Engineering CRISPR/Cas9 for Multiplexed Recombinant Coagulation Factor Production
Abstract
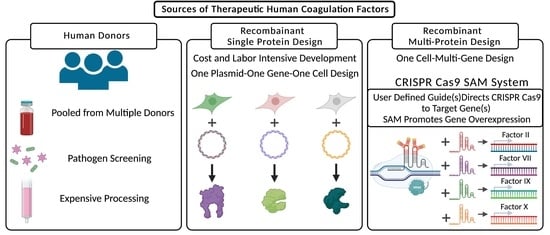
1. Introduction
2. Results
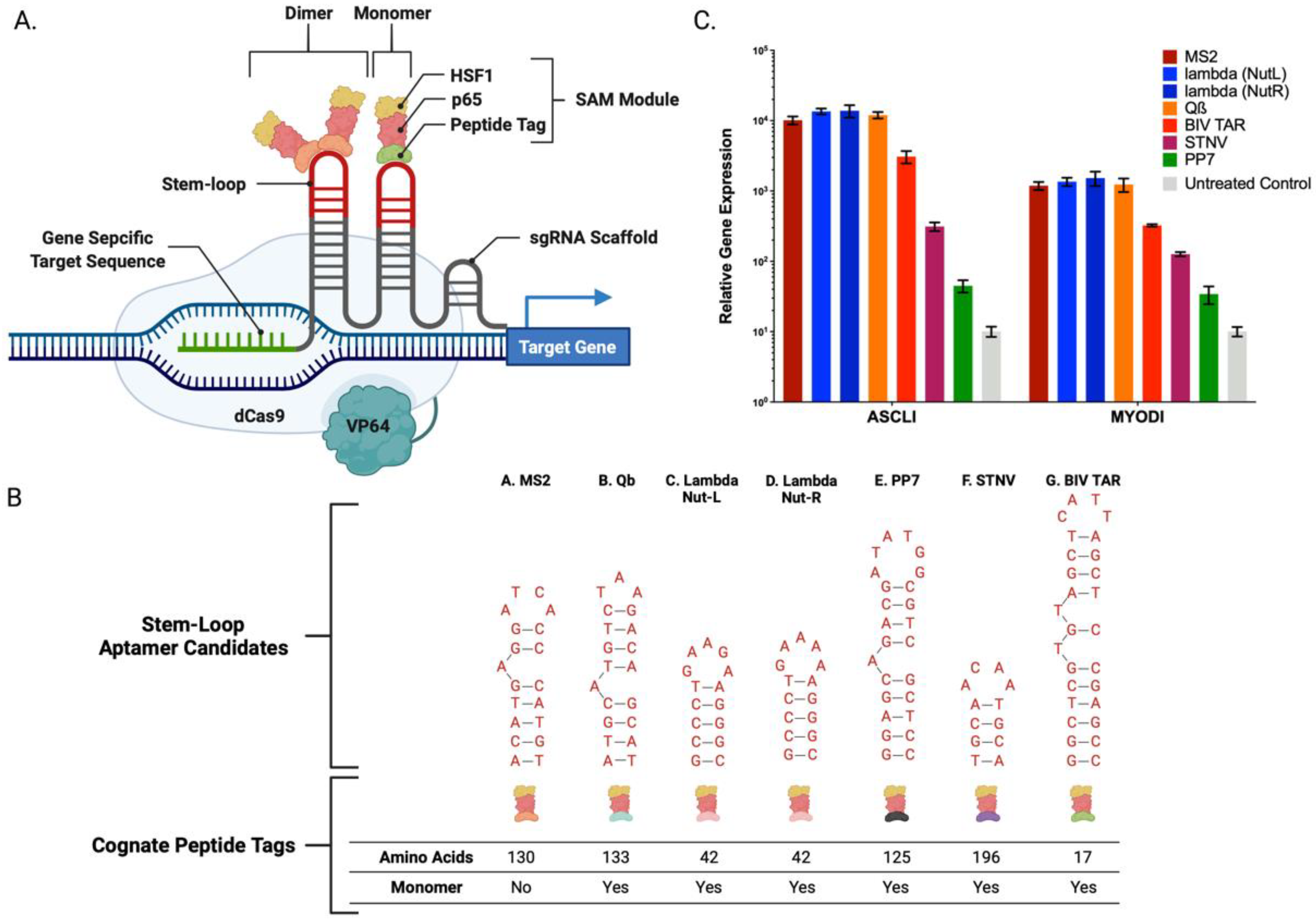
2.1. Clustered Regularly Interspaced Short Palindromic Repeats/Cas9-Synergistic Activation Module
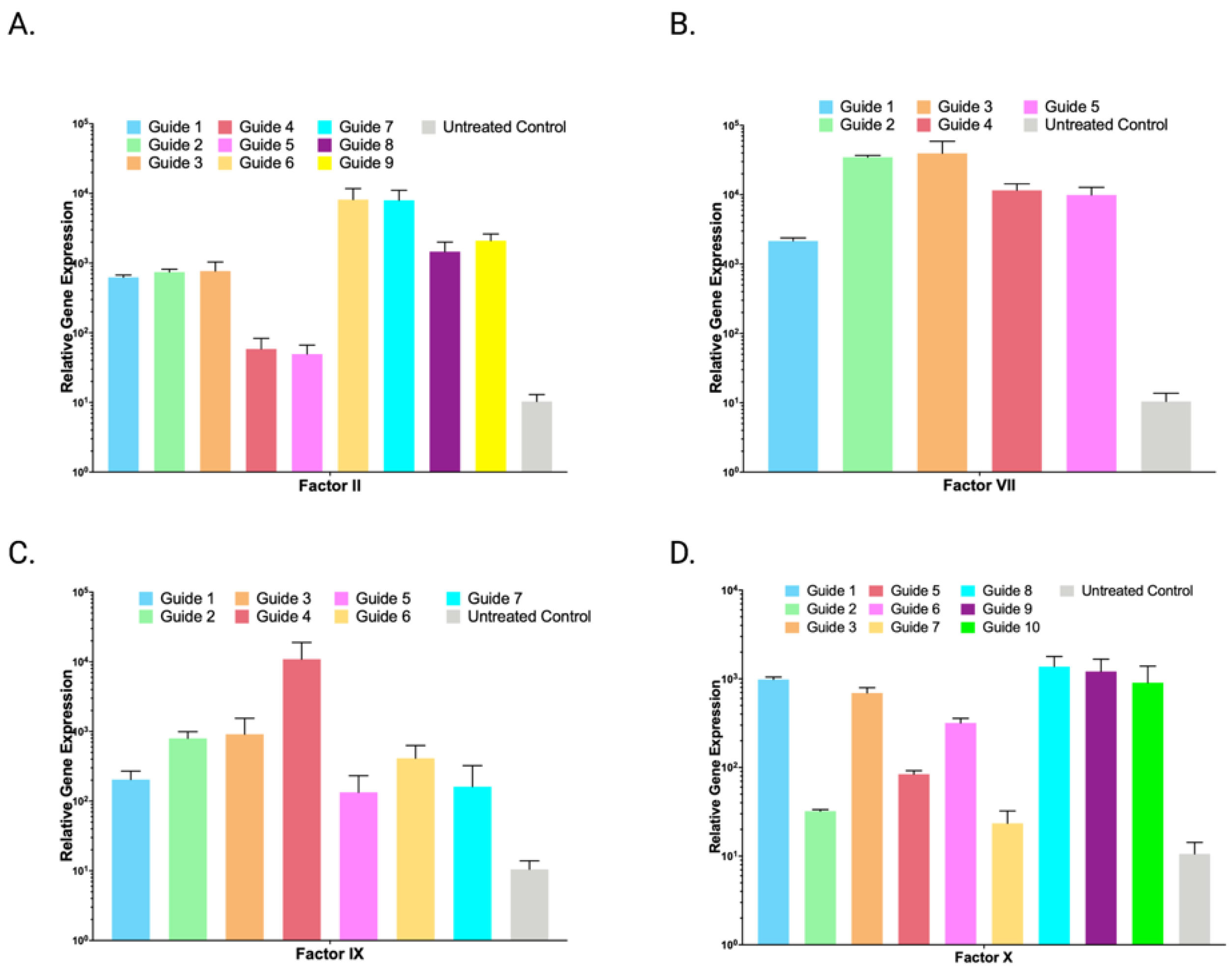
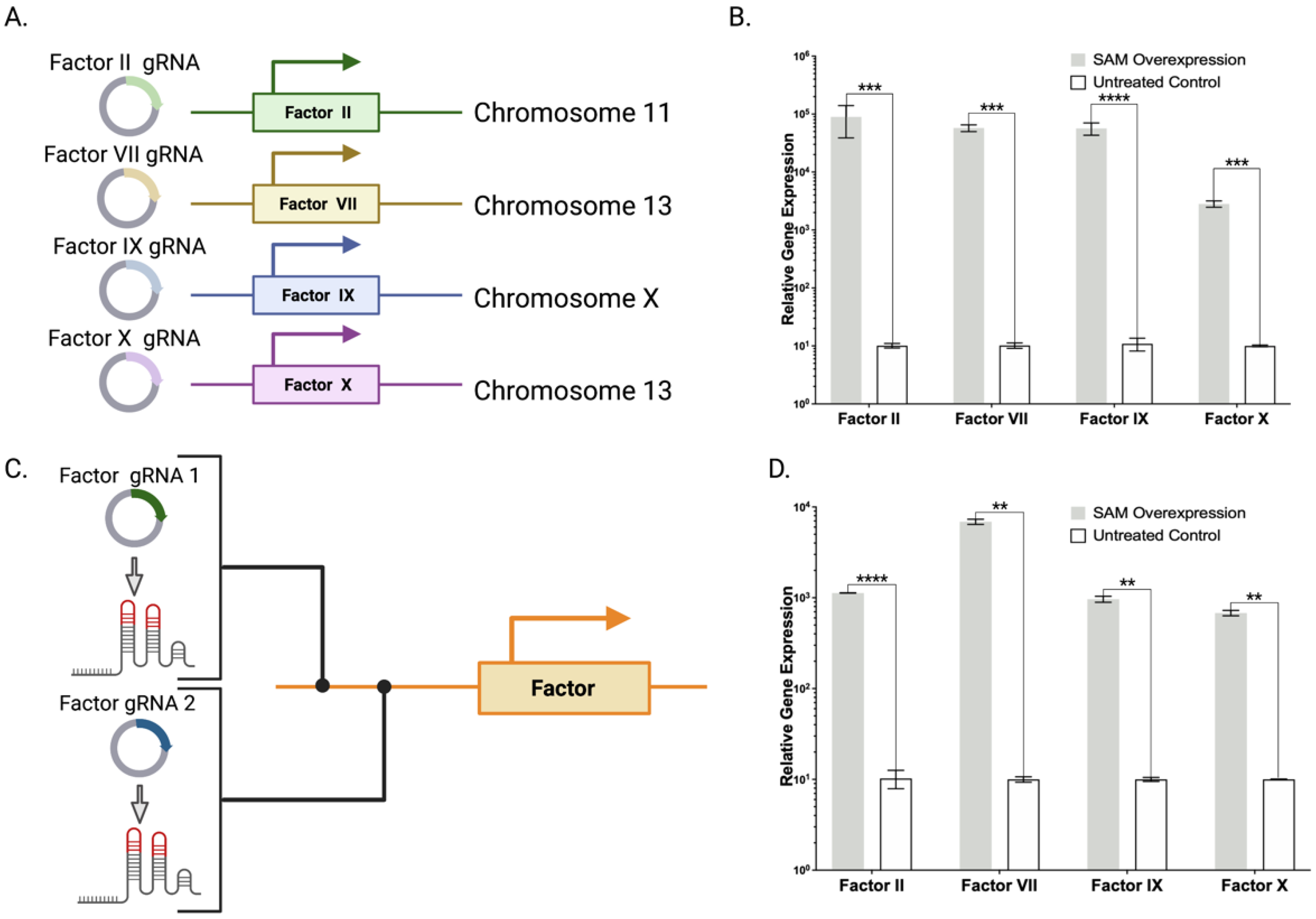
2.2. Lambda SAM Shows Potential for Prothrombin Complex Concentrate Production
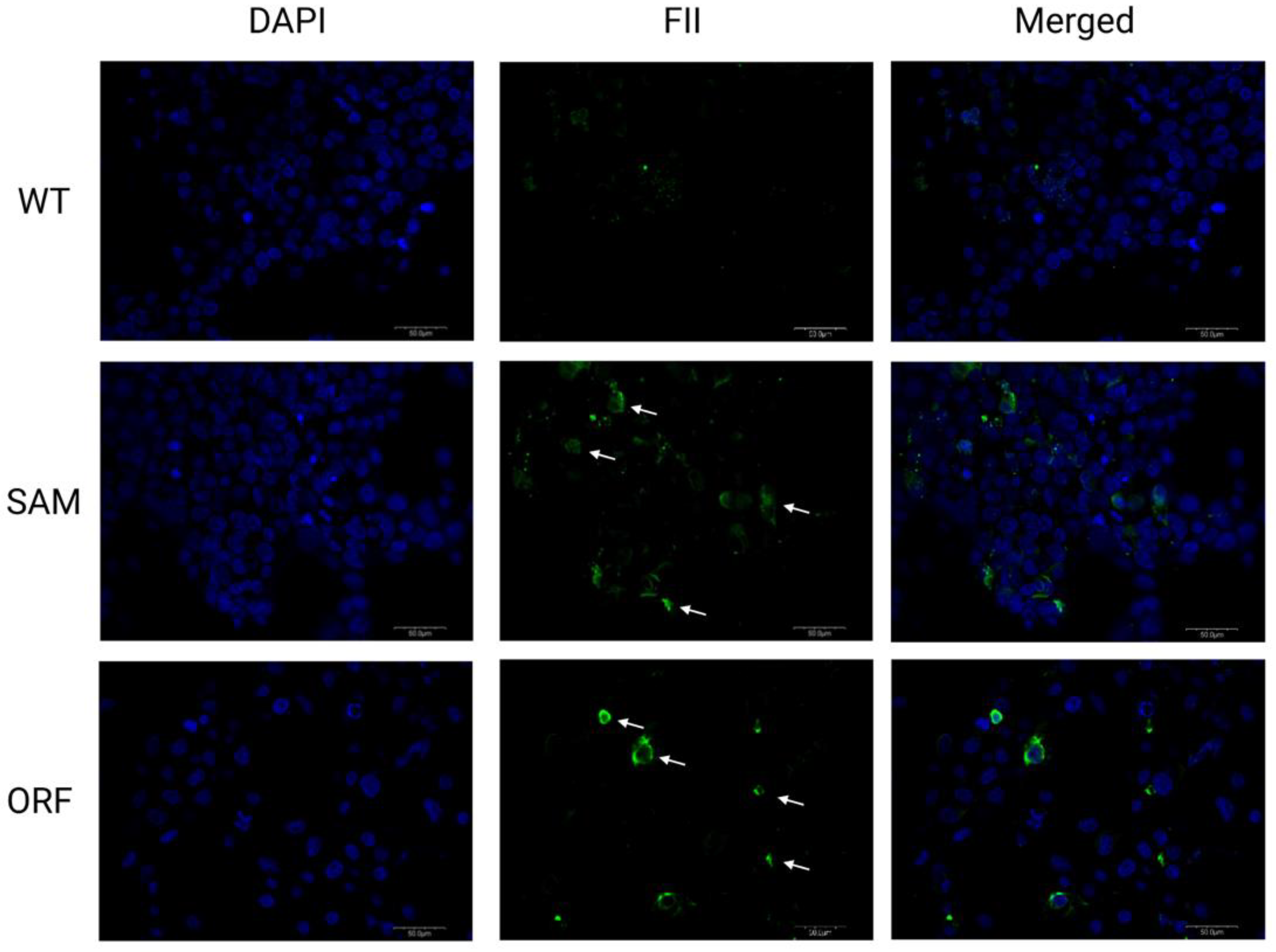
2.3. Lambda SAM Mediated Protein Overexpression
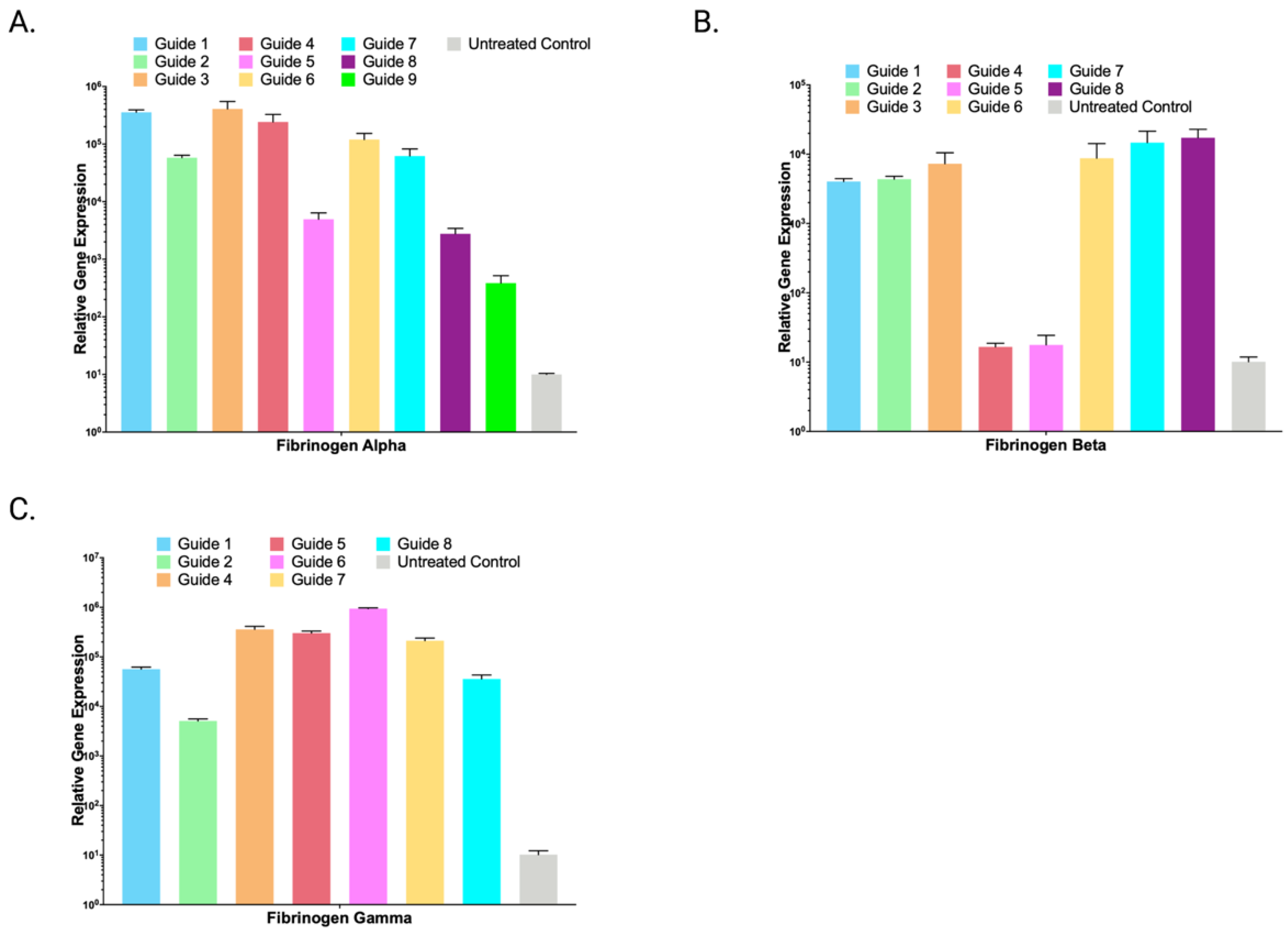
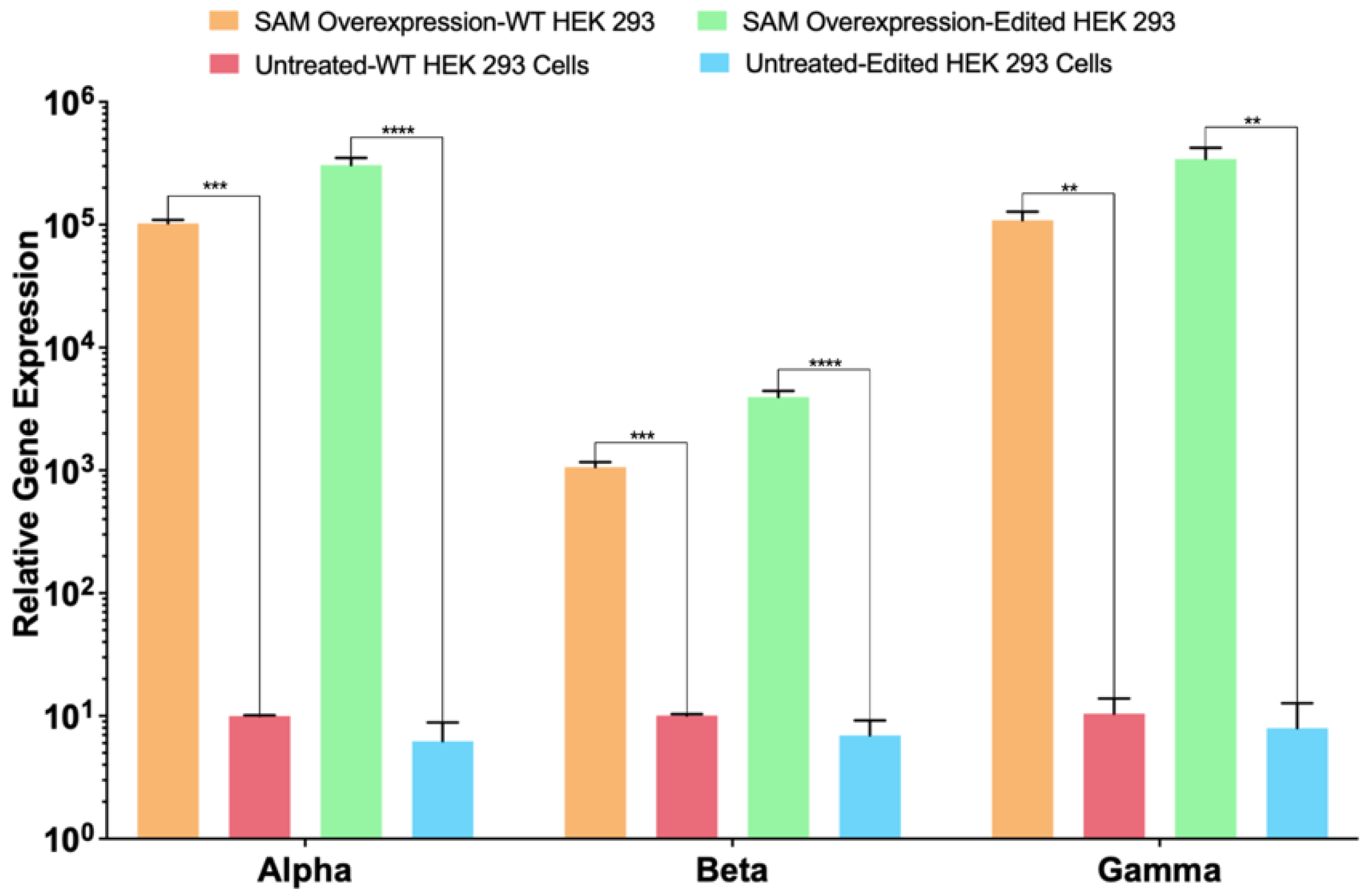
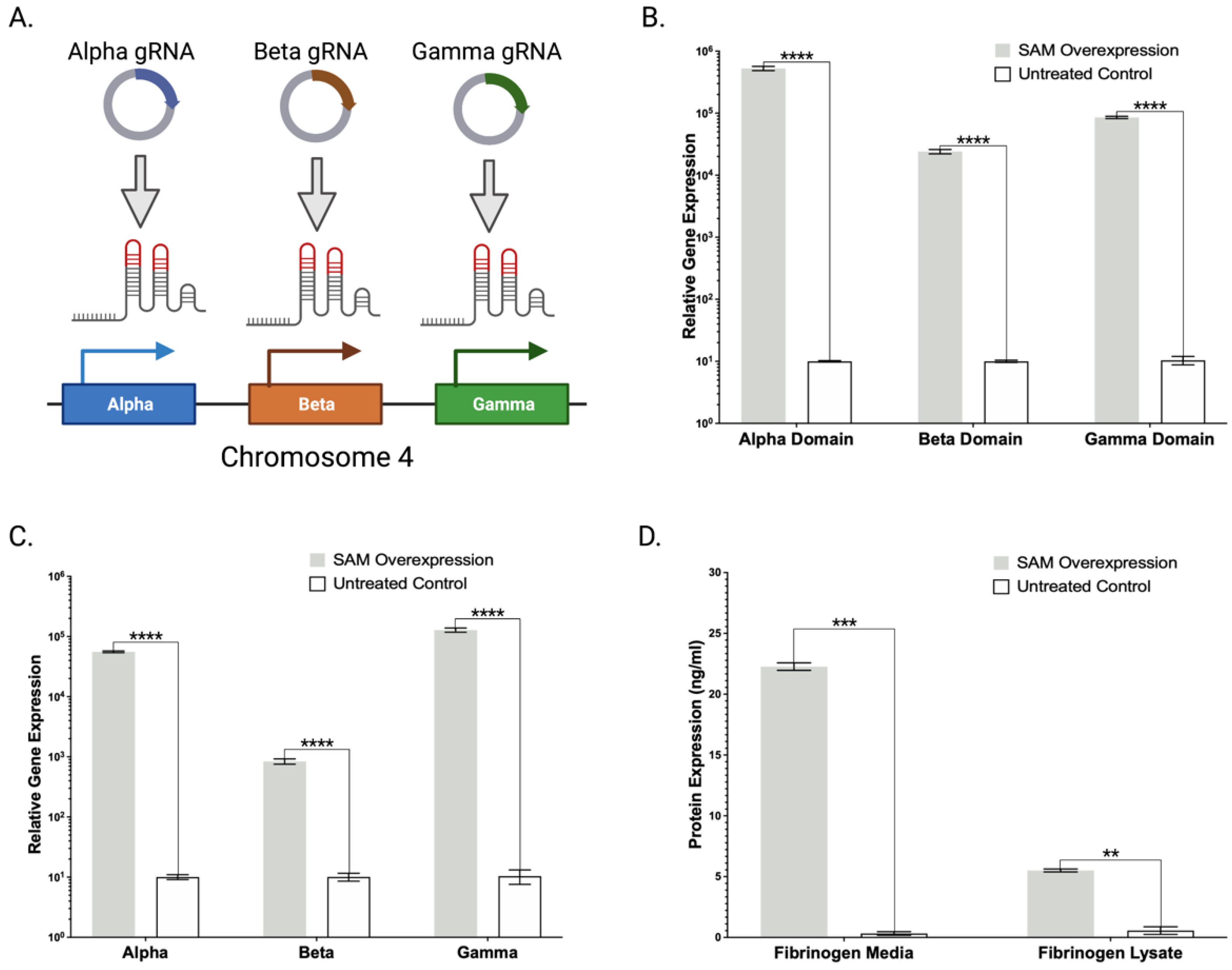
2.4. Lambda SAM Drives Activation of Multi-Domain Gene Fibrinogen
3. Discussion
4. Materials and Methods
4.1. SAM Vector Assembly
4.2. Cell Culture and Transfection Reagents
4.3. Quantitative RT-PCR
4.4. Protein Quantification
4.5. Immunofluorescent Staining
4.6. Graphing, Statistical Analysis, and Images
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Appendix A
| Factor/Guide | Sequence | Proximity to Initiating Methionine (Base Pairs) |
|---|---|---|
| Factor II Guide 1 | GGACAGGTCCTCCTGGGAGC | 161 |
| Factor II Guide 2 | TCCTGTCCATCTCCACCATC | 136 |
| Factor II Guide 3 | TCTAGGAGGAGTCACAAAAA | 193 |
| Factor II Guide 4 | CGCTATATGTTAGGATTCGA | 302 |
| Factor II Guide 5 | GGCGAGGGGCAACTTCCATC | 533 |
| Factor II Guide 6 | CACACTATGGCGCACGTCCG | −14 |
| Factor II Guide 7 | TATCTACCCACCCGTCCCCA | 112 |
| Factor II Guide 8 | AACATTAACCCAGAGGGGTC | 40 |
| Factor II Guide 9 | ACATGAAGAAATCAGCGGAG | 66 |
| Factor VII Guide 1 | ACCCCAGCTGTGCTGCAGAG | 221 |
| Factor VII Guide 2 | GCTCAGTGGCTGGGCAGCAG | 169 |
| Factor VII Guide 3 | TGCATGGCCACTGGCCGGCC | 145 |
| Factor VII Guide 4 | AACTTTGCCCGTCAGTCCCA | 44 |
| Factor VII Guide 5 | TTGCCAGCGTGCAGGTGTTA | 253 |
| Factor IX Guide 1 | AAATTTCACCTCTGATTTCG | 168 |
| Factor IX Guide 2 | CAGACTCAAATCAGCCACAG | 201 |
| Factor IX Guide 3 | CTTGGGACAAGTGAAGAGAA | 130 |
| Factor IX Guide 4 | CTCTCTGACAAAGATACGGT | 304 |
| Factor IX Guide 5 | TCATCAGGATCTAGTGTTAG | 607 |
| Factor IX Guide 6 | AATAGTCCAAAGACCCATTG | 99 |
| Factor IX Guide 7 | CTTTCACAATCTGCTAGCAA | 5 |
| Factor X Guide 1 | AGGCAGGGGAGTGACCACGC | 138 |
| Factor X Guide 2 | GATGAGGATGAGGTTCTGCG | 219 |
| Factor X Guide 3 | TCTGGCCGCCCAGCTCTGCC | 192 |
| Factor X Guide 4 1 | GTGGTCTCATGTCACGGGGG | 527 |
| Factor X Guide 5 | AGGTGGTCTCATGTCACGGG | 525 |
| Factor X Guide 6 | AGGCCGCTGGCAGTGAGCGT | 419 |
| Factor X Guide 7 | ACAGTACTCGGCCACACCAT | −2 |
| Factor X Guide 8 | AGCAGGTCTCGGCTCCAATC | 114 |
| Factor X Guide 9 | AGGTCTCGGCTCCAATCAGG | 111 |
| Factor X Guide 10 | CGCTGGGAACACATCCACTC | 155 |
| Fibrinogen Alpha Guide 1 | GGAGGAACAAAGGACGTAGA | 142 |
| Fibrinogen Alpha Guide 2 | TCGGAAGCAAACAAGCTGAG | 201 |
| Fibrinogen Alpha Guide 3 | TTCTGGGATACCAACAGCAT | 162 |
| Fibrinogen Alpha Guide 4 | CAAGAGGTTTGGTTAATCAT | 90 |
| Fibrinogen Alpha Guide 5 | TTTCAGCTGGAGTGCTCCTC | 26 |
| Fibrinogen Alpha Guide 6 | CTTCCGATAAGCTGTTGCAA | 214 |
| Fibrinogen Alpha Guide 7 | GAGATCTCCTAATTGTTGAC | 254 |
| Fibrinogen Alpha Guide 8 | TGATTCTCCAGTCAACAATT | 244 |
| Fibrinogen Alpha Guide 9 | GGAGGGTTGACTGTCTACAC | 122 |
| Fibrinogen Beta Guide 1 | AAGATACACATCTCTCTTTG | 264 |
| Fibrinogen Beta Guide 2 | GTAAATTTGACCTACTCACA | 354 |
| Fibrinogen Beta Guide 3 | TTAGTGGTTGCCTTGTGAGT | 367 |
| Fibrinogen Beta Guide 4 | TCTGAAGTCATTCCTAGCAG | 46 |
| Fibrinogen Beta Guide 5 | ACAAGTAAATAAGCTTTGCT | 149 |
| Fibrinogen Beta Guide 6 | CAGAGGACTCAGATATATAT | 29 |
| Fibrinogen Beta Guide 7 | TCAGTTAAGTCTACATGAAA | −6 |
| Fibrinogen Beta Guide 8 | GGACAAAATGATGGGAAGTT | 233 |
| Fibrinogen Gamma Guide 1 | AAGCATTATCCATTGCTGTG | 292 |
| Fibrinogen Gamma Guide 2 | CTCCTTTTTGGCCCAGCTCA | 231 |
| Fibrinogen Gamma Guide 3 1 | TGATCCAAAATTATCTGCAA | 263 |
| Fibrinogen Gamma Guide 4 | GGCCCCCAACCTATACTGTC | 178 |
| Fibrinogen Gamma Guide 5 | AATGGCTGGAGCTGATCACG | 103 |
| Fibrinogen Gamma Guide 6 | GGGAACCTGACAGTATAGGT | 186 |
| Fibrinogen Gamma Guide 7 | ACGGGGCCTCCTTACCAGAA | 120 |
| Fibrinogen Gamma Guide 8 | CACTACAAGGCTCGGAGCTC | 16 |
| Gene | qRT-PCR Probe |
|---|---|
| Factor II | Hs01011988_m1 |
| Factor VII | Hs01551992_m1 |
| Factor IX | Hs01592597_m1 |
| Factor X | Hs00984442_m1 |
| GAPDH | Hs02758991_g1 |
| Ascl1 | Hs00269932_m1 |
| MyoD1 | Hs00159528_m1 |
| Candidate | Aptamer Sequence | Peptide Size(aa) |
|---|---|---|
| MS2 | AACATGAGGATCACCCATGTC | 130 |
| Lambda Nut-L | GGGCCCTGAAGAAGGGCCC | 42 |
| Lambda Nut-R | GCCCTGAAAAAGGGC | 42 |
| Qb | AATGCATGTCTAAGACAGCAT | 133 |
| BIV TAR | GGCTCGTGTAGCTCATTAGCTCCGAGCC | 17 |
| STNV | CCTTTTCAAGACATGCAACAATGCACACAG | 196 |
| PP7 | GGAGCAGACGATATGGCGTCGCTCC | 125 |
Appendix B

References
- Swiech, K.; de Freitas, M.C.C.; Covas, D.T.; Picanço-Castro, V. Recombinant glycoprotein production in human cell lines. Methods Mol. Biol. 2015, 1258, 223–240. [Google Scholar] [CrossRef] [PubMed]
- Swiech, K.; Picanço-Castro, V.; Covas, D.T. Production of recombinant coagulation factors: Are humans the best host cells? Bioengineered 2017, 8, 462–470. [Google Scholar] [CrossRef] [PubMed]
- Popovic, G.; Kirby, N.C.; Dement, T.C.; Peterson, K.M.; Daub, C.E.; Belcher, H.A.; Guthold, M.; Offenbacher, A.R.; Hudson, N.E. Development of Transient Recombinant Expression and Affinity Chromatography Systems for Human Fibrinogen. Int. J. Mol. Sci. 2022, 23, 1054. [Google Scholar] [CrossRef] [PubMed]
- Hirashima, M.; Imamura, T.; Yano, K.; Kawamura, R.; Meta, A.; Tokieda, Y.; Nakashima, T. High-Level expression and preparation of recombinant human fibrinogen as biopharmaceuticals. J. Biochem. 2016, 159, 261–270. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Cong, L.; Ran, F.A.; Cox, D.; Lin, S.; Barretto, R.; Habib, N.; Hsu, P.D.; Wu, X.; Jiang, W.; Marraffini, L.A.; et al. Multiplex Genome Engineering Using CRISPR/Cas Systems. Science 2013, 339, 819–823. [Google Scholar] [CrossRef] [PubMed]
- Konermann, S.; Brigham, M.; Trevino, A.E.; Joung, J.; Abudayyeh, O.O.; Barcena, C.; Hsu, P.; Habib, N.; Gootenberg, J.; Nishimasu, H.; et al. Genome-Scale transcriptional activation by an engineered CRISPR-Cas9 complex. Nature 2015, 517, 583–588. [Google Scholar] [CrossRef] [PubMed]
- Lim, F.; Spingola, M.; Peabody, D.S. The RNA-binding Site of Bacteriophage Qβ Coat Protein. J. Biol. Chem. 1996, 271, 31839–31845. [Google Scholar] [CrossRef] [PubMed]
- Lim, F. RNA recognition site of PP7 coat protein. Nucleic Acids Res. 2002, 30, 4138–4144. [Google Scholar] [CrossRef] [PubMed]
- Legault, P.; Li, J.; Mogridge, J.; Kay, L.E.; Greenblatt, J. NMR Structure of the Bacteriophage λ N Peptide/boxB RNA Complex: Recognition of a GNRA Fold by an Arginine-Rich Motif. Cell 1998, 93, 289–299. [Google Scholar] [CrossRef]
- Patel, N.; Dykeman, E.C.; Coutts, R.H.A.; Lomonossoff, G.P.; Rowlands, D.J.; Phillips, S.E.V.; Ranson, N.; Twarock, R.; Tuma, R.; Stockley, P.G. Revealing the density of encoded functions in a viral RNA. Proc. Natl. Acad. Sci. USA 2015, 112, 2227–2232. [Google Scholar] [CrossRef] [PubMed]
- Wakiyama, M.; Kaitsu, Y.; Muramatsu, R.; Takimoto, K.; Yokoyama, S. Tethering of proteins to RNAs using the bovine immunodeficiency virus–Tat peptide and BIV–TAR RNA. Anal. Biochem. 2012, 427, 130–132. [Google Scholar] [CrossRef] [PubMed]
- Levy, J.H.; Welsby, I.; Goodnough, L.T. Fibrinogen as a therapeutic target for bleeding: A review of critical levels and replacement therapy. Transfusion 2014, 54, 1389–1405. [Google Scholar] [CrossRef] [PubMed]
- Durocher, Y.; Perret, S.; Kamen, A. High-Level and high-throughput recombinant protein production by transient transfection of suspension-growing human 293-EBNA1 cells. Nucleic Acids Res. 2002, 30, E9. [Google Scholar] [CrossRef] [PubMed]
- Nettleship, J.E.; Watson, P.J.; Rahman-Huq, N.; Fairall, L.; Posner, M.G.; Upadhyay, A.; Reddivari, Y.; Chamberlain, J.M.G.; Kolstoe, S.E.; Bagby, S.; et al. Transient expression in HEK 293 cells: An alternative to E. coli for the production of secreted and intracellular mammalian proteins. Methods Mol. Biol. 2015, 1258, 209–222. [Google Scholar] [CrossRef] [PubMed]
- Grottke, O.; Callum, J.; Cushing, M.M.; Haas, T. The use of coagulation factor concentrates for perioperative bleeding management—A global perspective. Transfusion 2020, 60, 663–666. [Google Scholar] [CrossRef] [PubMed]
- Gao, Z.; Herrera-Carrillo, E.; Berkhout, B. Delineation of the Exact Transcription Termination Signal for Type 3 Polymerase III. Mol. Ther.—Nucleic Acids 2018, 10, 36–44. [Google Scholar] [CrossRef] [PubMed]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Feser, C.J.; Lees, C.J.; Lammers, D.T.; Riddle, M.J.; Bingham, J.R.; Eckert, M.J.; Tolar, J.; Osborn, M.J. Engineering CRISPR/Cas9 for Multiplexed Recombinant Coagulation Factor Production. Int. J. Mol. Sci. 2022, 23, 5090. https://doi.org/10.3390/ijms23095090
Feser CJ, Lees CJ, Lammers DT, Riddle MJ, Bingham JR, Eckert MJ, Tolar J, Osborn MJ. Engineering CRISPR/Cas9 for Multiplexed Recombinant Coagulation Factor Production. International Journal of Molecular Sciences. 2022; 23(9):5090. https://doi.org/10.3390/ijms23095090
Chicago/Turabian StyleFeser, Colby J., Christopher J. Lees, Daniel T. Lammers, Megan J. Riddle, Jason R. Bingham, Matthew J. Eckert, Jakub Tolar, and Mark J. Osborn. 2022. "Engineering CRISPR/Cas9 for Multiplexed Recombinant Coagulation Factor Production" International Journal of Molecular Sciences 23, no. 9: 5090. https://doi.org/10.3390/ijms23095090
APA StyleFeser, C. J., Lees, C. J., Lammers, D. T., Riddle, M. J., Bingham, J. R., Eckert, M. J., Tolar, J., & Osborn, M. J. (2022). Engineering CRISPR/Cas9 for Multiplexed Recombinant Coagulation Factor Production. International Journal of Molecular Sciences, 23(9), 5090. https://doi.org/10.3390/ijms23095090