Quantification of 8-oxoG in Plant Telomeres
Abstract
1. Introduction
2. Results and Discussion
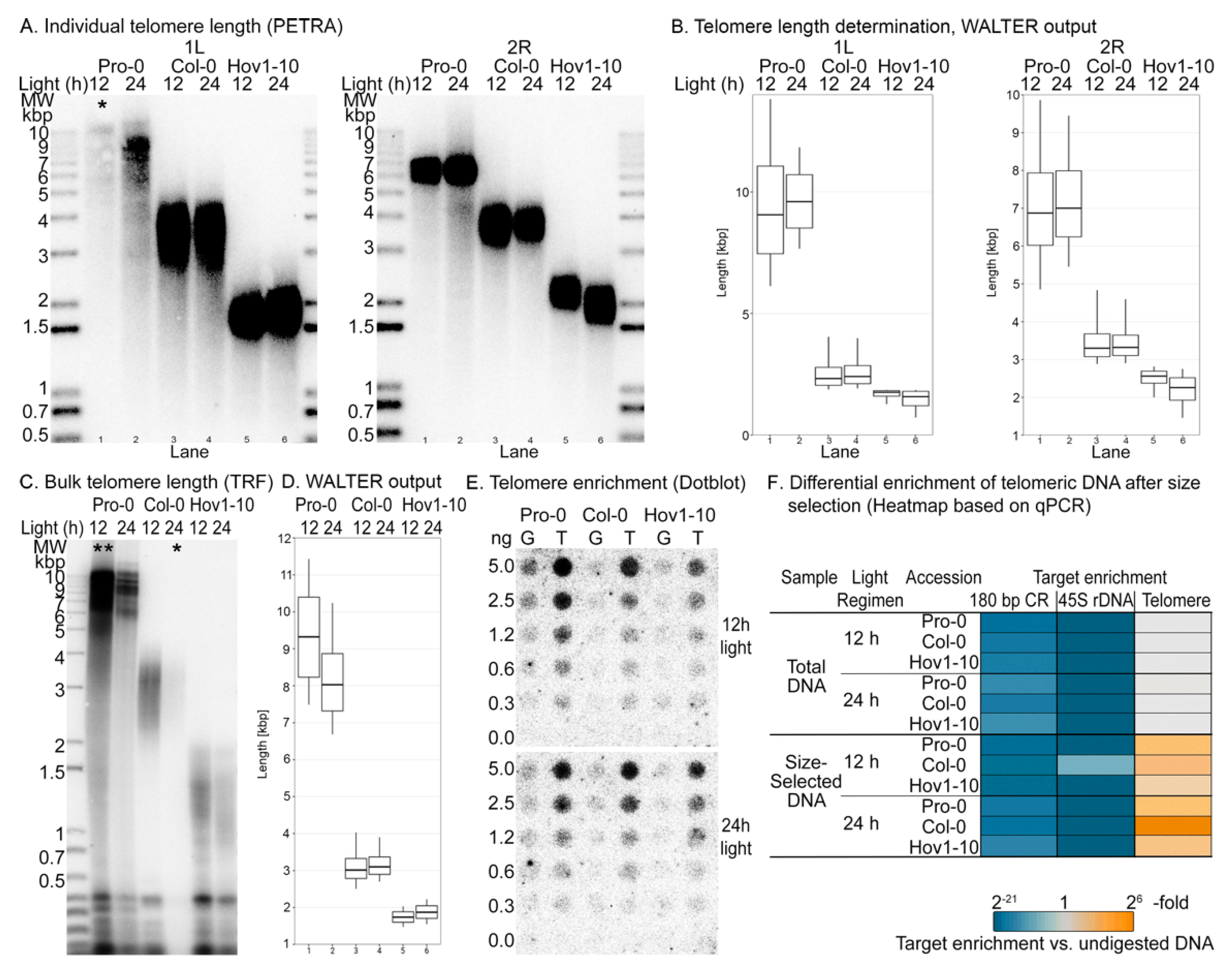
2.1. Differential Enrichment of Telomeric DNA
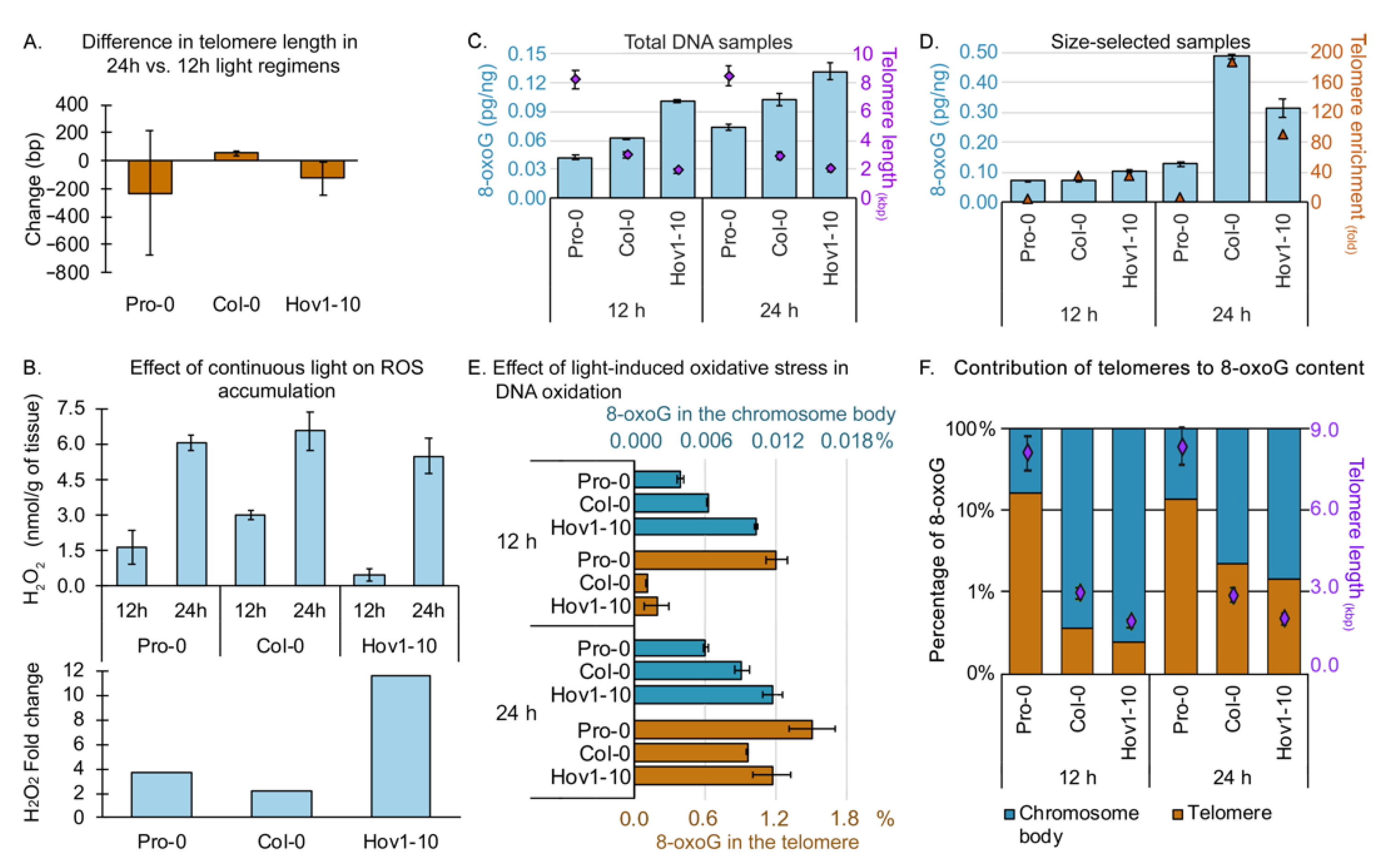
2.2. Quantification of Telomeric 8-oxoG
2.3. Telomeres Are Hotspots of 8-oxoG and Serve as Protectors of the Genome under Oxidative Stress
3. Materials and Methods
3.1. Plant Materials
3.2. Quantification of Hydrogen Peroxide
3.3. DNA Extraction
3.4. Telomere Length Assessment
3.5. qPCR for Quantification of Telomeric DNA, 45S rDNA and 180 bp Centromeric Repeats
3.6. Telomere Enrichment
3.7. 8-oxo-G Quantification
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Acknowledgments
Conflicts of Interest
References
- Lingner, J.; Cooper, J.; Cech, T. Telomerase and DNA end replication: No longer a lagging strand problem? Science 1995, 269, 1533–1534. [Google Scholar] [CrossRef]
- de Lange, T. How telomeres solve the end-protection problem. Science 2009, 326, 948–952. [Google Scholar] [CrossRef]
- Watson, J.D. Origin of concatemeric T7 DNA. Nat. New Biol. 1972, 239, 197–201. [Google Scholar] [CrossRef]
- Olovnikov, A.M. A theory of marginotomy. The incomplete copying of template margin in enzymic synthesis of polynucleotides and biological significance of the phenomenon. J. Theor. Biol. 1973, 41, 181–190. [Google Scholar] [CrossRef]
- Gong, Y.; Stock, A.J.; Liu, Y. The enigma of excessively long telomeres in cancer: Lessons learned from rare human POT1 variants. Curr. Opin. Genet. Dev. 2020, 60, 48–55. [Google Scholar] [CrossRef]
- Barnes, R.P.; Fouquerel, E.; Opresko, P.L. The impact of oxidative DNA damage and stress on telomere homeostasis. Mech. Ageing Dev. 2019, 177, 37–45. [Google Scholar] [CrossRef]
- Fleming, A.M.; Ding, Y.; Burrows, C.J. Oxidative DNA damage is epigenetic by regulating gene transcription via base excision repair. Proc. Natl. Acad. Sci. USA 2017, 114, 2604–2609. [Google Scholar] [CrossRef]
- Sanford, S.L.; Welfer, G.A.; Freudenthal, B.D.; Opresko, P.L. Mechanisms of telomerase inhibition by oxidized and therapeutic dNTPs. Nat. Commun. 2020, 11, 5288. [Google Scholar] [CrossRef]
- Blasco, M.A. The epigenetic regulation of mammalian telomeres. Nat. Rev. Genet. 2007, 8, 299–309. [Google Scholar] [CrossRef]
- Xie, X.; Shippen, D.E. DDM1 guards against telomere truncation in Arabidopsis. Plant Cell Rep. 2018, 37, 501–513. [Google Scholar] [CrossRef]
- Radak, Z.; Zhao, Z.; Goto, S.; Koltai, E. Age-associated neurodegeneration and oxidative damage to lipids, proteins and DNA. Mol. Aspects Med. 2011, 32, 305–315. [Google Scholar] [CrossRef] [PubMed]
- Mittler, R. ROS are good. Trends Plant Sci. 2017, 22, 11–19. [Google Scholar] [CrossRef] [PubMed]
- Dizdaroglu, M. Formation of 8-hydroxyguanine moiety in deoxyribonucleic acid on gamma-irradiation in aqueous solution. Biochemistry 2002, 24, 4476–4481. [Google Scholar] [CrossRef] [PubMed]
- Frenkel, K.; Goldstein, M.S.; Duker, N.J.; Teebor, G.W. Identification of the cis-thymine glycol moiety in oxidized deoxyribonucleic acid. Biochemistry 1981, 20, 750–754. [Google Scholar] [CrossRef]
- Su, D.G.T.; Fang, H.; Gross, M.L.; Taylor, J.S.A. Photocrosslinking of human telomeric G-quadruplex loops by anti cyclobutane thymine dimer formation. Proc. Natl. Acad. Sci. USA 2009, 106, 12861–12866. [Google Scholar] [CrossRef] [PubMed]
- Kawanishi, S.; Oikawa, S. Mechanism of telomere shortening by oxidative stress. Ann. N. Y. Acad. Sci. 2004, 1019, 278–284. [Google Scholar] [CrossRef] [PubMed]
- Oikawa, S.; Kawanishi, S. Site-specific DNA damage at GGG sequence by oxidative stress may accelerate telomere shortening. FEBS Lett. 1999, 453, 365–368. [Google Scholar] [CrossRef]
- Riha, K.; McKnight, T.D.; Griffing, L.R.; Shippen, D.E. Living with genome instability: Plant responses to telomere dysfunction. Science 2001, 291, 1797–1800. [Google Scholar] [CrossRef]
- Watson, J.M.; Trieb, J.; Troestl, M.; Renfrew, K.; Mandakova, T.; Shippen, D.E.; Riha, K. A hypomorphic allele of telomerase reverse transcriptase uncovers the minimal functional length of telomeres in Arabidopsis. Genetics 2021, 219, iyab126. [Google Scholar] [CrossRef]
- Derboven, E.; Ekker, H.; Kusenda, B.; Bulankova, P.; Riha, K. Role of STN1 and DNA polymerase α in telomere stability and genome-wide replication in Arabidopsis. PLoS Genet. 2014, 10, e1004682. [Google Scholar] [CrossRef]
- Shakirov, E.V.; Perroud, P.-F.; Nelson, A.D.; Cannell, M.E.; Quatrano, R.S.; Shippen, D.E. Protection of Telomeres 1 is required for telomere integrity in the moss Physcomitrella patens. Plant Cell 2010, 22, 1838–1848. [Google Scholar] [CrossRef] [PubMed]
- Fulcher, N.; Teubenbacher, A.; Kerdaffrec, E.; Farlow, A.; Nordborg, M.; Riha, K. Genetic architecture of natural variation of telomere length in Arabidopsis thaliana. Genetics 2014, 199, 625–635. [Google Scholar] [CrossRef] [PubMed]
- Abdulkina, L.R.; Kobayashi, C.; Lovell, J.T.; Chastukhina, I.B.; Aklilu, B.B.; Agabekian, I.A.; Suescún, A.V.; Valeeva, L.R.; Nyamsuren, C.; Aglyamova, G.V.; et al. Components of the ribosome biogenesis pathway underlie establishment of telomere length set point in Arabidopsis. Nat. Commun. 2019, 10, 5479. [Google Scholar] [CrossRef] [PubMed]
- Shakirov, E.V.; Shippen, D.E. Length regulation and dynamics of individual telomere tracts in wild-type Arabidopsis. Plant Cell 2004, 16, 1959–1967. [Google Scholar] [CrossRef]
- Miga, K.H.; Koren, S.; Rhie, A.; Vollger, M.R.; Gershman, A.; Bzikadze, A.; Brooks, S.; Howe, E.; Porubsky, D.; Logsdon, G.A.; et al. Telomere-to-telomere assembly of a complete human X chromosome. Nature 2020, 585, 79–84. [Google Scholar] [CrossRef]
- An, N.; Fleming, A.M.; White, H.S.; Burrows, C.J. Nanopore detection of 8-oxoguanine in the human telomere repeat sequence. ACS Nano 2015, 9, 4296–4307. [Google Scholar] [CrossRef]
- Farrell, C.; Vaquero-Sedas, M.I.; Cubiles, M.D.; Thompson, M.; Vega-Vaquero, A.; Pellegrini, M.; Vega-Palas, M.A. A complex network of interactions governs DNA methylation at telomeric regions. Nucleic Acids Res. 2022, 50, 1449–1464. [Google Scholar] [CrossRef]
- Arabidopsis Genome Initiative Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature 2000, 408, 796–815. [CrossRef]
- Riha, K.; Watson, J.M.; Parkey, J.; Shippen, D.E. Telomere length deregulation and enhanced sensitivity to genotoxic stress in Arabidopsis mutants deficient in Ku70. EMBO J. 2002, 21, 2819–2826. [Google Scholar] [CrossRef]
- Vaquero-Sedas, M.I.; Vega-Palas, M.A. Determination of Arabidopsis thaliana telomere length by PCR. Sci. Rep. 2014, 4, 5540. [Google Scholar] [CrossRef]
- Muchová, V.; Amiard, S.; Mozgová, I.; Dvořáčková, M.; Gallego, M.E.; White, C.; Fajkus, J. Homology-dependent repair is involved in 45S rDNA loss in plant CAF-1 mutants. Plant J. 2015, 81, 198–209. [Google Scholar] [CrossRef] [PubMed]
- Davison, J.; Tyagi, A.; Comai, L. Large-scale polymorphism of heterochromatic repeats in the DNA of Arabidopsis thaliana. BMC Plant Biol. 2007, 7, 44. [Google Scholar] [CrossRef] [PubMed]
- Sharma, P.; Jha, A.B.; Dubey, R.S.; Pessarakli, M. Reactive oxygen species, oxidative damage, and antioxidative defense mechanism in plants under stressful conditions. J. Bot. 2012, 2012, 217037. [Google Scholar] [CrossRef]
- Fleming, A.M.; Ding, Y.; Burrows, C.J. Sequencing DNA for the Oxidatively Modified Base 8-Oxo-7,8-Dihydroguanine. Methods Enzymol. 2017, 591, 187–210. [Google Scholar] [CrossRef]
- Yoshihara, M.; Jiang, L.; Akatsuka, S.; Suyama, M.; Toyokuni, S. Genome-wide profiling of 8-oxoguanine reveals its association with spatial positioning in nucleus. DNA Res. 2014, 21, 603–612. [Google Scholar] [CrossRef]
- Mishra, A.K.; Agarwal, S.; Jain, C.K.; Rani, V. High GC content: Critical parameter for predicting stress regulated miRNAs in Arabidopsis thaliana. Bioinformation 2009, 4, 151–154. [Google Scholar] [CrossRef][Green Version]
- Marsico, G.; Chambers, V.S.; Sahakyan, A.B.; McCauley, P.; Boutell, J.M.; Di Antonio, M.; Balasubramanian, S. Whole genome experimental maps of DNA G-quadruplexes in multiple species. Nucleic Acids Res. 2019, 47, 3862–3874. [Google Scholar] [CrossRef]
- Fleming, A.M.; Burrows, C.J. Oxidative stress-mediated epigenetic regulation by G-quadruplexes. NAR Cancer 2021, 3, zcab038. [Google Scholar] [CrossRef]
- Yadav, V.; Hemansi; Kim, N.; Tuteja, N.; Yadav, P. G quadruplex in plants: A ubiquitous regulatory element and its biological relevance. Front. Plant Sci. 2017, 8, 1163. [Google Scholar] [CrossRef]
- Moye, A.L.; Porter, K.C.; Cohen, S.B.; Phan, T.; Zyner, K.G.; Sasaki, N.; Lovrecz, G.O.; Beck, J.L.; Bryan, T.M. Telomeric G-quadruplexes are a substrate and site of localization for human telomerase. Nat. Commun. 2015, 6, 7643. [Google Scholar] [CrossRef]
- Barbero Barcenilla, B.; Shippen, D.E. Back to the future: The intimate and evolving connection between telomere-related factors and genotoxic stress. J. Biol. Chem. 2019, 294, 14803–14813. [Google Scholar] [CrossRef] [PubMed]
- Ahmed, W.; Lingner, J. Impact of oxidative stress on telomere biology. Differentiation 2018, 99, 21–27. [Google Scholar] [CrossRef] [PubMed]
- Piovesan, A.; Chiara Pelleri, M.; Antonaros, F.; Strippoli, P.; Caracausi, M.; Vitale, L. On the length, weight and GC content of the human genome. BMC Res. Notes 2019, 12, 106. [Google Scholar] [CrossRef] [PubMed]
- Abuelsoud, W.; Cortleven, A.; Schmülling, T. Photoperiod stress induces an oxidative burst-like response and is associated with increased apoplastic peroxidase and decreased catalase activities. J. Plant Physiol. 2020, 253, 153252. [Google Scholar] [CrossRef]
- Loreto, F.; Velikova, V. Isoprene produced by leaves protects the photosynthetic apparatus against ozone damage, quenches ozone products, and reduces lipid peroxidation of cellular membranes. Plant Physiol. 2001, 127, 1781–1787. [Google Scholar] [CrossRef]
- Doyle, J.; Doyle, J. A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochemistry 1987, 19, 11–15. [Google Scholar]
- Fitzgerald, M.S.; Riha, K.; Gao, F.; Ren, S.; McKnight, T.D.; Shippen, D.E. Disruption of the telomerase catalytic subunit gene from Arabidopsis inactivates telomerase and leads to a slow loss of telomeric DNA. Proc. Natl. Acad. Sci. USA 1999, 96, 14813–14818. [Google Scholar] [CrossRef]
- Heacock, M.; Spangler, E.; Riha, K.; Puizina, J.; Shippen, D.E. Molecular analysis of telomere fusions in Arabidopsis: Multiple pathways for chromosome end-joining. EMBO J. 2004, 23, 2304–2313. [Google Scholar] [CrossRef]
- Lyčka, M.; Peska, V.; Demko, M.; Spyroglou, I.; Kilar, A.; Fajkus, J.; Fojtovaá, M. WALTER: An easy way to online evaluate telomere lengths from terminal restriction fragment analysis. BMC Bioinform. 2021, 22, 145. [Google Scholar] [CrossRef]
- Fouquerel, E.; Barnes, R.P.; Uttam, S.; Watkins, S.C.; Bruchez, M.P.; Opresko, P.L. Targeted and persistent 8-oxoguanine base damage at telomeres promotes telomere loss and crisis. Mol. Cell 2019, 75, 117–130. [Google Scholar] [CrossRef]
- Lee, H.-T.; Bose, A.; Lee, C.-Y.; Opresko, P.L.; Myong, S. Molecular mechanisms by which oxidative DNA damage promotes telomerase activity. Nucleic Acids Res. 2017, 45, 11752–11765. [Google Scholar] [CrossRef] [PubMed]
- Coluzzi, E.; Leone, S.; Sgura, A. Oxidative stress induces telomere dysfunction and senescence by replication fork arrest. Cells 2019, 8, 19. [Google Scholar] [CrossRef] [PubMed]
- Zhou, J.; Liu, M.; Fleming, A.M.; Burrows, C.J.; Wallace, S.S. Neil3 and NEIL1 DNA glycosylases remove oxidative damages from quadruplex DNA and exhibit preferences for lesions in the telomeric sequence context. J. Biol. Chem. 2013, 288, 27263–27272. [Google Scholar] [CrossRef] [PubMed]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Castillo-González, C.; Barbero Barcenilla, B.; Young, P.G.; Hall, E.; Shippen, D.E. Quantification of 8-oxoG in Plant Telomeres. Int. J. Mol. Sci. 2022, 23, 4990. https://doi.org/10.3390/ijms23094990
Castillo-González C, Barbero Barcenilla B, Young PG, Hall E, Shippen DE. Quantification of 8-oxoG in Plant Telomeres. International Journal of Molecular Sciences. 2022; 23(9):4990. https://doi.org/10.3390/ijms23094990
Chicago/Turabian StyleCastillo-González, Claudia, Borja Barbero Barcenilla, Pierce G. Young, Emily Hall, and Dorothy E. Shippen. 2022. "Quantification of 8-oxoG in Plant Telomeres" International Journal of Molecular Sciences 23, no. 9: 4990. https://doi.org/10.3390/ijms23094990
APA StyleCastillo-González, C., Barbero Barcenilla, B., Young, P. G., Hall, E., & Shippen, D. E. (2022). Quantification of 8-oxoG in Plant Telomeres. International Journal of Molecular Sciences, 23(9), 4990. https://doi.org/10.3390/ijms23094990





