Epigenomic Profiling of Epithelial Ovarian Cancer Stem-Cell Differentiation Reveals GPD1 Associated Immune Suppressive Microenvironment and Poor Prognosis
Abstract
:1. Introduction
2. Results
2.1. Establishment of an Epithelial Ovarian Cancer Differentiation Model
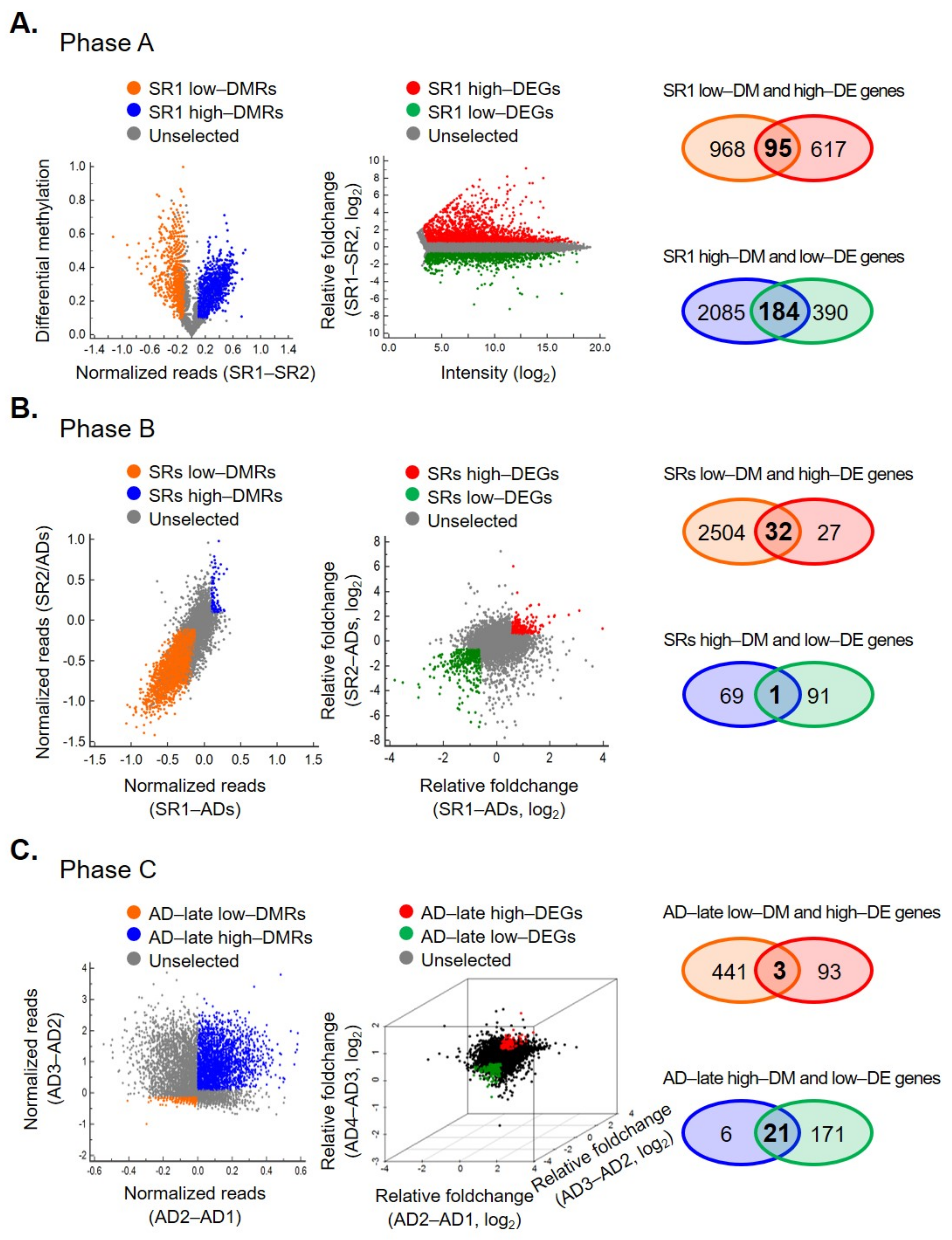
2.2. Integrated Transcriptomics and Methylomics Analysis of OCSCs Differentiation
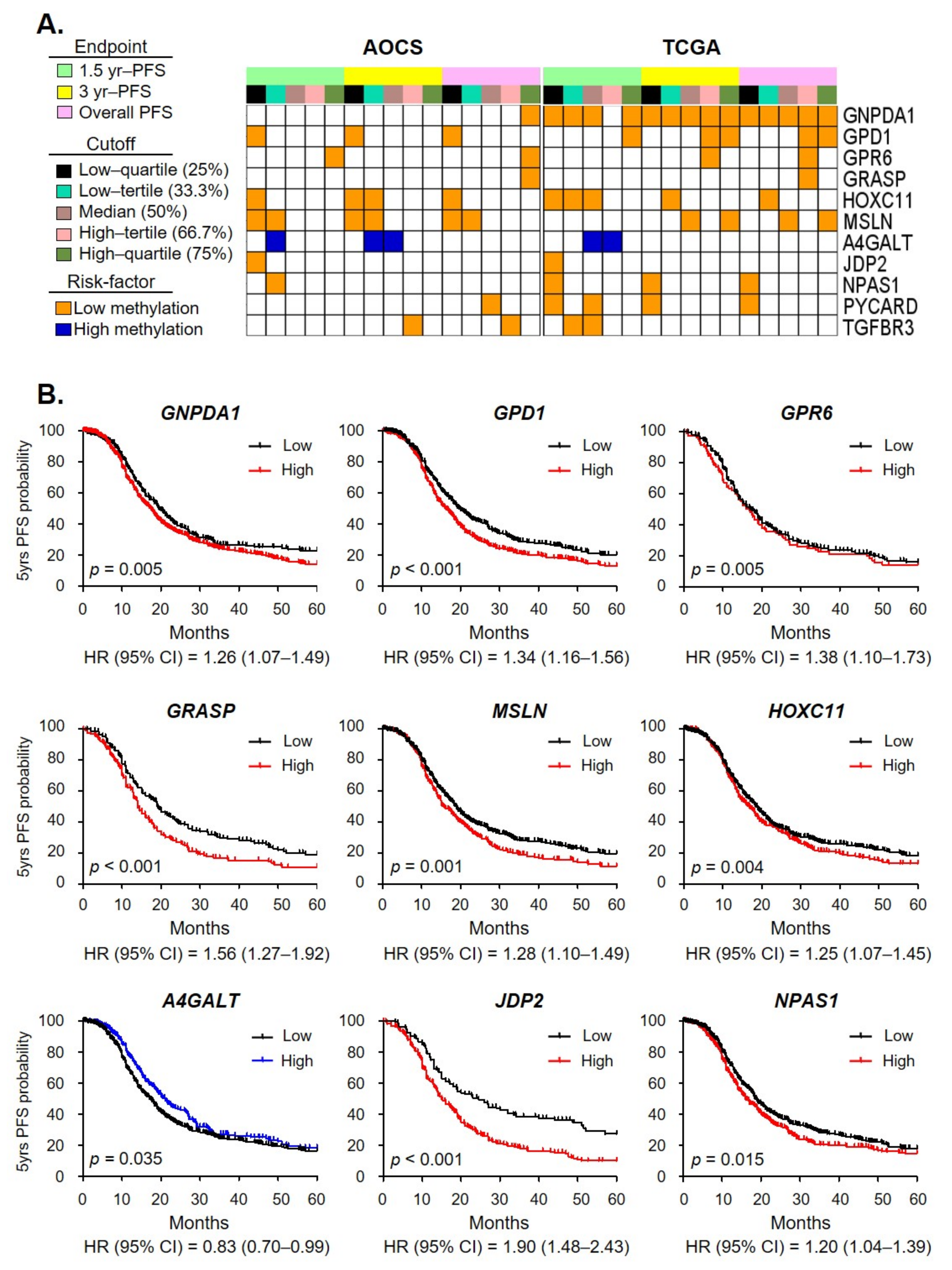
2.3. Clinical Relevance of OCSC Differentiation Genes
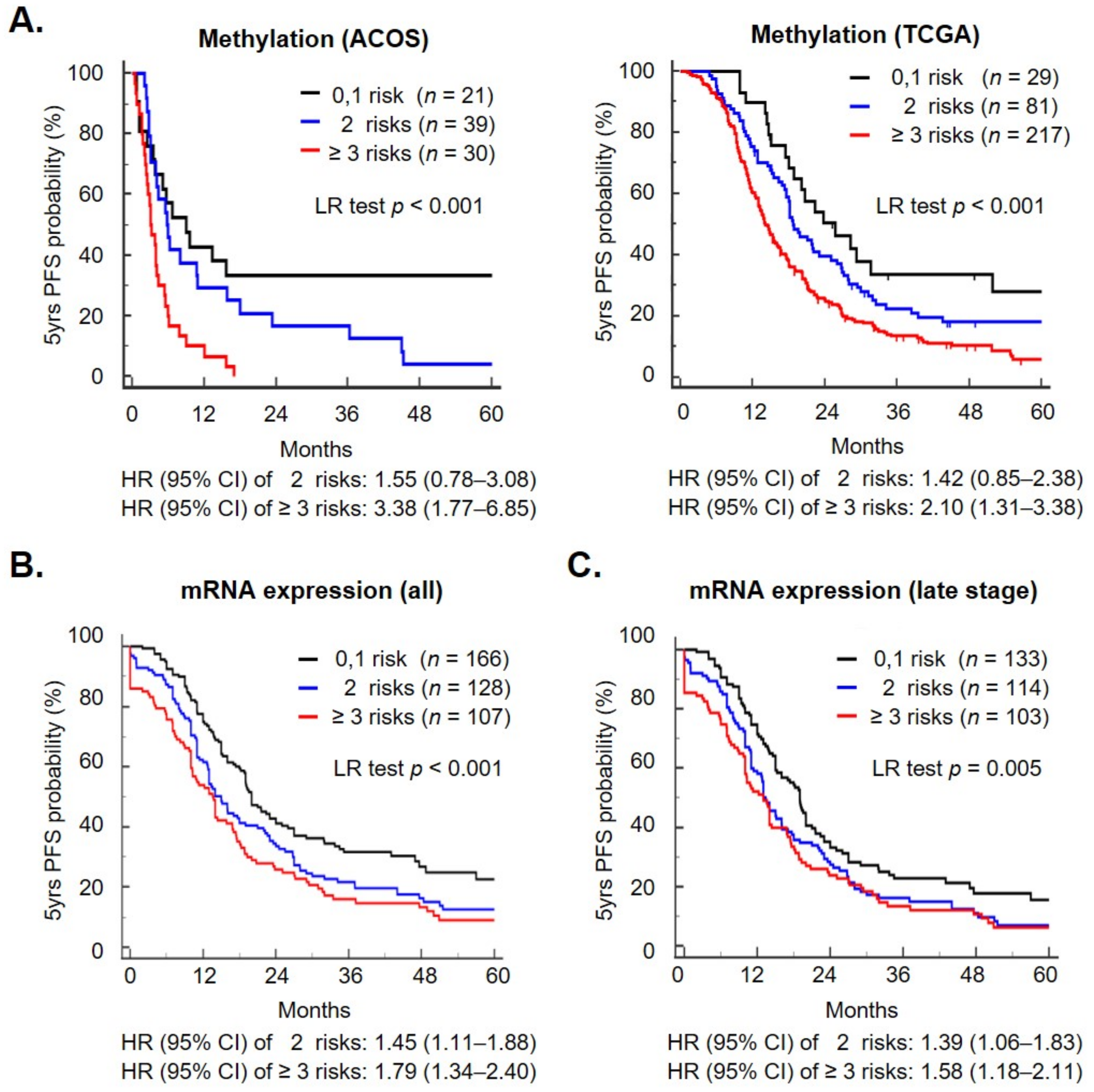
2.4. A Methylation Signature including Metabolism Genes as a Prognostic Biomarker
2.5. A Metabolism Shift during OCSC Differentiation
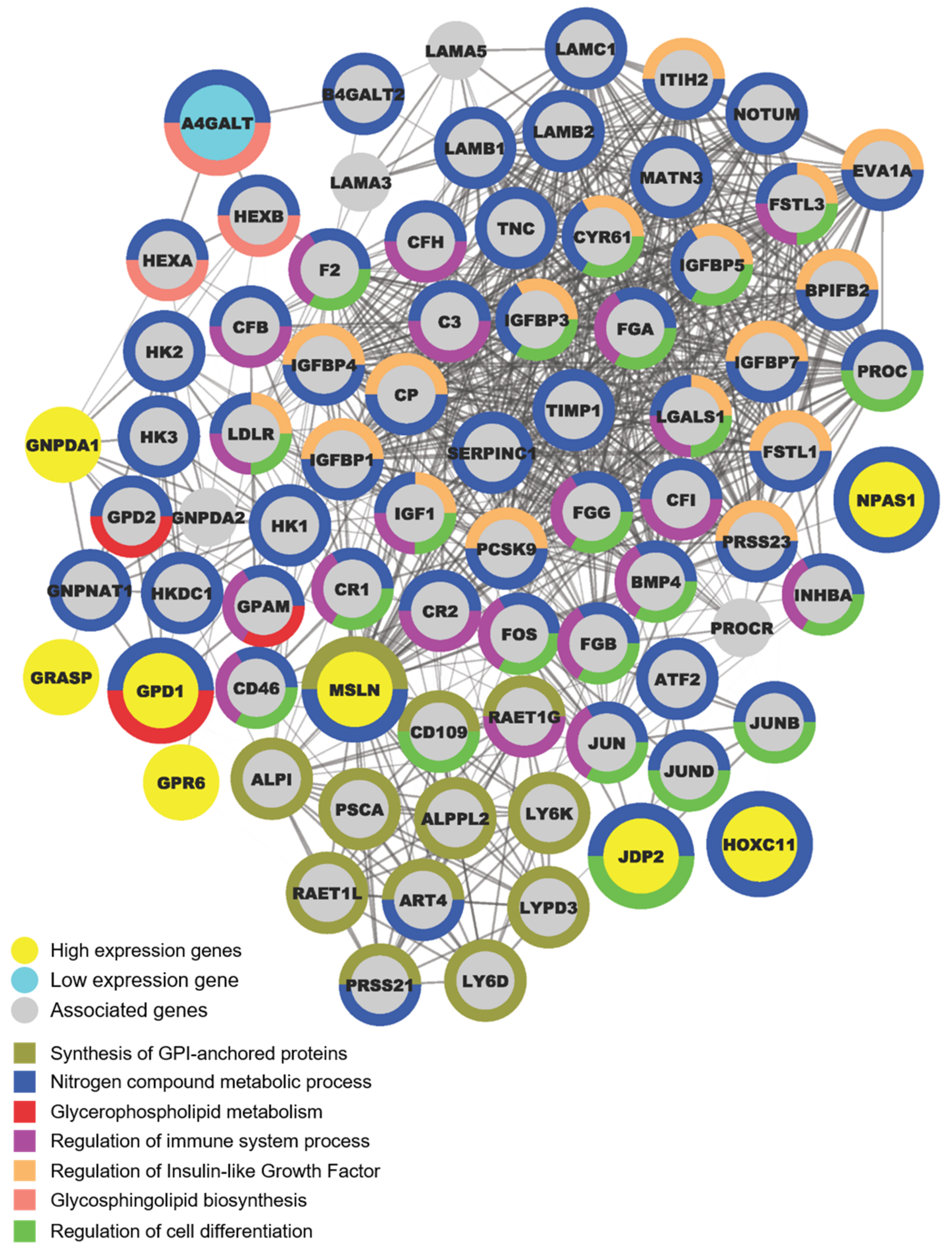
2.6. OCSC Differentiation Genes Were Involved in Regulating Suppressive Immune Microenvironment
3. Discussion
4. Materials and Methods
4.1. Cell Lines, CSCs and Enrichment of Spheroid Cells
4.2. Methylation Profiling and Gene Expression Array Analysis
4.3. Recurrent Risk Score and Survival Analysis
4.4. Protein–Protein Interaction (PPI) Networks Functional Enrichment Analysis
4.5. Estimation of the Infiltration Level of Immune Cells in Tumor Tissues
4.6. Statistical Analysis of Methylation Signature
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| AOCS | Australian Ovarian Cancer Study |
| CI | Confidence interval |
| EMT | Epithelial-to-mesenchymal transition |
| HR | Hazard ratio |
| HGSOC | High-grade serous ovarian cancer |
| EOC | Epithelial ovarian cancer |
| OCSC | Ovarian cancer stem cell |
| PFS | Progression-free survival |
| PPI | Protein–protein interaction |
| RRS | Recurrent risk score |
| STRING | Search tool for the retrieval of interacting genes/proteins |
| TCGA | The Cancer Genome Atlas |
References
- Chao, K.C.; Chen, Y.J.; Juang, C.M.; Lau, H.Y.; Wen, K.C.; Sung, P.L.; Fang, F.Y.; Twu, N.F.; Yen, M.S. Prognosis for advanced-stage primary peritoneal serous papillary carcinoma and serous ovarian cancer in Taiwan. Taiwan J. Obstet. Gynecol. 2013, 52, 81–84. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Landen, C.N., Jr.; Birrer, M.J.; Sood, A.K. Early events in the pathogenesis of epithelial ovarian cancer. J. Clin. Oncol. 2008, 26, 995–1005. [Google Scholar] [CrossRef] [PubMed]
- Novak, C.; Horst, E.; Mehta, G. Review: Mechanotransduction in ovarian cancer: Shearing into the unknown. APL Bioeng. 2018, 2, 031701. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Davis, A.; Tinker, A.V.; Friedlander, M. “Platinum resistant” ovarian cancer: What is it, who to treat and how to measure benefit? Gynecol. Oncol. 2014, 133, 624–631. [Google Scholar] [CrossRef] [PubMed]
- Tan, D.S.; Agarwal, R.; Kaye, S.B. Mechanisms of transcoelomic metastasis in ovarian cancer. Lancet Oncol. 2006, 7, 925–934. [Google Scholar] [CrossRef]
- Pereira, A.; Mendizabal, E.; de Leon, J.; Perez-Medina, T.; Magrina, J.F.; Magtibay, P.M.; Rodriguez-Tapia, A.; Lizarraga, S.; Ortiz-Quintana, L. Peritoneal carcinomatosis: A malignant disease with an embryological origin? Surg. Oncol. 2015, 24, 305–311. [Google Scholar] [CrossRef]
- Lengyel, E. Ovarian cancer development and metastasis. Am. J. Pathol. 2010, 177, 1053–1064. [Google Scholar] [CrossRef]
- Kenny, H.A.; Chiang, C.Y.; White, E.A.; Schryver, E.M.; Habis, M.; Romero, I.L.; Ladanyi, A.; Penicka, C.V.; George, J.; Matlin, K.; et al. Mesothelial cells promote early ovarian cancer metastasis through fibronectin secretion. J. Clin. Investig. 2014, 124, 4614–4628. [Google Scholar] [CrossRef] [Green Version]
- Yeung, T.L.; Leung, C.S.; Yip, K.P.; Au Yeung, C.L.; Wong, S.T.; Mok, S.C. Cellular and molecular processes in ovarian cancer metastasis. A Review in the Theme: Cell and Molecular Processes in Cancer Metastasis. Am. J. Physiol. Cell Physiol. 2015, 309, C444–C456. [Google Scholar] [CrossRef] [Green Version]
- Jordan, C.T.; Guzman, M.L.; Noble, M. Cancer stem cells. N. Engl. J. Med. 2006, 355, 1253–1261. [Google Scholar] [CrossRef]
- Alvero, A.B.; Chen, R.; Fu, H.H.; Montagna, M.; Schwartz, P.E.; Rutherford, T.; Silasi, D.A.; Steffensen, K.D.; Waldstrom, M.; Visintin, I.; et al. Molecular phenotyping of human ovarian cancer stem cells unravels the mechanisms for repair and chemoresistance. Cell Cycle 2009, 8, 158–166. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bregenzer, M.E.; Horst, E.N.; Mehta, P.; Novak, C.M.; Repetto, T.; Mehta, G. The Role of Cancer Stem Cells and Mechanical Forces in Ovarian Cancer Metastasis. Cancers 2019, 11, 1008. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dalerba, P.; Cho, R.W.; Clarke, M.F. Cancer stem cells: Models and concepts. Annu. Rev. Med. 2007, 58, 267–284. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhou, S.; Schuetz, J.D.; Bunting, K.D.; Colapietro, A.M.; Sampath, J.; Morris, J.J.; Lagutina, I.; Grosveld, G.C.; Osawa, M.; Nakauchi, H.; et al. The ABC transporter Bcrp1/ABCG2 is expressed in a wide variety of stem cells and is a molecular determinant of the side-population phenotype. Nat. Med. 2001, 7, 1028–1034. [Google Scholar] [CrossRef] [PubMed]
- Challen, G.A.; Little, M.H. A side order of stem cells: The SP phenotype. Stem Cells 2006, 24, 3–12. [Google Scholar] [CrossRef]
- Goodell, M.A.; Rosenzweig, M.; Kim, H.; Marks, D.F.; DeMaria, M.; Paradis, G.; Grupp, S.A.; Sieff, C.A.; Mulligan, R.C.; Johnson, R.P. Dye efflux studies suggest that hematopoietic stem cells expressing low or undetectable levels of CD34 antigen exist in multiple species. Nat. Med. 1997, 3, 1337–1345. [Google Scholar] [CrossRef]
- He, Q.Z.; Luo, X.Z.; Wang, K.; Zhou, Q.; Ao, H.; Yang, Y.; Li, S.X.; Li, Y.; Zhu, H.T.; Duan, T. Isolation and characterization of cancer stem cells from high-grade serous ovarian carcinomas. Cell Physiol. Biochem. 2014, 33, 173–184. [Google Scholar] [CrossRef]
- Silva, I.A.; Bai, S.; McLean, K.; Yang, K.; Griffith, K.; Thomas, D.; Ginestier, C.; Johnston, C.; Kueck, A.; Reynolds, R.K.; et al. Aldehyde dehydrogenase in combination with CD133 defines angiogenic ovarian cancer stem cells that portend poor patient survival. Cancer Res. 2011, 71, 3991–4001. [Google Scholar] [CrossRef] [Green Version]
- Liu, K.C.; Yo, Y.T.; Huang, R.L.; Wang, Y.C.; Liao, Y.P.; Huang, T.S.; Chao, T.K.; Lin, C.K.; Weng, S.J.; Ma, K.H.; et al. Ovarian cancer stem-like cells show induced translineage-differentiation capacity and are suppressed by alkaline phosphatase inhibitor. Oncotarget 2013, 4, 2366–2382. [Google Scholar] [CrossRef] [Green Version]
- Weiss, S.; Reynolds, B.A.; Vescovi, A.L.; Morshead, C.; Craig, C.G.; van der Kooy, D. Is there a neural stem cell in the mammalian forebrain? Trends Neurosci. 1996, 19, 387–393. [Google Scholar] [CrossRef]
- Reynolds, B.A.; Weiss, S. Clonal and population analyses demonstrate that an EGF-responsive mammalian embryonic CNS precursor is a stem cell. Dev. Biol. 1996, 175, 1–13. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dontu, G.; Abdallah, W.M.; Foley, J.M.; Jackson, K.W.; Clarke, M.F.; Kawamura, M.J.; Wicha, M.S. In vitro propagation and transcriptional profiling of human mammary stem/progenitor cells. Genes Dev. 2003, 17, 1253–1270. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fang, D.; Nguyen, T.K.; Leishear, K.; Finko, R.; Kulp, A.N.; Hotz, S.; Van Belle, P.A.; Xu, X.; Elder, D.E.; Herlyn, M. A tumorigenic subpopulation with stem cell properties in melanomas. Cancer Res. 2005, 65, 9328–9337. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, S.; Balch, C.; Chan, M.W.; Lai, H.C.; Matei, D.; Schilder, J.M.; Yan, P.S.; Huang, T.H.; Nephew, K.P. Identification and characterization of ovarian cancer-initiating cells from primary human tumors. Cancer Res. 2008, 68, 4311–4320. [Google Scholar] [CrossRef] [Green Version]
- Gyorffy, B.; Lanczky, A.; Szallasi, Z. Implementing an online tool for genome-wide validation of survival-associated biomarkers in ovarian-cancer using microarray data from 1287 patients. Endocr. Relat. Cancer 2012, 19, 197–208. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Singh, R.K.; Dhadve, A.; Sakpal, A.; De, A.; Ray, P. An active IGF-1R-AKT signaling imparts functional heterogeneity in ovarian CSC population. Sci. Rep. 2016, 6, 36612. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Motohara, T.; Masuda, K.; Morotti, M.; Zheng, Y.; El-Sahhar, S.; Chong, K.Y.; Wietek, N.; Alsaadi, A.; Karaminejadranjbar, M.; Hu, Z.; et al. An evolving story of the metastatic voyage of ovarian cancer cells: Cellular and molecular orchestration of the adipose-rich metastatic microenvironment. Oncogene 2019, 38, 2885–2898. [Google Scholar] [CrossRef] [Green Version]
- Jiang, P.; Gu, S.; Pan, D.; Fu, J.; Sahu, A.; Hu, X.; Li, Z.; Traugh, N.; Bu, X.; Li, B.; et al. Signatures of T cell dysfunction and exclusion predict cancer immunotherapy response. Nat. Med. 2018, 24, 1550–1558. [Google Scholar] [CrossRef]
- Li, T.; Fu, J.; Zeng, Z.; Cohen, D.; Li, J.; Chen, Q.; Li, B.; Liu, X.S. TIMER2.0 for analysis of tumor-infiltrating immune cells. Nucleic Acids Res. 2020, 48, W509–W514. [Google Scholar] [CrossRef]
- Malanchi, I.; Santamaria-Martinez, A.; Susanto, E.; Peng, H.; Lehr, H.A.; Delaloye, J.F.; Huelsken, J. Interactions between cancer stem cells and their niche govern metastatic colonization. Nature 2011, 481, 85–89. [Google Scholar] [CrossRef]
- Fishman, A.; Shalom-Paz, E.; Fejgin, M.; Gaber, E.; Altaras, M.; Amiel, A. Comparing the genetic changes detected in the primary and secondary tumor sites of ovarian cancer using comparative genomic hybridization. Int. J. Gynecol. Cancer 2005, 15, 261–266. [Google Scholar] [CrossRef] [PubMed]
- Cao, Z.; Livas, T.; Kyprianou, N. Anoikis and EMT: Lethal “Liaisons” during Cancer Progression. Crit. Rev. Oncog. 2016, 21, 155–168. [Google Scholar] [CrossRef] [Green Version]
- Li, S.S.; Ip, C.K.M.; Tang, M.Y.H.; Tang, M.K.S.; Tong, Y.; Zhang, J.; Hassan, A.A.; Mak, A.S.C.; Yung, S.; Chan, T.M.; et al. Sialyl Lewis(x)-P-selectin cascade mediates tumor-mesothelial adhesion in ascitic fluid shear flow. Nat. Commun. 2019, 10, 2406. [Google Scholar] [CrossRef] [Green Version]
- Ajani, J.A.; Song, S.; Hochster, H.S.; Steinberg, I.B. Cancer stem cells: The promise and the potential. Semin. Oncol. 2015, 42 (Suppl. 1), S3–S17. [Google Scholar] [CrossRef] [PubMed]
- Adorno-Cruz, V.; Kibria, G.; Liu, X.; Doherty, M.; Junk, D.J.; Guan, D.; Hubert, C.; Venere, M.; Mulkearns-Hubert, E.; Sinyuk, M.; et al. Cancer stem cells: Targeting the roots of cancer, seeds of metastasis, and sources of therapy resistance. Cancer Res. 2015, 75, 924–929. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, M.; Yang, X.; Chen, S.; Jia, W.; Ma, X.; Wang, J.; Qian, Y.; Lei, D.; Liu, H.; Pan, X. GPR12 inhibits migration and promotes apoptosis in esophageal cancer and hypopharyngeal cancer cells. Thorac. Cancer 2021, 12, 1525–1535. [Google Scholar] [CrossRef] [PubMed]
- Das, B.; Pal, B.; Bhuyan, R.; Li, H.; Sarma, A.; Gayan, S.; Talukdar, J.; Sandhya, S.; Bhuyan, S.; Gogoi, G.; et al. MYC Regulates the HIF2alpha Stemness Pathway via Nanog and Sox2 to Maintain Self-Renewal in Cancer Stem Cells versus Non-Stem Cancer Cells. Cancer Res. 2019, 79, 4015–4025. [Google Scholar] [CrossRef]
- Lathia, J.D.; Liu, H. Overview of Cancer Stem Cells and Stemness for Community Oncologists. Target Oncol. 2017, 12, 387–399. [Google Scholar] [CrossRef] [Green Version]
- Malta, T.M.; Sokolov, A.; Gentles, A.J.; Burzykowski, T.; Poisson, L.; Weinstein, J.N.; Kaminska, B.; Huelsken, J.; Omberg, L.; Gevaert, O.; et al. Machine Learning Identifies Stemness Features Associated with Oncogenic Dedifferentiation. Cell 2018, 173, 338–354. [Google Scholar] [CrossRef] [Green Version]
- Lu, T.Y.; Lu, R.M.; Liao, M.Y.; Yu, J.; Chung, C.H.; Kao, C.F.; Wu, H.C. Epithelial cell adhesion molecule regulation is associated with the maintenance of the undifferentiated phenotype of human embryonic stem cells. J. Biol. Chem. 2010, 285, 8719–8732. [Google Scholar] [CrossRef] [Green Version]
- Tayama, S.; Motohara, T.; Narantuya, D.; Li, C.; Fujimoto, K.; Sakaguchi, I.; Tashiro, H.; Saya, H.; Nagano, O.; Katabuchi, H. The impact of EpCAM expression on response to chemotherapy and clinical outcomes in patients with epithelial ovarian cancer. Oncotarget 2017, 8, 44312–44325. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Moffitt, L.; Karimnia, N.; Stephens, A.; Bilandzic, M. Therapeutic Targeting of Collective Invasion in Ovarian Cancer. Int. J. Mol. Sci. 2019, 20, 1466. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Attar, M.A.; Santy, L.C. The scaffolding protein GRASP/Tamalin directly binds to Dock180 as well as to cytohesins facilitating GTPase crosstalk in epithelial cell migration. BMC Cell Biol. 2013, 14, 9. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, Z.; Du, R.; Long, J.; Dong, A.; Fan, J.; Guo, K.; Xu, Y. JDP2 inhibits the epithelial-to-mesenchymal transition in pancreatic cancer BxPC3 cells. Tumour Biol. 2012, 33, 1527–1534. [Google Scholar] [CrossRef] [PubMed]
- Li, D.; Cheng, X.; Zheng, W.; Chen, J. Glucosamine-6-Phosphate Isomerase 1 Promotes Tumor Progression and Indicates Poor Prognosis in Hepatocellular Carcinoma. Cancer Manag. Res. 2020, 12, 4923–4935. [Google Scholar] [CrossRef]
- Pavlova, N.N.; Thompson, C.B. The Emerging Hallmarks of Cancer Metabolism. Cell Metab. 2016, 23, 27–47. [Google Scholar] [CrossRef] [Green Version]
- Maccio, A.; Gramignano, G.; Cherchi, M.C.; Tanca, L.; Melis, L.; Madeddu, C. Role of M1-polarized tumor-associated macrophages in the prognosis of advanced ovarian cancer patients. Sci. Rep. 2020, 10, 6096. [Google Scholar] [CrossRef] [Green Version]
- Maccio, A.; Madeddu, C. Blocking inflammation to improve immunotherapy of advanced cancer. Immunology 2020, 159, 357–364. [Google Scholar] [CrossRef] [Green Version]
- Bodmer, M.; Becker, C.; Meier, C.; Jick, S.S.; Meier, C.R. Use of metformin and the risk of ovarian cancer: A case-control analysis. Gynecol. Oncol. 2011, 123, 200–204. [Google Scholar] [CrossRef]
- Wen, K.C.; Sung, P.L.; Wu, A.T.H.; Chou, P.C.; Lin, J.H.; Huang, C.F.; Yeung, S.J.; Lee, M.H. Neoadjuvant metformin added to conventional chemotherapy synergizes anti-proliferative effects in ovarian cancer. J. Ovarian Res. 2020, 13, 95. [Google Scholar] [CrossRef]
- Deshmukh, A.; Deshpande, K.; Arfuso, F.; Newsholme, P.; Dharmarajan, A. Cancer stem cell metabolism: A potential target for cancer therapy. Mol. Cancer 2016, 15, 69. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pasto, A.; Bellio, C.; Pilotto, G.; Ciminale, V.; Silic-Benussi, M.; Guzzo, G.; Rasola, A.; Frasson, C.; Nardo, G.; Zulato, E.; et al. Cancer stem cells from epithelial ovarian cancer patients privilege oxidative phosphorylation, and resist glucose deprivation. Oncotarget 2014, 5, 4305–4319. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rusu, P.; Shao, C.; Neuerburg, A.; Acikgoz, A.A.; Wu, Y.; Zou, P.; Phapale, P.; Shankar, T.S.; Doring, K.; Dettling, S.; et al. GPD1 Specifically Marks Dormant Glioma Stem Cells with a Distinct Metabolic Profile. Cell Stem Cell 2019, 25, 241–257. [Google Scholar] [CrossRef]
- Chang, P.Y.; Liao, Y.P.; Wang, H.C.; Chen, Y.C.; Huang, R.L.; Wang, Y.C.; Yuan, C.C.; Lai, H.C. An epigenetic signature of adhesion molecules predicts poor prognosis of ovarian cancer patients. Oncotarget 2017, 8, 53432–53449. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pisanic, T.R., 2nd; Cope, L.M.; Lin, S.F.; Yen, T.T.; Athamanolap, P.; Asaka, R.; Nakayama, K.; Fader, A.N.; Wang, T.H.; Shih, I.M.; et al. Methylomic Analysis of Ovarian Cancers Identifies Tumor-Specific Alterations Readily Detectable in Early Precursor Lesions. Clin. Cancer Res. 2018, 24, 6536–6547. [Google Scholar] [CrossRef] [Green Version]
- Bodelon, C.; Killian, J.K.; Sampson, J.N.; Anderson, W.F.; Matsuno, R.; Brinton, L.A.; Lissowska, J.; Anglesio, M.S.; Bowtell, D.D.L.; Doherty, J.A.; et al. Molecular Classification of Epithelial Ovarian Cancer Based on Methylation Profiling: Evidence for Survival Heterogeneity. Clin. Cancer Res. 2019, 25, 5937–5946. [Google Scholar] [CrossRef]
- Oza, A.M.; Matulonis, U.A.; Alvarez Secord, A.; Nemunaitis, J.; Roman, L.D.; Blagden, S.P.; Banerjee, S.; McGuire, W.P.; Ghamande, S.; Birrer, M.J.; et al. A Randomized Phase II Trial of Epigenetic Priming with Guadecitabine and Carboplatin in Platinum-resistant, Recurrent Ovarian Cancer. Clin. Cancer Res. 2020, 26, 1009–1016. [Google Scholar] [CrossRef]
- Cardenas, H.; Fang, F.; Jiang, G.; Perkins, S.M.; Zhang, C.; Emerson, R.E.; Hutchins, G.; Keer, H.N.; Liu, Y.; Matei, D.; et al. Methylomic Signatures of High Grade Serous Ovarian Cancer. Epigenetics 2021, 16, 1201–1216. [Google Scholar] [CrossRef]

| Univariate | Multivariate | ||||
|---|---|---|---|---|---|
| Characteristics | Crude HR (95% CI) | p-Value | Adjusted HR 1 (95% CI) | p-Value | |
| Risk group | 0–1 gene | 1 | 1 | ||
| 2 genes | 1.45 (1.10–1.90) | 0.008 | 1.46 (1.11–1.92) | 0.007 | |
| ≥3 genes | 1.79 (1.35–2.38) | <0.001 | 1.55 (1.17–2.06) | 0.002 | |
| Stage | Early | 1 | 1 | ||
| Late | 5.36 (3.06–9.36) | <0.001 | 3.66 (2.06–6.52) | <0.001 | |
| Grade | G1 | 1 | 1 | ||
| G2 | 3.81 (1.54–9.39) | 0.004 | 1.85 (0.74–4.67) | 0.191 | |
| G3 | 4.06 (1.67–9.87) | 0.002 | 1.76 0.70–4.40) | 0.227 | |
| Debulk | Optimal | 1 | 1 | ||
| Suboptimal | 2.17 (1.72–2.74) | <0.001 | 1.77 (1.38–2.26) | <0.001 | |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chen, L.-Y.; Huang, R.-L.; Su, P.-H.; Chu, L.-H.; Weng, Y.-C.; Wang, H.-C.; Lai, H.-C.; Wen, K.-C. Epigenomic Profiling of Epithelial Ovarian Cancer Stem-Cell Differentiation Reveals GPD1 Associated Immune Suppressive Microenvironment and Poor Prognosis. Int. J. Mol. Sci. 2022, 23, 5120. https://doi.org/10.3390/ijms23095120
Chen L-Y, Huang R-L, Su P-H, Chu L-H, Weng Y-C, Wang H-C, Lai H-C, Wen K-C. Epigenomic Profiling of Epithelial Ovarian Cancer Stem-Cell Differentiation Reveals GPD1 Associated Immune Suppressive Microenvironment and Poor Prognosis. International Journal of Molecular Sciences. 2022; 23(9):5120. https://doi.org/10.3390/ijms23095120
Chicago/Turabian StyleChen, Lin-Yu, Rui-Lan Huang, Po-Hsuan Su, Ling-Hui Chu, Yu-Chun Weng, Hui-Chen Wang, Hung-Cheng Lai, and Kuo-Chang Wen. 2022. "Epigenomic Profiling of Epithelial Ovarian Cancer Stem-Cell Differentiation Reveals GPD1 Associated Immune Suppressive Microenvironment and Poor Prognosis" International Journal of Molecular Sciences 23, no. 9: 5120. https://doi.org/10.3390/ijms23095120
APA StyleChen, L.-Y., Huang, R.-L., Su, P.-H., Chu, L.-H., Weng, Y.-C., Wang, H.-C., Lai, H.-C., & Wen, K.-C. (2022). Epigenomic Profiling of Epithelial Ovarian Cancer Stem-Cell Differentiation Reveals GPD1 Associated Immune Suppressive Microenvironment and Poor Prognosis. International Journal of Molecular Sciences, 23(9), 5120. https://doi.org/10.3390/ijms23095120






