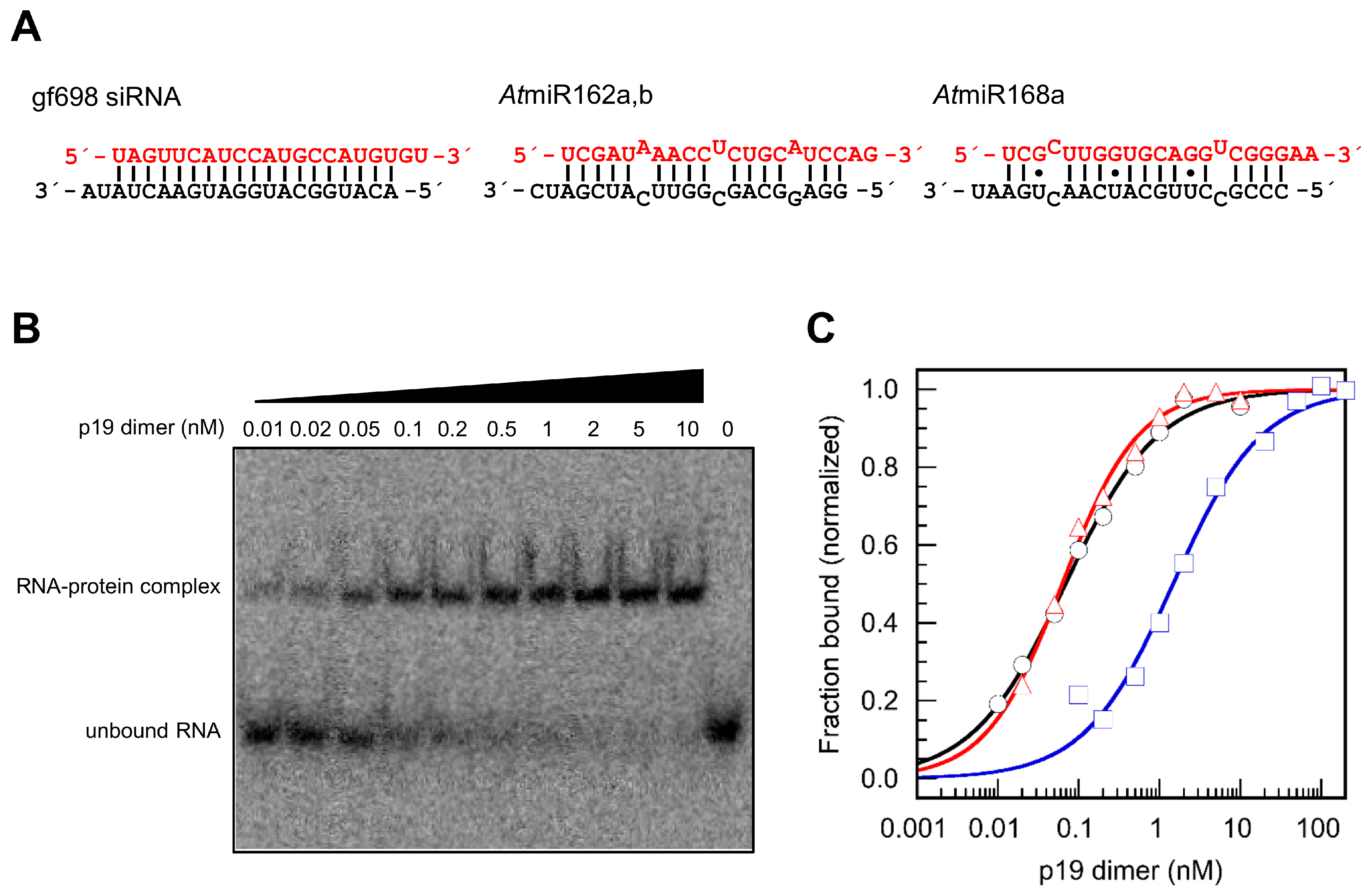
2. Results
3. Discussion
4. Materials and Methods
4.1. Plasmid Constructs
4.2. Protein Expression and Purification
4.3. Origin, Generation and Modification of Examined Small RNAs (sRNAs)
4.4. Determination of Binding Affinities
4.5. Cell Culture and Preparation of BYL
4.6. In Vitro Translation of p19 Variants
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Ding, S.W. RNA-based antiviral immunity. Nat. Rev. Immunol. 2010, 10, 632–644. [Google Scholar] [CrossRef] [PubMed]
- Pantaleo, V. Plant RNA silencing in viral defence. Adv. Exp. Med. Biol. 2011, 722, 39–58. [Google Scholar] [PubMed]
- Blevins, T.; Rajeswaran, R.; Shivaprasad, P.V.; Beknazariants, D.; Si-Ammour, A.; Park, H.S.; Vazquez, F.; Robertson, F.C.; Meins, F., Jr.; Hohn, H.; et al. Four plant Dicers mediate viral small RNA biogenesis and DNA virus induced silencing. Nucleic Acids Res. 2006, 34, 6233–6246. [Google Scholar] [CrossRef] [Green Version]
- Deleris, A.; Gallego-Bartolome, J.; Bao, J.; Kasschau, K.D.; Carrington, J.C.; Voinnet, O. Hierarchical action and inhibition of plant Dicer-like proteins in antiviral defense. Science 2006, 313, 68–71. [Google Scholar] [CrossRef]
- Weiberg, A.; Jin, H. Small RNAs—The secret agents in the plant-pathogen interaction. Curr. Opin. Plant Biol. 2015, 26, 87–94. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Garcia-Ruiz, H.; Takeda, A.; Chapman, E.J.; Sullivan, C.M.; Fahlgren, N.; Brempelis, K.J.; Carrington, J.C. Arabidopsis RNA-dependent RNA polymerases and Dicer-like proteins in antiviral defense and small interfering RNA biogenesis during Turnip Mosaic Virus infection. Plant Cell 2010, 22, 481–496. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Qu, F.; Ye, X.; Morris, T.J. Arabidopsis DRB4, AGO1, AGO7, and RDR6 participate in a DCL4-initiated antiviral RNA silencing pathway negatively regulated by DCL1. Proc. Natl. Acad. Sci. USA 2008, 105, 14732–14737. [Google Scholar] [CrossRef] [Green Version]
- Kalantidis, K.; Schumacher, H.T.; Alexiadis, T.; Helm, J.M. RNA silencing movement in plants. Biol. Cell 2008, 100, 13–26. [Google Scholar] [CrossRef] [Green Version]
- Wang, X.B.; Jovel, J.; Udomporn, P.; Wang, Y.; Wu, Q.; Li, W.X.; Gasciolli, V.; Vaucheret, H.; Ding, S.W. The 21-nucleotide, but not 22-nucleotide, viral secondary small interfering RNAs direct potent antiviral defense by two cooperative argonautes in Arabidopsis thaliana. Plant Cell 2011, 23, 1625–1638. [Google Scholar] [CrossRef] [Green Version]
- Szittya, G.; Molnár, A.; Silhavy, D.; Hornyik, C.; Burgyán, J. Short defective interfering RNAs of tombusviruses are not targeted but trigger post-transcriptional gene silencing against their helper virus. Plant Cell 2002, 14, 359–372. [Google Scholar] [CrossRef]
- Meister, G. Argonaute proteins: Functional insights and emerging roles. Nat. Rev. Genet. 2013, 14, 447–459. [Google Scholar] [CrossRef] [PubMed]
- Mi, S.; Cai, T.; Hu, Y.; Chen, Y.; Hodges, E.; Ni, F.; Wu, L.; Li, S.; Zhou, H.; Long, C.; et al. Sorting of small RNAs into Arabidopsis argonaute complexes is directed by the 5′-terminal nucleotide. Cell 2008, 133, 116–127. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Omarov, R.T.; Ciomperlik, J.J.; Scholthof, H.B. RNAi-associated ssRNA-specific ribonucleases in Tombusvirus P19 mutant-infected plants and evidence for a discrete siRNA-containing effector complex. Proc. Natl. Acad. Sci. USA 2007, 104, 1714–1719. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pantaleo, V.; Szittya, G.; Burgyán, J. Molecular bases of viral RNA targeting by viral small interfering RNA-programmed RISC. J. Virol. 2007, 81, 3797–3806. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Carbonell, A.; Carrington, J.C. Antiviral roles of plant ARGONAUTES. Curr. Opin. Plant Biol. 2015, 27, 111–117. [Google Scholar] [CrossRef] [Green Version]
- Harvey, J.J.; Lewsey, M.G.; Patel, K.; Westwood, J.; Heimstädt, S.; Carr, J.P.; Baulcombe, D.C. An antiviral defense role of AGO2 in plants. PLoS ONE 2011, 6, e14639. [Google Scholar] [CrossRef] [PubMed]
- Fátyol, K.; Ludman, M.; Burgyán, J. Functional dissection of a plant argonaute. Nucleic Acids Res. 2016, 44, 1384–1397. [Google Scholar] [CrossRef] [Green Version]
- Schuck, J.; Gursinsky, T.; Pantaleo, V.; Burgyán, J.; Behrens, S.E. AGO/RISC-mediated antiviral RNA silencing in a plant in vitro system. Nucleic Acids Res. 2013, 41, 5090–5103. [Google Scholar] [CrossRef] [Green Version]
- Carbonell, A.; Fahlgren, N.; Garcia-Ruiz, H.; Gilbert, K.B.; Montgomery, T.A.; Nguyen, T.; Cuperus, J.T.; Carrington, J.C. Functional analysis of three Arabidopsis ARGONAUTES using slicer-defective mutants. Plant Cell 2012, 24, 3613–3629. [Google Scholar] [CrossRef] [Green Version]
- Csorba, T.; Kontra, L.; Burgyán, J. Viral silencing suppressors: Tools forged to fine-tune host-pathogen coexistence. Virology 2015, 479–480, 85–103. [Google Scholar] [CrossRef] [Green Version]
- Silhavy, D.; Molnár, A.; Lucioli, A.; Szittya, G.; Hornyik, C.; Tavazza, M. A viral protein suppresses RNA silencing and binds silencing-generated, 21- to 25-nucleotide double-stranded RNAs. EMBO J. 2002, 21, 3070–3080. [Google Scholar] [CrossRef] [PubMed]
- Vargason, J.M.; Szittya, G.; Burgyán, J.; Hall, T.M.T. Size selective recognition of siRNA by an RNA silencing suppressor. Cell 2003, 115, 799–811. [Google Scholar] [CrossRef] [Green Version]
- Ye, K.; Malinina, L.; Patel, D.J. Recognition of small interfering RNA by a viral suppressor of RNA silencing. Nature 2003, 426, 874–878. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lakatos, L.; Csorba, T.; Pantaleo, V.; Chapman, E.J.; Carrington, J.C.; Liu, Y.P.; Dolja, V.V.; Calvino, L.F.; López-Moya, J.J.; Burgyán, J. Small RNA binding is a common strategy to suppress RNA silencing by several viral suppressors. EMBO J. 2006, 25, 2768–2780. [Google Scholar] [CrossRef] [Green Version]
- Omarov, R.T.; Sparks, K.; Smith, L.; Zindovic, J.; Scholthof, H.B. Biological Relevance of a Stable Biochemical Interaction between the Tombusvirus-encoded P19 and short interfering RNAs. J. Virol. 2006, 80, 3000–3008. [Google Scholar] [CrossRef] [Green Version]
- Rogers, K.; Chen, X. Biogenesis, turnover, and mode of action of plant microRNAs. Plant Cell 2013, 25, 2383–2399. [Google Scholar] [CrossRef] [Green Version]
- Iwakawa, H.O.; Tomari, Y. Molecular insights into microRNA-mediated translational repression in plants. Mol. Cell 2013, 52, 591–601. [Google Scholar] [CrossRef] [Green Version]
- Iwakawa, H.O.; Tomari, Y. The functions of microRNAs: mRNA decay and translational repression. Trends Cell Biol. 2015, 25, 651–665. [Google Scholar] [CrossRef]
- Lanet, E.; Delannoy, E.; Sormani, R.; Floris, M.; Brodersen, P.; Créte, P.; Voinnet, O.; Robaglia, C. Biochemical evidence for translational repression by Arabidopsis microRNAs. Plant Cell 2009, 21, 1762–1768. [Google Scholar] [CrossRef] [Green Version]
- Xie, Z.; Kasschau, K.D.; Carrington, J.C. Negative feedback regulation of Dicer-like1 in Arabidopsis by microRNA-guided mRNA degradation. Curr. Biol. 2003, 13, 784–789. [Google Scholar] [CrossRef] [Green Version]
- Vaucheret, H.; Vazquez, F.; Créte, P.; Bartel, D.P. The action of ARGONAUTE1 in the miRNA pathway and its regulation by the miRNA pathway are crucial for plant development. Genes Dev. 2004, 18, 1187–1197. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Várallyay, É.; Válóczi, A.; Agyi, A.; Burgyán, J.; Havelda, Z. Plant virus-mediated induction of miR168 is associated with repression of ARGONAUTE1 accumulation. EMBO J. 2010, 29, 3507–3519. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vaucheret, H.; Mallory, A.C.; Bartel, D.P. AGO1 homeostasis entails coexpression of MIR168 and AGO1 and preferential stabilization of miR168 by AGO1. Mol. Cell 2006, 22, 129–136. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gursinsky, T.; Pirovano, W.; Gambino, G.; Friedrich, S.; Behrens, S.E.; Pantaleo, V. Homeologs of the Nicotiana benthamiana antiviral ARGONAUTE1 show different susceptibilities to microRNA168-mediated control. Plant Physiol. 2015, 168, 938–952. [Google Scholar] [CrossRef] [Green Version]
- Iki, T.; Cléry, A.; Bologna, N.G.; Sarazin, A.; Brosnan, C.A.; Pumplin, N.; Allain, F.H.T.; Voinnet, O. Structural Flexibility Enables Alternative Maturation. ARGONAUTE Sorting and Activities of miR168, a Global Gene Silencing Regulator in Plants. Mol. Plant 2018, 11, 1008–1023. [Google Scholar] [CrossRef] [Green Version]
- Pertermann, R.; Tamilarasan, S.; Gursinsky, T.; Gambino, G.; Schuck, J.; Weinholdt, C.; Lilie, H.; Grosse, I.; Golbik, R.P.; Pantaleo, V.; et al. A Viral Suppressor Modulates the Plant Immune Response Early in Infection by Regulating MicroRNA Activity. mBio 2018, 9, e00419-18. [Google Scholar] [CrossRef] [Green Version]
- Reich, S.; Golbik, R.P.; Geissler, R.; Lilie, H.; Behrens, S.-E. Mechanisms of activity and inhibition of the hepatitis C virus RNA-dependent RNA polymerase. J. Biol. Chem. 2010, 285, 13685–13693. [Google Scholar] [CrossRef] [Green Version]
- Iki, T.; Yoshikawa, M.; Nishikiori, M.; Jaudal, M.C.; Matsumoto-Yokoyama, E.; Mitsuhara, I.; Meshi, T.; Ishikawa, M. In vitro assembly of plant RNA-induced silencing complexes facilitated by molecular chaperone HSP90. Mol. Cell 2010, 39, 282–291. [Google Scholar] [CrossRef]
- Komoda, K.; Naito, S.; Ishikawa, M. Replication of plant RNA virus genomes in a cell-free extract of evacuolated plant protoplasts. Proc. Natl. Acad. Sci. USA 2004, 101, 1863–1867. [Google Scholar] [CrossRef] [Green Version]
- Gursinsky, T.; Schulz, B.; Behrens, S.E. Replication of Tomato bushy stunt virus RNA in a plant in vitro system. Virology 2009, 390, 250–260. [Google Scholar] [CrossRef] [Green Version]
- Várallyay, É.; Oláh, E.; Havelda, Z. Independent parallel functions of p19 plant viral suppressor of RNA silencing required for effective suppressor activity. Nucleic Acids Res. 2013, 42, 599–608. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lózsa, R.; Csorba, T.; Lakatos, L.; Burgyán, J. Inhibition of 3′ modification of small RNAs in virus-infected plants require spatial and temporal co-expression of small RNAs and viral silencing-suppressor proteins. Nucleic Acids Res. 2008, 36, 4099–4107. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cheng, J.; Danielson, D.C.; Nasheri, N.; Singaravelu, R.; Pezacki, J.P. Enhanced specificity of the viral suppressor of RNA silencing protein p19 toward sequestering of human microRNA-122. Biochemistry 2011, 50, 7745–7755. [Google Scholar] [CrossRef]
- Huang, H.; Qiao, R.; Zhao, D.; Zhang, T.; Li, Y.; Yi, F.; Lai, F.; Hong, J.; Ding, X.; Yang, Z.; et al. Profiling of mismatch discrimination in RNAi enabled rational design of allele-specific siRNAs. Nucleic Acids Res. 2009, 37, 7560–7569. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Varani, G.; McClain, W.H. The G·U wobble base pair. EMBO Rep. 2000, 1, 18–23. [Google Scholar] [CrossRef]
- Foss, D.V.; Schirle, N.T.; MacRae, I.J.; Pezacki, J.P. Structural insights into interactions between viral suppressor of RNA silencing protein p19 mutants and small RNAs. FEBS Open Bio 2019, 9, 1042–1051. [Google Scholar] [CrossRef] [Green Version]
- Jay, F.; Wang, Y.; Yu, A.; Taconnat, L.; Pelletier, S.; Colot, V.; Renou, P.; Voinnet, O. Misregulation of AUXIN RESPONSE FACTOR 8 underlies the developmental abnormalities caused by three distinct viral silencing suppressors in Arabidopsis. PLoS Pathog. 2011, 7, e1002035. [Google Scholar] [CrossRef] [Green Version]
- Lewsey, M.; Surette, M.; Robertson, F.C.; Ziebell, H.; Choi, S.H.; Ryu, K.H.; Canto, T.; Palukaitis, P.; Payne, T.; Walsh, J.A.; et al. The role of the Cucumber mosaic virus 2b protein in viral movement and symptom induction. Mol. Plant Microbe Interact. 2009, 22, 642–654. [Google Scholar] [CrossRef] [Green Version]
- Chapman, E.J.; Prokhnevsky, A.I.; Gopinath, K.; Dolja, V.V.; Carrington, J.C. Viral RNA silencing suppressors inhibit the microRNA pathway at an intermediate step. Genes Dev. 2004, 18, 1179–1186. [Google Scholar] [CrossRef] [Green Version]
- Kasschau, K.D.; Xie, Z.; Allen, E.; Llave, C.; Chapman, E.J.; Krizan, K.A.; Carrington, J.C. P1/HC-Pro, a Viral Suppressor of RNA Silencing, Interferes with Arabidopsis Development and miRNA Function. Dev. Cell 2003, 4, 205–217. [Google Scholar] [CrossRef] [Green Version]
- Csorba, T.; Bovi, A.; Dalmay, T.; Burgyán, J. The p122 subunit of Tobacco mosaic virus replicase is a potent silencing suppressor and compromises both small interfering RNA- and microRNA-mediated pathways. J. Virol. 2007, 81, 11768–11780. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schott, G.; Mari-Ordonez, A.; Himber, C.; Alioua, A.; Voinnet, O.; Dunoyer, P. Differential effects of viral silencing suppressors on siRNA and miRNA loading support the existence of two distinct cellular pools of ARGONAUTE1. EMBO J. 2012, 31, 2553–2565. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Azevedo, J.; Garcia, D.; Pontier, D.; Ohnesorge, S.; Yu, A.; Garcia, S.; Braun, L.; Bergdoll, M.; Hakimi, M.A.; Lagrange, T.; et al. Argonaute quenching and global changes in Dicer homeostasis caused by a pathogen-encoded GW repeat protein. Genes Dev. 2010, 24, 904–915. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Várallyay, É.; Havelda, Z. Unrelated viral suppressors of RNA silencing mediate the control of ARGONAUTE1 level. Mol. Plant Pathol. 2013, 14, 567–575. [Google Scholar] [CrossRef] [PubMed]
- Allison, J.R.; Lechner, M.; Hoeppner, M.P.; Poole, A.M. Positive Selection or Free to Vary? Assessing the Functional Significance of Sequence Change Using Molecular Dynamics. PLoS ONE 2016, 11, e0147619. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jagtap, S.; Shivaprasad, P.V. Diversity, expression and mRNA targeting abilities of Argonaute-targeting miRNAs among selected vascular plants. BMC Genom. 2014, 15, 1049. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Humphrey, W.; Dalke, A.; Schulten, K. VMD: Visual molecular dynamics. J. Mol. Graph. 1996, 14, 33–38. [Google Scholar] [CrossRef]






| sRNA Name | Property | KD (nM) | aKrel |
|---|---|---|---|
| gf698 (siRNA) | siRNA control | 0.06 ± 0.04 | - |
| AtmiR162 | Wild-type of AtmiR162 | 0.06 ± 0.02 | 1.00 ± 0.31 |
| miR162-0 | miR162 variant with Watson–Crick base pairing terminus | 0.15 ± 0.06 | 2.31 ± 0.92 |
| miR162-19gC/1*C | miR162 variant with C/C mismatch at 1*/19g | 0.34 ± 0.15 | 5.24 ± 2.31 |
| miR162-18gC/2*C | miR162 variant with C/C mismatch at 2*/18g | 0.12 ± 0.08 | 1.85 ± 1.23 |
| miR162-17gU/3*C | miR162 variant with C/U mismatch at 3*/17g | 0.30 ± 0.17 | 4.62 ± 2.62 |
| miR162-16gA/4*G | miR162 variant with G/A mismatch at 4*/16g | 0.38 ± 0.13 | 5.86 ± 2.00 |
| miR162-15gC/5*C | miR162 variant with C/C mismatch at 5*/15g | 0.23 ± 0.08 | 3.55 ± 1.23 |
| miR162-14gG/6*A | miR162 variant with A/G mismatch at 6*/14g | 0.86 ± 0.49 | 13.26 ± 7.55 |
| miR162-13gU/7*C | miR162 variant with C/U mismatch at 7*/13g | 1.01 ± 0.10 | 15.57 ± 1.54 |
| miR162-12gC/8*C | miR162 variant with C/C mismatch at 8*/12g | 0.28 ± 0.24 | 4.32 ± 3.70 |
| miR162-11gU/9*C | miR162 variant with C/U mismatch at 9*/11g | 0.22 ± 0.03 | 3.39 ± 0.46 |
| miR162-10gC/10*C | miR162 variant with C/C mismatch at 10*/10g | 1.73 ± 0.57 | 26.67 ± 8.79 |
| miR162-19gU/1*G | miR162 variant with G•U wobble at 1*/19g | 0.15 ± 0.12 | 2.31 ± 1.85 |
| miR162-18gU/2*G | miR162 variant with G•U wobble at 2*/18g | 0.13 ± 0.03 | 2.00 ± 0.31 |
| miR162-17gU/3*G | miR162 variant with G•U wobble at 3*/17g | 0.09 ± 0.04 | 1.39 ± 0.62 |
| miR162-16gU/4*G | miR162 variant with G•U wobble at 4*/16g | 0.17 ± 0.05 | 2.62 ± 0.77 |
| miR162-15gU/5*G | miR162 variant with G•U wobble at 5*/15g | 0.10 ± 0.01 | 1.54 ± 0.15 |
| miR162-14gU/6*G | miR162 variant with G•U wobble at 6*/14g | 0.45 ± 0.08 | 6.94 ± 1.23 |
| miR162-13gU/7*G | miR162 variant with G•U wobble at 7*/13g | 1.82 ± 0.38 | 28.05 ± 5.86 |
| miR162-12gU/8*G | miR162 variant with G•U wobble at 8*/12g | 1.93 ± 0.13 | 29.75 ± 2.00 |
| miR162-11gU/9*G | miR162 variant with G•U wobble at 9*/11g | 0.14 ± 0.12 | 2.16 ± 1.85 |
| miR162-10gU/10*G | miR162 variant with G•U wobble at 10*/10g | 0.63 ± 0.21 | 9.71 ± 3.24 |
| miR162-14gG/6*U | miR162 variant with U•G wobble at 6*/14g | 0.15 ± 0.09 | 2.31 ± 1.39 |
| miR162-13gG/7*U | miR162 variant with U•G wobble at 7*/13g | 1.53 ± 0.49 | 23.58 ± 7.55 |
| miR162-12gG/8*U | miR162 variant with U•G wobble at 8*/12g | 0.31 ± 0.08 | 4.78 ± 1.23 |
| miR162-12gC/8*A | miR162 variant with A/C mismatch at 8*/12g | 0.16 ± 0.03 | 2.47 ± 0.46 |
| miR162-12gG/8*G | miR162 variant with G/G mismatch at 8*/12g | 0.07 ± 0.04 | 1.08 ± 0.62 |
| miR162-10gC/10*A | miR162 variant with A/C mismatch at 10*/10g | 0.35 ± 0.24 | 5.39 ± 3.70 |
| AtmiR168a | Wild-type of AtmiR168a | 2.49 ± 0.88 | 1.00 ± 2.22 |
| AtmiR168a variant 1 | AtmiR168a variant without G•U wobble at 7*/13g | 3.13 ± 1.67 | 1.26 ± 2.84 |
| AtmiR168a variant 2 | AtmiR168a variant without G•U wobble at 8g/12* | 0.71 ± 0.28 | 0.29 ± 0.64 |
| AtmiR168a variant 3 | AtmiR168a variant without wobbles at 8g/12* and 17g/3* | 1.00 ± 0.24 | 0.40 ± 0.89 |
| AtmiR168a variant 4 | AtmiR168a variant without G•U wobble at 7*/13g and mismatches | 2.65 ± 0.49 | 1.07 ± 2.34 |
| AtmiR168a variant 5 | AtmiR168a variant without G•U wobble at 8g/12* and mismatches | 0.48 ± 0.17 | 0.19 ± 0.43 |
| AtmiR168a variant 6 | siRNA variant of AtmiR168a | 0.34 ± 0.16 | 0.14 ± 0.31 |
| Competitor RNA | KC (nM) | aKrel |
|---|---|---|
| gf698 (siRNA) | 0.53 ± 0.08 | 1.00 ± 0.16 |
| AtmiR162 | 0.67 ± 0.09 | 1.26 ± 0.18 |
| AtmiR168a | 12.10 ± 0.70 | 22.83 ± 1.64 |
| AtmiR168a variant 2 | 1.10 ± 0.20 | 2.08 ± 0.39 |
| AtmiR168a variant 3 | 2.20 ± 0.40 | 4.15 ± 0.77 |
| AtmiR168a variant 5 | 1.40 ± 0.20 | 2.64 ± 0.39 |
| AtmiR168a variant 6 | 0.38 ± 0.05 | 0.72 ± 0.10 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Pertermann, R.; Golbik, R.P.; Tamilarasan, S.; Gursinsky, T.; Gago-Zachert, S.; Pantaleo, V.; Thondorf, I.; Behrens, S.-E. RNA and Protein Determinants Mediate Differential Binding of miRNAs by a Viral Suppressor of RNA Silencing Thus Modulating Antiviral Immune Responses in Plants. Int. J. Mol. Sci. 2022, 23, 4977. https://doi.org/10.3390/ijms23094977
Pertermann R, Golbik RP, Tamilarasan S, Gursinsky T, Gago-Zachert S, Pantaleo V, Thondorf I, Behrens S-E. RNA and Protein Determinants Mediate Differential Binding of miRNAs by a Viral Suppressor of RNA Silencing Thus Modulating Antiviral Immune Responses in Plants. International Journal of Molecular Sciences. 2022; 23(9):4977. https://doi.org/10.3390/ijms23094977
Chicago/Turabian StylePertermann, Robert, Ralph Peter Golbik, Selvaraj Tamilarasan, Torsten Gursinsky, Selma Gago-Zachert, Vitantonio Pantaleo, Iris Thondorf, and Sven-Erik Behrens. 2022. "RNA and Protein Determinants Mediate Differential Binding of miRNAs by a Viral Suppressor of RNA Silencing Thus Modulating Antiviral Immune Responses in Plants" International Journal of Molecular Sciences 23, no. 9: 4977. https://doi.org/10.3390/ijms23094977
APA StylePertermann, R., Golbik, R. P., Tamilarasan, S., Gursinsky, T., Gago-Zachert, S., Pantaleo, V., Thondorf, I., & Behrens, S.-E. (2022). RNA and Protein Determinants Mediate Differential Binding of miRNAs by a Viral Suppressor of RNA Silencing Thus Modulating Antiviral Immune Responses in Plants. International Journal of Molecular Sciences, 23(9), 4977. https://doi.org/10.3390/ijms23094977






