Comparative Transcriptomics Reveals the Molecular Mechanism of the Parental Lines of Maize Hybrid An’nong876 in Response to Salt Stress
Abstract
:1. Introduction
2. Results
2.1. Characteristics of Maize Inbred Lines CM37 and cmh15
2.2. RNA-seq Analysis and Identification of DEGs
2.3. Gene Ontology and Kyoto Encyclopedia of Genes and Genomes Pathway Functional Enrichment Analysis of DEGs
2.4. Analysis of DEGs Encoding Transcription Factors
2.5. Analysis of DEGs Associated with Plant Hormone
2.6. Analysis of DEGs Associated with Photosynthesis
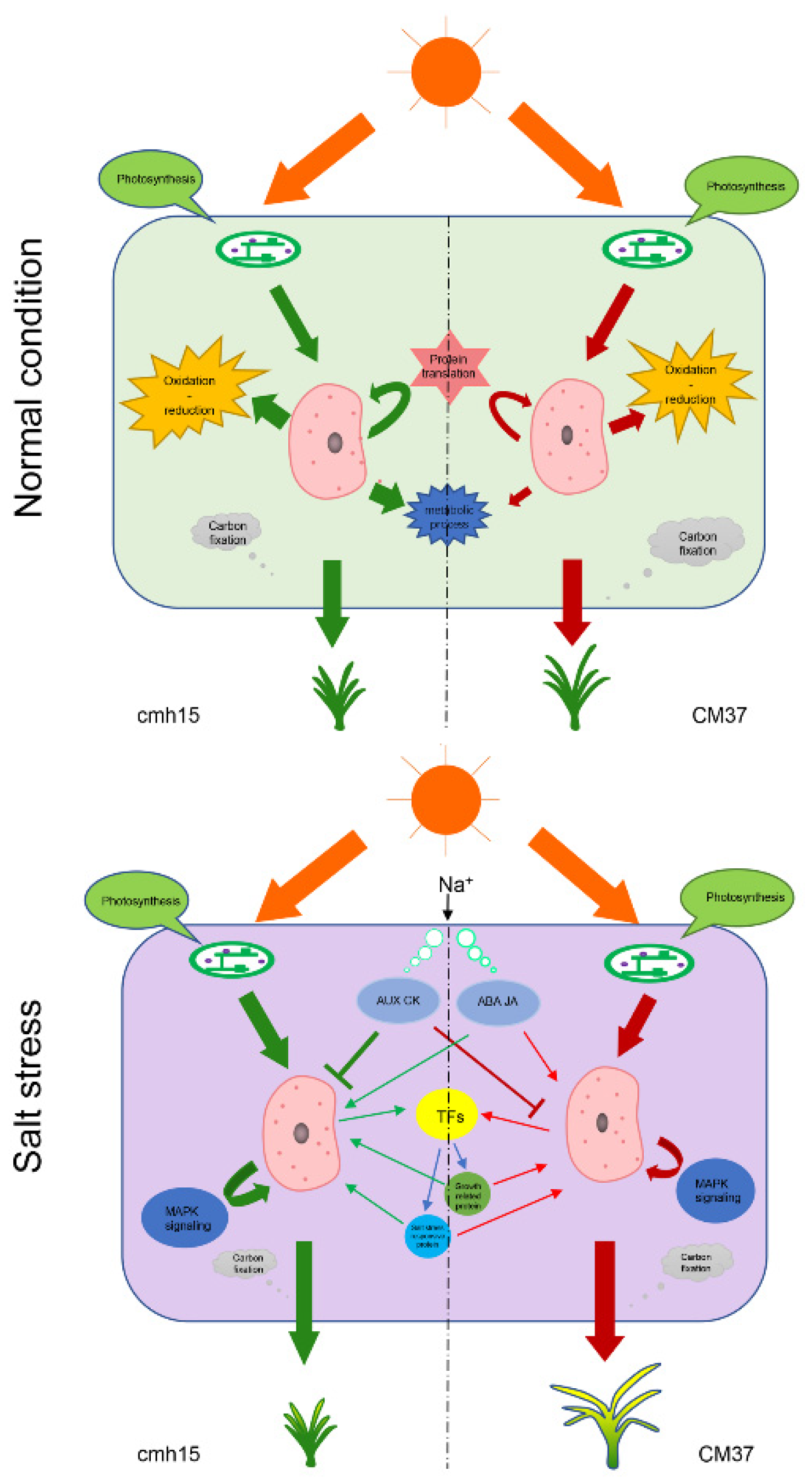
3. Discussion
4. Materials and Methods
4.1. Plant Growth and Salt Treatment
4.2. Determination of RWC, REL MDA and Protocol of DAB Staining
4.3. Extraction of RNA and Construction of cDNA Library
4.4. Sequence Assembly and Data Analysis
4.5. Validation of RNA-seq Data by qRT-PCR
4.6. GO and KEGG Enrichment Analysis
4.7. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| ABA | abscisic acid |
| ABF | ABA-responsive element binding factor |
| AUX | auxin |
| CK | cytokinin |
| CKF | control group of cmh15 |
| CKM | control group of CM37 |
| DAB | diaminobenzidine |
| DEG | differential expressed gene |
| GO | gene Ontology |
| HKT1 | high affinity potassium transporter1 |
| JA | jasmonic acid |
| KEGG | Kyoto Encyclopedia of Genes and Genomes |
| MAPK | mitogen-activated protein kinase |
| MDA | malondialdehyde |
| PCA | principal component analysis |
| POD | peroxidase |
| qRT-PCR | quantitative real-time polymerase chain reaction |
| REL | relative electrolyte leakage |
| RNA-seq | RNA sequencing |
| ROS | reactive oxygen species |
| RWC | relative water content |
| SA | salicylic acid |
| SOD | superoxide dismutase |
| SOS | salt overly sensitive; |
| UVF | salt treatment group of cmh15 |
| UVM | salt treatment of CM37 |
References
- Zhang, M.; Cao, Y.B.; Wang, Z.P.; Wang, Z.Q.; Shi, J.P.; Liang, X.Y.; Song, W.B.; Chen, Q.J.; Lai, J.S.; Jiang, C.F. A retrotransposon in an HKT1 family sodium transporter causes variation of leaf Na+ exclusion and salt tolerance in maize. New Phytol. 2018, 217, 1161–1176. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Luo, X.; Wang, B.C.; Gao, S.; Zhang, F.; Terzaghi, W.; Dai, M.Q. Genome-wide association study dissects the genetic bases of salt tolerance in maize seedlings. J. Integr. Plant Biol. 2019, 61, 658–674. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhu, J.K. Abiotic Stress Signaling and Responses in Plants. Cell 2016, 167, 313–324. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Endler, A.; Kesten, C.; Schneider, R.; Zhang, Y.; Ivakov, A.; Froehlich, A.; Funke, N.; Persson, S. A Mechanism for Sustained Cellulose Synthesis during Salt Stress. Cell 2015, 162, 1353–1364. [Google Scholar] [CrossRef] [Green Version]
- Yang, Y.Q.; Guo, Y. Unraveling salt stress signaling in plants. J. Integr. Plant Biol. 2018, 60, 796–804. [Google Scholar] [CrossRef] [Green Version]
- Yang, Y.Q.; Guo, Y. Elucidating the molecular mechanisms mediating plant salt-stress responses. New Phytol. 2018, 217, 523–539. [Google Scholar] [CrossRef] [Green Version]
- Munns, R.; Tester, M. Mechanisms of salinity tolerance. Annu. Rev. Plant Biol. 2008, 59, 651–681. [Google Scholar] [CrossRef] [Green Version]
- Deinlein, U.; Stephan, A.B.; Horie, T.; Luo, W.; Xu, G.H.; Schroeder, J.I. Plant salt-tolerance mechanisms. Trends Plant Sci. 2014, 19, 371–379. [Google Scholar] [CrossRef] [Green Version]
- Kudla, J.; Becker, D.; Grill, E.; Hedrich, R.; Hippler, M.; Kummer, U.; Parniske, M.; Romeis, T.; Schumacher, K. Advances and current challenges in calcium signaling. New Phytol. 2018, 218, 414–431. [Google Scholar] [CrossRef]
- Jiang, Z.H.; Zhou, X.P.; Tao, M.; Yuan, F.; Liu, L.L.; Wu, F.H.; Wu, X.M.; Xiang, Y.; Niu, Y.; Liu, F.; et al. Plant cell-surface GIPC sphingolipids sense salt to trigger Ca2+ influx. Nature 2019, 572, 341–346. [Google Scholar] [CrossRef]
- Quan, R.D.; Lin, H.X.; Mendoza, I.; Zhang, Y.G.; Cao, W.H.; Yang, Y.Q.; Shang, M.; Chen, S.Y.; Pardo, J.M.; Guo, Y. SCABP8/CBL10, a putative calcium sensor, interacts with the protein kinase SOS2 to protect Arabidopsis shoots from salt stress. Plant Cell 2007, 19, 1415–1431. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shi, H.Z.; Quintero, F.J.; Pardo, J.M.; Zhu, J.K. The putative plasma membrane Na+/H+ antiporter SOS1 controls long-distance Na+ transport in plants. Plant Cell 2002, 14, 465–477. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Davenport, R.J.; Munoz-Mayor, A.; Jha, D.; Essah, P.A.; Rus, A.; Tester, M. The Na+ transporter AtHKT1;1 controls retrieval of Na+ from the xylem in Arabidopsis. Plant Cell Environ. 2007, 30, 497–507. [Google Scholar] [CrossRef] [PubMed]
- An, D.; Chen, J.G.; Gao, Y.Q.; Li, X.; Chao, Z.F.; Chen, Z.R.; Li, Q.Q.; Han, M.L.; Wang, Y.L.; Wang, Y.F.; et al. AtHKT1 drives adaptation of Arabidopsis thaliana to salinity by reducing floral sodium content. PLoS Genet. 2017, 13, e1007086. [Google Scholar] [CrossRef] [Green Version]
- Hill, C.B.; Jha, D.; Bacic, A.; Tester, M.; Roessner, U. Characterization of Ion Contents and Metabolic Responses to Salt Stress of Different Arabidopsis AtHKT1;1 Genotypes and Their Parental Strains. Mol. Plant 2013, 6, 350–368. [Google Scholar] [CrossRef] [Green Version]
- Liang, W.J.; Ma, X.L.; Wan, P.; Liu, L.Y. Plant salt-tolerance mechanism: A review. Biochem. Biophys. Res. Commun. 2018, 495, 286–291. [Google Scholar] [CrossRef]
- Stadtman, E.R. Protein oxidation and aging. Free Radic. Res. 2006, 40, 1250–1258. [Google Scholar] [CrossRef] [Green Version]
- Zong, N.; Li, X.J.; Wang, L.; Wang, Y.; Wen, H.T.; Li, L.; Zhang, X.; Fan, Y.L.; Zhao, J. Maize ABP2 enhances tolerance to drought and salt stress in transgenic Arabidopsis. J. Integr. Agric. 2018, 17, 2379–2393. [Google Scholar] [CrossRef] [Green Version]
- Chen, Y.E.; Mao, J.J.; Sun, L.Q.; Huang, B.; Ding, C.B.; Gu, Y.; Liao, J.Q.; Hu, C.; Zhang, Z.W.; Yuan, S.; et al. Exogenous melatonin enhances salt stress tolerance in maize seedlings by improving antioxidant and photosynthetic capacity. Physiol. Plant. 2018, 164, 349–363. [Google Scholar] [CrossRef]
- Yu, Z.P.; Duan, X.B.; Luo, L.; Dai, S.J.; Ding, Z.J.; Xia, G.M. How Plant Hormones Mediate Salt Stress Responses. Trends Plant Sci. 2020, 25, 1117–1130. [Google Scholar] [CrossRef]
- Verma, V.; Ravindran, P.; Kumar, P.P. Plant hormone-mediated regulation of stress responses. BMC Plant Biol. 2016, 16, 86. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, K.; Li, G.J.; Bressan, R.A.; Song, C.P.; Zhu, J.K.; Zhao, Y. Abscisic acid dynamics, signaling, and functions in plants. J. Integr. Plant Biol. 2020, 62, 25–54. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Niu, M.L.; Xie, J.J.; Chen, C.; Cao, H.S.; Sun, J.Y.; Kong, Q.S.; Shabala, S.; Shabala, L.; Huang, Y.; Bie, Z.L. An early ABA-induced stomatal closure, Na+ sequestration in leaf vein and K+ retention in mesophyll confer salt tissue tolerance in Cucurbita species. J. Exp. Bot. 2018, 69, 4945–4960. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Farhangi-Abriz, S.; Ghassemi-Golezani, K. How can salicylic acid and jasmonic acid mitigate salt toxicity in soybean plants? Ecotoxicol. Environ. Saf. 2018, 147, 1010–1016. [Google Scholar] [CrossRef]
- Hazman, M.; Hause, B.; Eiche, E.; Nick, P.; Riemann, M. Increased tolerance to salt stress in OPDA-deficient rice ALLENE OXIDE CYCLASE mutants is linked to an increased ROS-scavenging activity. J. Exp. Bot. 2015, 66, 3339–3352. [Google Scholar] [CrossRef] [Green Version]
- Ahmad, R.M.; Cheng, C.; Sheng, J.; Wang, W.; Ren, H.; Aslam, M.; Yan, Y.X. Interruption of Jasmonic Acid Biosynthesis Causes Differential Responses in the Roots and Shoots of Maize Seedlings against Salt Stress. Int. J. Mol. Sci. 2019, 20, 6202. [Google Scholar] [CrossRef] [Green Version]
- Wang, J.; Qin, H.; Zhou, S.R.; Wei, P.C.; Zhang, H.W.; Zhou, Y.; Miao, Y.C.; Huang, R.F. The Ubiquitin-Binding Protein OsDSK2a Mediates Seedling Growth and Salt Responses by Regulating Gibberellin Metabolism in Rice. Plant Cell 2020, 32, 414–428. [Google Scholar] [CrossRef] [Green Version]
- Zhou, J.H.; Li, Z.Y.; Xiao, G.Q.; Zhai, M.J.; Pan, X.W.; Huang, R.F.; Zhang, H.W. CYP71D8L is a key regulator involved in growth and stress responses by mediating gibberellin homeostasis in rice. J. Exp. Bot. 2020, 71, 1160–1170. [Google Scholar]
- Hyoung, S.; Cho, S.H.; Chung, J.H.; So, W.M.; Cui, M.H.; Shin, J.S. Cytokinin oxidase PpCKX1 plays regulatory roles in development and enhances dehydration and salt tolerance in Physcomitrella patens. Plant Cell Rep. 2020, 39, 419–430. [Google Scholar] [CrossRef]
- Iglesias, M.J.; Terrile, M.C.; Bartoli, C.G.; D’Ippolito, S.; Casalongue, C.A. Auxin signaling participates in the adaptative response against oxidative stress and salinity by interacting with redox metabolism in Arabidopsis. Plant Mol. Biol. 2010, 74, 215–222. [Google Scholar] [CrossRef]
- Li, Z.Y.; Xu, Z.S.; He, G.Y.; Yang, G.X.; Chen, M.; Li, L.C.; Ma, Y.Z. A mutation in Arabidopsis BSK5 encoding a brassinosteroid-signaling kinase protein affects responses to salinity and abscisic acid. Biochem. Biophys. Res. Commun. 2012, 426, 522–527. [Google Scholar] [CrossRef] [PubMed]
- Fu, J.J.; Cheng, Y.B.; Linghu, J.J.; Yang, X.H.; Kang, L.; Zhang, Z.X.; Zhang, J.; He, C.; Du, X.M.; Peng, Z.Y.; et al. RNA sequencing reveals the complex regulatory network in the maize kernel. Nat. Commun. 2013, 4, 2832. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fernandez-Garcia, N.; Hernandez, M.; Casado-Vela, J.; Bru, R.; Elortza, F.; Hedden, P.; Olmos, E. Changes to the proteome and targeted metabolites of xylem sap in Brassica oleracea in response to salt stress. Plant Cell Environ. 2011, 34, 821–836. [Google Scholar] [CrossRef] [PubMed]
- Kakumanu, A.; Ambavaram, M.M.R.; Klumas, C.; Krishnan, A.; Batlang, U.; Myers, E.; Grene, R.; Pereira, A. Effects of Drought on Gene Expression in Maize Reproductive and Leaf Meristem Tissue Revealed by RNA-Seq. Plant Physiol. 2012, 160, 846–867. [Google Scholar] [CrossRef] [Green Version]
- Yu, F.; Liang, K.; Fang, T.; Zhao, H.L.; Han, X.S.; Cai, M.J.; Qiu, F.Z. A group VII ethylene response factor gene, ZmEREB180, coordinates waterlogging tolerance in maize seedlings. Plant Biotechnol. J. 2019, 17, 2286–2298. [Google Scholar] [CrossRef] [Green Version]
- Li, C.H.; Sun, B.C.; Li, Y.X.; Liu, C.; Wu, X.; Zhang, D.F.; Shi, Y.S.; Song, Y.C.; Buckler, E.S.; Zhang, Z.W.; et al. Numerous genetic loci identified for drought tolerance in the maize nested association mapping populations. BMC Genom. 2016, 17, 894. [Google Scholar] [CrossRef] [Green Version]
- Wang, C.T.; Yang, Q.A.; Yang, Y.M. Characterization of the ZmDBP4 gene encoding a CRT/DRE-binding protein responsive to drought and cold stress in maize. Acta Physiol. Plant. 2011, 33, 575–583. [Google Scholar] [CrossRef]
- Bo, C.; Chen, H.W.; Luo, G.W.; Li, W.; Zhang, X.G.; Ma, Q.; Cheng, B.J.; Cai, R.H. Maize WRKY114 gene negatively regulates salt-stress tolerance in transgenic rice. Plant Cell Rep. 2020, 39, 135–148. [Google Scholar] [CrossRef]
- Delgado, C.; Mora-Poblete, F.; Ahmar, S.; Chen, J.T.; Figueroa, C.R. Jasmonates and Plant Salt Stress: Molecular Players, Physiological Effects, and Improving Tolerance by Using Genome-Associated Tools. Int. J. Mol. Sci. 2021, 22, 3082. [Google Scholar]
- Black, C.C.; Brown, R.H.; Moore, R.C. Plant Photosynthesis; Springer: New York, NY, USA, 1978. [Google Scholar]
- van Zelm, E.; Zhang, Y.X.; Testerink, C. Salt Tolerance Mechanisms of Plants. Annu. Rev. Plant Biol. 2020, 71, 403–433. [Google Scholar] [CrossRef] [Green Version]
- Wu, T.Y.; Wu, X.Q.; Xu, X.Q.; Kong, W.L.; Wu, F. Salt Tolerance Mechanism and Species Identification of the Plant Rhizosphere Bacterium JYZ-SD2. Curr. Microbiol. 2020, 77, 388–395. [Google Scholar] [CrossRef] [PubMed]
- Abbasi, H.; Jamil, M.; Haq, A.; Ali, S.; Ahmad, R.; Malik, Z.; Parveen, Z. Salt stress manifestation on plants, mechanism of salt tolerance and potassium role in alleviating it: A review. Zemdirbyste 2016, 103, 229–238. [Google Scholar] [CrossRef] [Green Version]
- Wang, Y.; Huang, L.; Du, F.; Wang, J.; Zhao, X.; Li, Z.; Wang, W.; Xu, J.; Fu, B. Comparative transcriptome and metabolome profiling reveal molecular mechanisms underlying OsDRAP1-mediated salt tolerance in rice. Sci. Rep. 2021, 11, 5166. [Google Scholar] [CrossRef] [PubMed]
- Duarte-Delgado, D.; Said, D.; Schoof, H.; Oyiga, B.C.; Schneider, M.; Mathew, B.; Léon, J.; Ballvora, A. Transcriptome profiling at osmotic and ionic phases of salt stress response in bread wheat uncovers trait-specific candidate genes. BMC Plant Biol. 2020, 20, 428. [Google Scholar] [CrossRef]
- Umezawa, T.; Sugiyama, N.; Mizoguchi, M.; Hayashi, S.; Myouga, F.; Yamaguchi-Shinozaki, K.; Ishihama, Y.; Hirayama, T.; Shinozaki, K. Type 2C protein phosphatases directly regulate abscisic acid-activated protein kinases in Arabidopsis. Proc. Natl. Acad. Sci. USA 2009, 106, 17588–17593. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Thalmann, M.; Pazmino, D.; Seung, D.; Horrer, D.; Nigro, A.; Meier, T.; Kolling, K.; Pfeifhofer, H.W.; Zeeman, S.C.; Santelia, D. Regulation of Leaf Starch Degradation by Abscisic Acid Is Important for Osmotic Stress Tolerance in Plants. Plant Cell 2016, 28, 1860–1878. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ye, H.Y.; Du, H.; Tang, N.; Li, X.H.; Xiong, L.Z. Identification and expression profiling analysis of TIFY family genes involved in stress and phytohormone responses in rice. Plant Mol. Biol. 2009, 71, 291–305. [Google Scholar] [CrossRef]
- Valenzuela, C.E.; Acevedo-Acevedo, O.; Miranda, G.S.; Vergara-Barros, P.; Holuigue, L.; Figueroa, C.R.; Figueroa, P.M. Salt stress response triggers activation of the jasmonate signaling pathway leading to inhibition of cell elongation in Arabidopsis primary root. J. Exp. Bot. 2016, 67, 4209–4220. [Google Scholar] [CrossRef] [Green Version]
- Iglesias, M.J.; Terrile, M.C.; Windels, D.; Lombardo, M.C.; Bartoli, C.G.; Vazquez, F.; Estelle, M.; Casalongue, C.A. MiR393 regulation of auxin signaling and redox-related components during acclimation to salinity in Arabidopsis. PLoS ONE 2014, 9, e107678. [Google Scholar] [CrossRef] [Green Version]
- Wang, Y.; Shen, W.; Chan, Z.; Wu, Y. Endogenous Cytokinin Overproduction Modulates ROS Homeostasis and Decreases Salt Stress Resistance in Arabidopsis Thaliana. Front. Plant Sci. 2015, 6, 1004. [Google Scholar] [CrossRef] [Green Version]
- Jiang, K.; Moe-Lange, J.; Hennet, L.; Feldman, L.J. Salt Stress Affects the Redox Status of Arabidopsis Root Meristems. Front. Plant Sci. 2016, 7, 81. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cai, R.H.; Dai, W.; Zhang, C.S.; Wang, Y.; Wu, M.; Zhao, Y.; Ma, Q.; Xiang, Y.; Cheng, B.J. The maize WRKY transcription factor ZmWRKY17 negatively regulates salt stress tolerance in transgenic Arabidopsis plants. Planta 2017, 246, 1215–1231. [Google Scholar] [CrossRef] [PubMed]
- Sun, J.Q.; Jiang, H.L.; Xu, Y.X.; Li, H.M.; Wu, X.Y.; Xie, Q.; Li, C.Y. The CCCH-type zinc finger proteins AtSZF1 and AtSZF2 regulate salt stress responses in Arabidopsis. Plant Cell Physiol. 2007, 48, 1148–1158. [Google Scholar] [CrossRef]
- Lee, S.K.; Kim, B.G.; Kwon, T.R.; Jeong, M.J.; Park, S.R.; Lee, J.W.; Byun, M.O.; Kwon, H.B.; Matthews, B.F.; Hong, C.B.; et al. Overexpression of the mitogen-activated protein kinase gene OsMAPK33 enhances sensitivity to salt stress in rice (Oryza sativa L.). J. Biosci. 2011, 36, 139–151. [Google Scholar] [CrossRef] [PubMed]
- Mehlmer, N.; Wurzinger, B.; Stael, S.; Hofmann-Rodrigues, D.; Csaszar, E.; Pfister, B.; Bayer, R.; Teige, M. The Ca2+-dependent protein kinase CPK3 is required for MAPK-independent salt-stress acclimation in Arabidopsis. Plant J. 2010, 63, 484–498. [Google Scholar] [CrossRef] [Green Version]
- Liu, L.; Chen, J.; Tan, Y.N.; Zhou, T.S.; Ouyang, N.; Zeng, J.; Yuan, D.Y.; Duan, M.J. Increasing Fatty Acids in Rice Root Improves Silence of Rice Seedling to Salt Stress. Rice Sci. 2019, 26, 339–342. [Google Scholar]
- Zhang, X.G.; Cai, H.L.; Lu, M.; Wei, Q.Y.; Xu, L.J.; Bo, C.; Ma, Q.; Zhao, Y.; Cheng, B.J. A maize stress-responsive Di19 transcription factor, ZmDi19-1, confers enhanced tolerance to salt in transgenic Arabidopsis. Plant Cell Rep. 2019, 38, 1563–1578. [Google Scholar] [CrossRef]
- Kim, D.; Landmead, B.; Salzberg, S.L. HISAT: A fast spliced aligner with low memory requirements. Nat. Methods 2015, 12, 357–360. [Google Scholar] [CrossRef] [Green Version]
- Anders, S.; Pyl, P.T.; Huber, W. HTSeq-a Python framework to work with high-throughput sequencing data. Bioinformatics 2015, 31, 166–169. [Google Scholar] [CrossRef]
- Marioni, J.C.; Mason, C.E.; Mane, S.M.; Stephens, M.; Gilad, Y. RNA-seq: An assessment of technical reproducibility and comparison with gene expression arrays. Genome Res. 2008, 18, 1509–1517. [Google Scholar] [CrossRef] [Green Version]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhao, Y.; Hu, F.X.; Zhang, X.G.; Wei, Q.Y.; Dong, J.L.; Bo, C.; Cheng, B.J.; Ma, Q. Comparative transcriptome analysis reveals important roles of nonadditive genes in maize hybrid An’nong 591 under heat stress. BMC Plant Biol. 2019, 19, 273. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(T)(-Delta Delta C) method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]








Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, X.; Liu, J.; Huang, Y.; Wu, H.; Hu, X.; Cheng, B.; Ma, Q.; Zhao, Y. Comparative Transcriptomics Reveals the Molecular Mechanism of the Parental Lines of Maize Hybrid An’nong876 in Response to Salt Stress. Int. J. Mol. Sci. 2022, 23, 5231. https://doi.org/10.3390/ijms23095231
Zhang X, Liu J, Huang Y, Wu H, Hu X, Cheng B, Ma Q, Zhao Y. Comparative Transcriptomics Reveals the Molecular Mechanism of the Parental Lines of Maize Hybrid An’nong876 in Response to Salt Stress. International Journal of Molecular Sciences. 2022; 23(9):5231. https://doi.org/10.3390/ijms23095231
Chicago/Turabian StyleZhang, Xingen, Jing Liu, Yuanxiang Huang, Hongying Wu, Xiaolin Hu, Beijiu Cheng, Qing Ma, and Yang Zhao. 2022. "Comparative Transcriptomics Reveals the Molecular Mechanism of the Parental Lines of Maize Hybrid An’nong876 in Response to Salt Stress" International Journal of Molecular Sciences 23, no. 9: 5231. https://doi.org/10.3390/ijms23095231





