Presence of Ceramidase Activity in Electronegative LDL
Abstract
1. Introduction
2. Results
2.1. Sph and Cer Content in LDL(−) Increase at 37 °C
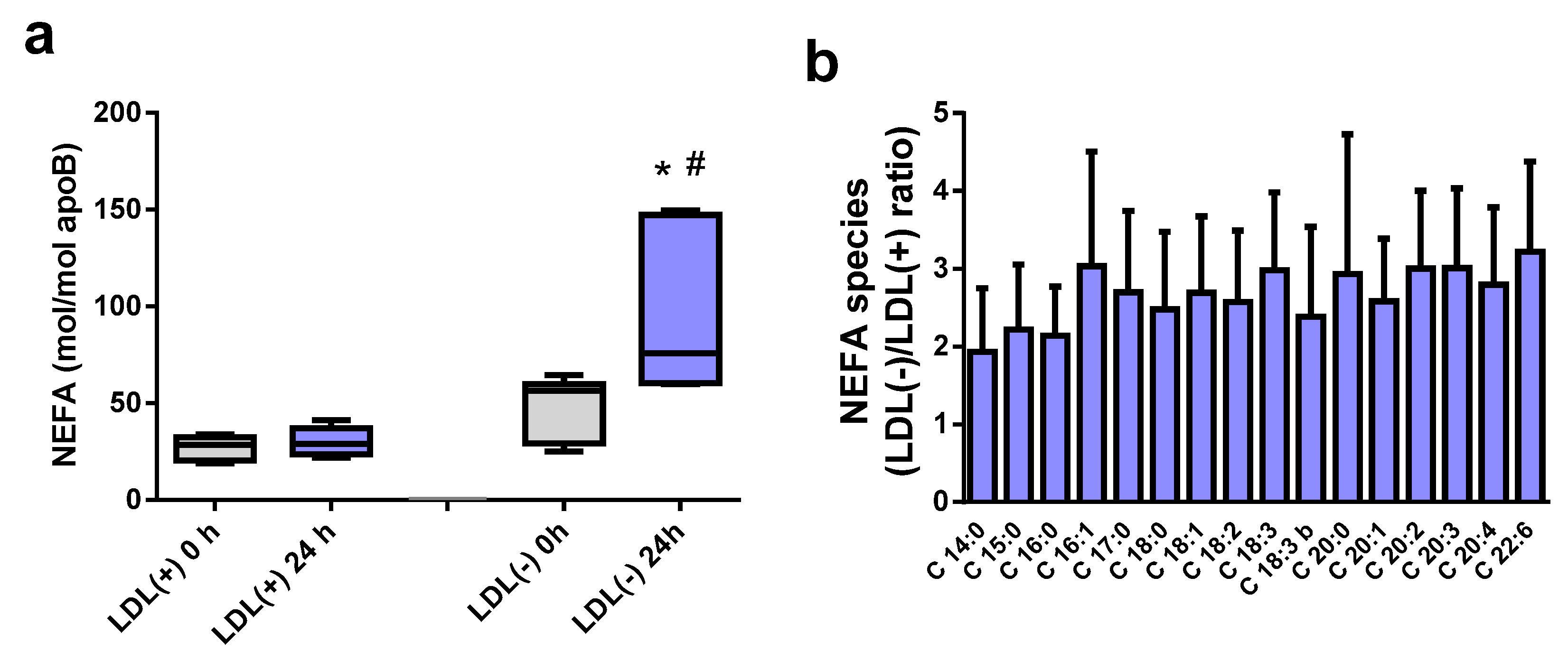
2.2. NEFA Species Increase at 37 °C in LDL(−)
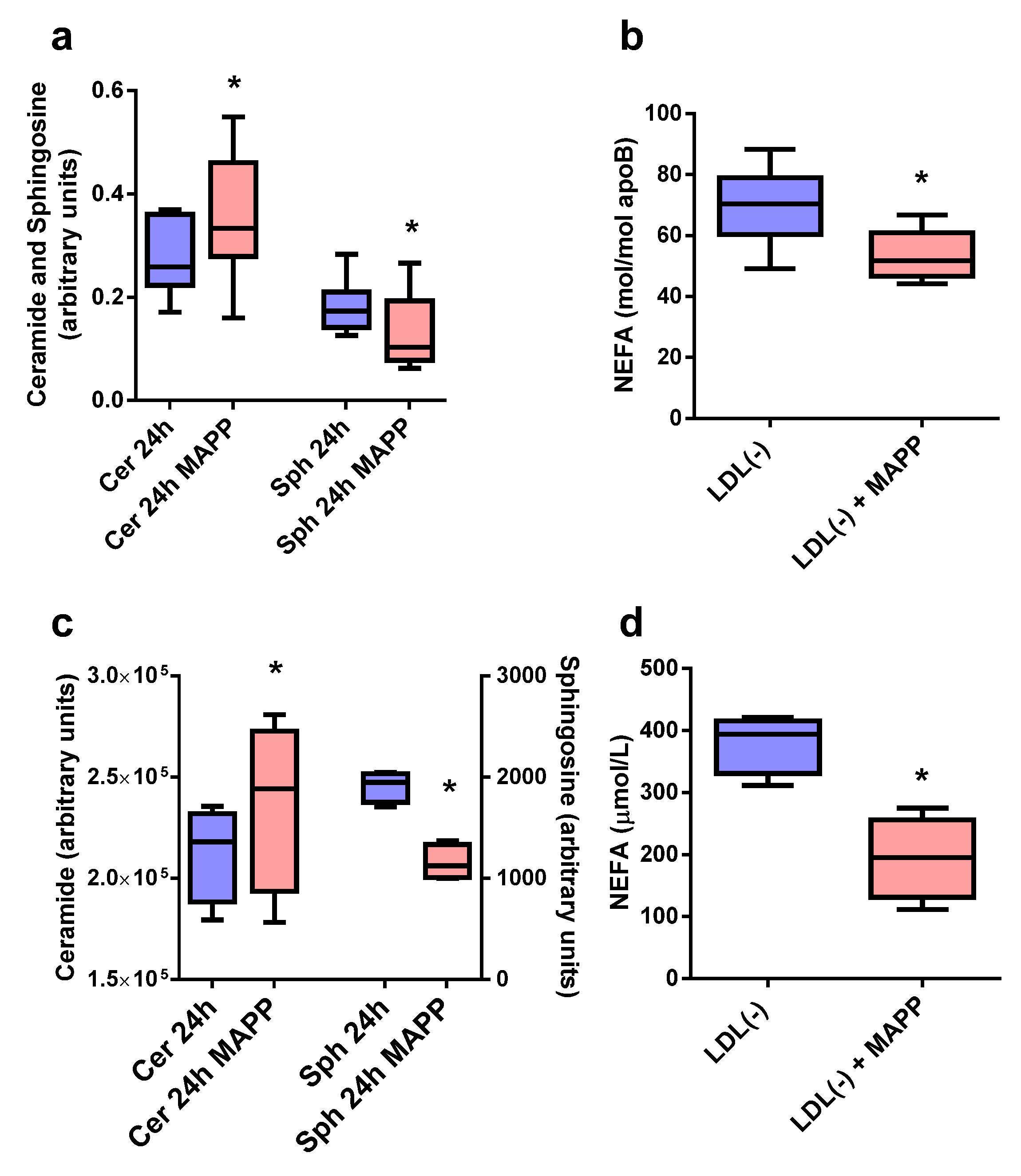
2.3. Effect of CDase Inhibitor on Sph, Cer, and NEFA Content in LDL(−)
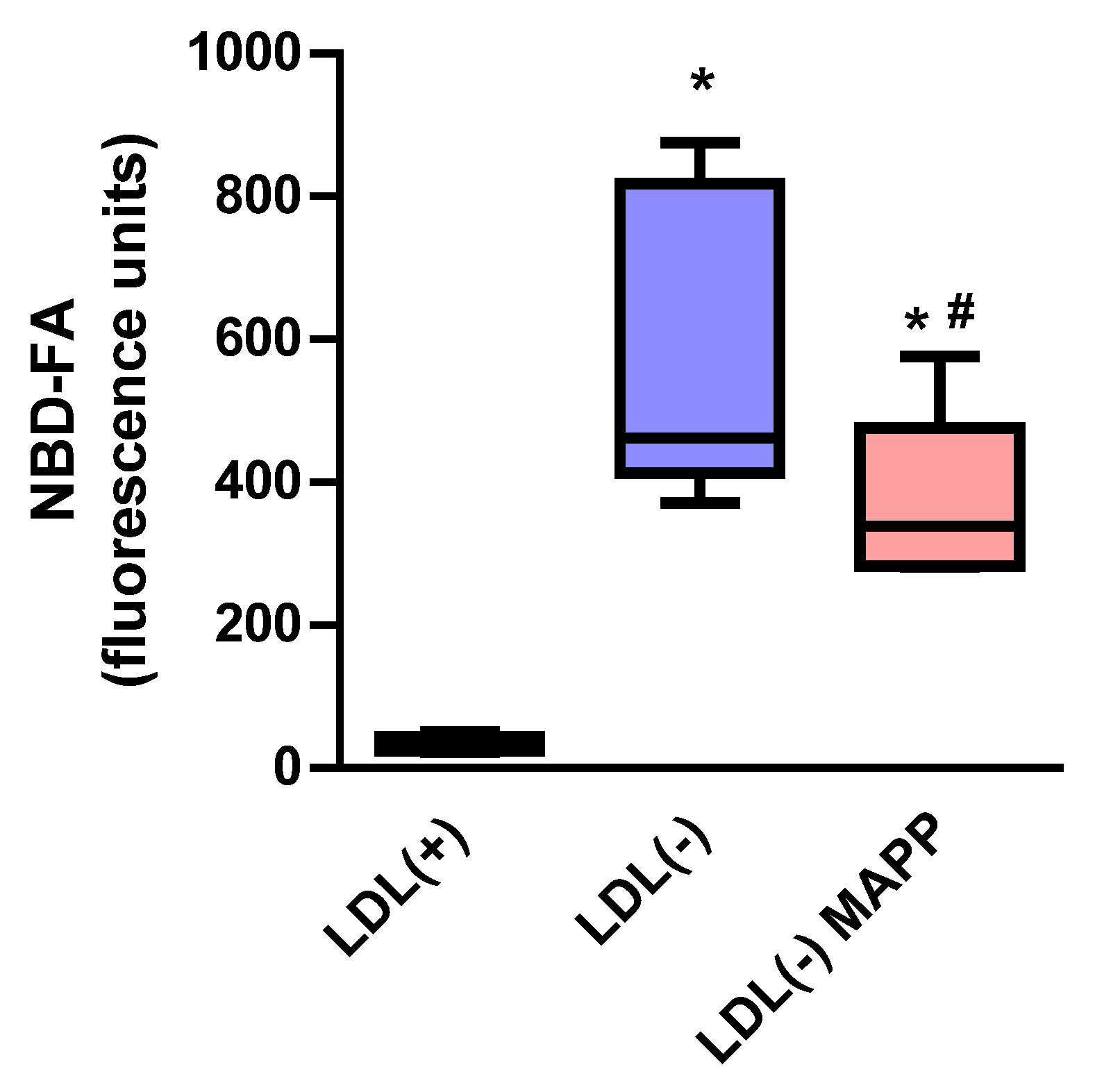
2.4. LDL(−) Is Able to Degrade Cer from an External Source
2.5. Proteomic Analysis of LDL(−)
2.6. Western Blot of Neutral CDase
3. Discussion
4. Materials and Methods
4.1. LDL Isolation
4.2. LDL Incubation at 37 °C
4.3. TLC Lipid Determination
4.4. Lipid Determination via Liquid Chromatography-Mass Spectrometry
4.4.1. Sphingolipid Analysis
4.4.2. NEFA Analysis
4.5. Hydrolysis of External Cer by LDL(−)
4.6. Proteomic Analysis
4.7. Western Blot of Neutral CDase
4.8. Statistical Analyses
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Tabas, I.; Williams, K.J.; Borén, J. Subendothelial Lipoprotein Retention as the Initiating Process in Atherosclerosis. Circulation 2007, 116, 1832–1844. [Google Scholar] [CrossRef] [PubMed]
- Sánchez-Quesada, J.L.; Estruch, M.; Benítez, S.; Ordóñez-Llanos, J. Electronegative LDL: A useful biomarker of cardiovascular risk? Clin. Lipidol. 2012, 7, 345–359. [Google Scholar] [CrossRef]
- Sanchez-Quesada, J.L.; Benítez, S.; Ordonez-Llanos, J. Electronegative low-density lipoprotein. Curr. Opin. Lipidol. 2004, 15, 329–335. [Google Scholar] [CrossRef] [PubMed]
- Ke, L.-Y.; Law, S.H.; Mishra, V.K.; Parveen, F.; Chan, H.-C.; Lu, Y.-H.; Chu, C.-S. Molecular and Cellular Mechanisms of Electronegative Lipoproteins in Cardiovascular Diseases. Biomedicines 2020, 8, 550. [Google Scholar] [CrossRef]
- Benítez, S.; Bancells, C.; Ordóñez-Llanos, J.; Sánchez-Quesada, J.L. Pro-inflammatory action of LDL(−) on mononuclear cells is counteracted by increased IL10 production. Biochim. Biophys. Acta 2007, 1771, 613–622. [Google Scholar] [CrossRef] [PubMed]
- Puig, N.; Montolio, L.; Camps-Renom, P.; Navarra, L.; Jiménez-Altayó, F.; Jiménez-Xarrié, E.; Sánchez-Quesada, J.L.; Benitez, S. Electronegative LDL Promotes Inflammation and Triglyceride Accumulation in Macrophages. Cells 2020, 9, 583. [Google Scholar] [CrossRef] [PubMed]
- Chu, C.-S.; Law, S.H.; Lenzen, D.; Tan, Y.-H.; Weng, S.-F.; Ito, E.; Wu, J.-C.; Chen, C.-H.; Chan, H.-C.; Ke, L.-Y. Clinical Significance of Electronegative Low-Density Lipoprotein Cholesterol in Atherothrombosis. Biomedicines 2020, 8, 254. [Google Scholar] [CrossRef] [PubMed]
- Mello, A.P.Q.; da Silva, I.T.; Abdalla, D.S.P.; Damasceno, N.R.T. Electronegative low-density lipoprotein: Origin and impact on health and disease. Atherosclerosis 2011, 215, 257–265. [Google Scholar] [CrossRef]
- Benítez, S.; Camacho, M.; Arcelus, R.; Vila, L.; Bancells, C.; Ordonez-Llanos, J.; Sánchez-Quesada, J.L. Increased lysophosphatidylcholine and non-esterified fatty acid content in LDL induces chemokine release in endothelial cells: Relationship with electronegative LDL. Atherosclerosis 2004, 177, 299–305. [Google Scholar] [CrossRef]
- Benítez, S.; Villegas, V.; Bancells, C.; Jorba, O.; González-Sastre, F.; Ordóñez-Llanos, J.; Sánchez-Quesada, J.L. Impaired Binding Affinity of Electronegative Low-Density Lipoprotein (LDL) to the LDL Receptor Is Related to Nonesterified Fatty Acids and Lysophosphatidylcholine Content. Biochemistry 2004, 43, 15863–15872. [Google Scholar] [CrossRef]
- Bancells, C.; Sánchez-Quesada, J.L.; Birkelund, R.; Ordóñez-Llanos, J.; Benítez, S. HDL and electronegative LDL exchange anti- and pro-inflammatory properties. J. Lipid Res. 2010, 51, 2947–2956. [Google Scholar] [CrossRef] [PubMed]
- Estruch, M.; Sanchez-Quesada, J.L.; Beloki, L.; Ordoñez-Llanos, J.; Benitez, S. The Induction of Cytokine Release in Monocytes by Electronegative Low-Density Lipoprotein (LDL) Is Related to Its Higher Ceramide Content than Native LDL. Int. J. Mol. Sci. 2013, 14, 2601–2616. [Google Scholar] [CrossRef]
- Puig, N.; Estruch, M.; Jin, L.; Sanchez-Quesada, J.L.; Benitez, S. The Role of Distinctive Sphingolipids in the Inflammatory and Apoptotic Effects of Electronegative LDL on Monocytes. Biomolecules 2019, 9, 300. [Google Scholar] [CrossRef] [PubMed]
- Song, J.-H.; Kim, G.-T.; Park, K.-H.; Park, W.-J.; Park, T.-S. Bioactive Sphingolipids as Major Regulators of Coronary Artery Disease. Biomol. Ther. 2021, 29, 373–383. [Google Scholar] [CrossRef] [PubMed]
- Maceyka, M.; Spiegel, S. Sphingolipid metabolites in inflammatory disease. Nature 2014, 510, 58–67. [Google Scholar] [CrossRef] [PubMed]
- Havulinna, A.S.; Sysi-Aho, M.; Hilvo, M.; Kauhanen, D.; Hurme, R.; Ekroos, K.; Salomaa, V.; Laaksonen, R. Circulating Ceramides Predict Cardiovascular Outcomes in the Population-Based FINRISK 2002 Cohort. Arter. Thromb. Vasc. Biol. 2016, 36, 2424–2430. [Google Scholar] [CrossRef]
- Arana, L.; Gangoiti, P.; Ouro, A.; Trueba, M.; Gómez-Muñoz, A. Ceramide and ceramide 1-phosphate in health and disease. Lipids Health Dis. 2010, 9, 15. [Google Scholar] [CrossRef]
- Woodcock, J. Sphingosine and ceramide signalling in apoptosis. IUBMB Life 2006, 58, 462–466. [Google Scholar] [CrossRef]
- Luheshi, N.M.; Giles, J.; Lopez-Castejon, G.; Brough, D. Sphingosine regulates the NLRP3-inflammasome and IL-1β release from macrophages. Eur. J. Immunol. 2012, 42, 716–725. [Google Scholar] [CrossRef]
- Benítez, S.; Sanchez-Quesada, J.L.; Ribas, V.; Jorba, O.; Blanco-Vaca, F.; González-Sastre, F.; Ordóñez-Llanos, J. Platelet-Activating Factor Acetylhydrolase Is Mainly Associated With Electronegative Low-Density Lipoprotein Subfraction. Circulation 2003, 108, 92–96. [Google Scholar] [CrossRef]
- Bancells, C.; Benítez, S.; Villegas, S.; Jorba, O.; Ordóñez-Llanos, J.; Sánchez-Quesada, J.L. Novel Phospholipolytic Activities Associated with Electronegative Low-Density Lipoprotein Are Involved in Increased Self-Aggregation. Biochemistry 2008, 47, 8186–8194. [Google Scholar] [CrossRef] [PubMed]
- Bancells, C.; Villegas, S.; Blanco, F.; Benítez, S.; Gállego, I.; Beloki, L.; Pérez-Cuellar, M.; Ordonez-Llanos, J.; Sánchez-Quesada, J.L. Aggregated Electronegative Low Density Lipoprotein in Human Plasma Shows a High Tendency toward Phospholipolysis and Particle Fusion. J. Biol. Chem. 2010, 285, 32425–32435. [Google Scholar] [CrossRef] [PubMed]
- Ke, L.-Y.; Chan, H.-C.; Chen, C.-C.; Lu, J.; Marathe, G.K.; Chu, C.-S.; Chan, H.-C.; Wang, C.-Y.; Tung, Y.-C.; McIntyre, T.M.; et al. Enhanced Sphingomyelinase Activity Contributes to the Apoptotic Capacity of Electronegative Low-Density Lipoprotein. J. Med. Chem. 2016, 59, 1032–1040. [Google Scholar] [CrossRef]
- Oörni, K.; Pentikäinen, M.O.; Ala-Korpela, M.; Kovanen, P.T. Aggregation, fusion, and vesicle formation of modified low density lipoprotein particles: Molecular mechanisms and effects on matrix interactions. J. Lipid Res. 2000, 41, 1703–1714. [Google Scholar] [CrossRef] [PubMed]
- Schissel, S.L.; Tweedie-Hardman, J.; Rapp, J.H.; Graham, G.; Williams, K.J.; Tabas, I. Rabbit aorta and human atherosclerotic lesions hydrolyze the sphingomyelin of retained low-density lipoprotein. Proposed role for arterial-wall sphingomyelinase in subendothelial retention and aggregation of atherogenic lipoproteins. J. Clin. Investig. 1996, 98, 1455–1464. [Google Scholar] [CrossRef]
- Duarte, C.; Akkaoui, J.; Yamada, C.; Ho, A.; Mao, C.; Movila, A. Elusive Roles of the Different Ceramidases in Human Health, Pathophysiology, and Tissue Regeneration. Cells 2020, 9, 1379. [Google Scholar] [CrossRef] [PubMed]
- Parveen, F.; Bender, D.; Law, S.-H.; Mishra, V.K.; Chen, C.-C.; Ke, L.-Y. Role of Ceramidases in Sphingolipid Metabolism and Human Diseases. Cells 2019, 8, 1573. [Google Scholar] [CrossRef]
- Bancells, C.; Canals, F.; Benítez, S.; Colomé, N.; Julve, J.; Ordonez-Llanos, J.; Sánchez-Quesada, J.L. Proteomic analysis of electronegative low-density lipoprotein. J. Lipid Res. 2010, 51, 3508–3515. [Google Scholar] [CrossRef]
- Bourgeois, R.; Girard, A.; Perrot, N.; Guertin, J.; Mitchell, P.L.; Couture, C.; Gotti, C.; Bourassa, S.; Poggio, P.; Mass, E.; et al. A Comparative Analysis of the Lipoprotein(a) and Low-Density Lipoprotein Proteomic Profiles Combining Mass Spectrometry and Mendelian Randomization. CJC Open 2021, 3, 450–459. [Google Scholar] [CrossRef]
- Diffenderfer, M.R.; Schaefer, E.J. The composition and metabolism of large and small LDL. Curr. Opin. Lipidol. 2014, 25, 221–226. [Google Scholar] [CrossRef]
- Iqbal, J.; Walsh, M.T.; Hammad, S.M.; Hussain, M.M. Sphingolipids and Lipoproteins in Health and Metabolic Disorders. Trends Endocrinol. Metab. 2017, 28, 506–518. [Google Scholar] [CrossRef] [PubMed]
- Kurano, M.; Tsukamoto, K.; Sakai, E.; Yatomi, Y. Differences in the Distribution of Ceramides and Sphingosine among Lipoprotein and Lipoprotein-Depleted Fractions in Patients with Type 2 Diabetes Mellitus. J. Atheroscler. Thromb. 2022, 29, 1727–1758. [Google Scholar] [CrossRef] [PubMed]
- Estruch, M.; Bancells, C.; Beloki, L.; Sanchez-Quesada, J.L.; Ordóñez-Llanos, J.; Benitez, S. CD14 and TLR4 mediate cytokine release promoted by electronegative LDL in monocytes. Atherosclerosis 2013, 229, 356–362. [Google Scholar] [CrossRef] [PubMed]
- Estruch, M.; Sánchez-Quesada, J.L.; Ordóñez-Llanos, J.; Benítez, S. Ceramide-enriched LDL induces cytokine release through TLR4 and CD14 in monocytes. Similarities with electronegative LDL. Clin. Investig. Arter. 2014, 26, 131–137. [Google Scholar] [CrossRef]
- Kinnunen, P.K.J. Sphingomyelinase Activity of LDL A Link between Atherosclerosis, Ceramide, and Apoptosis? Trends Cardiovasc. Med. 2002, 12, 37–42. [Google Scholar] [CrossRef]
- Holopainen, J.M.; Medina, O.P.; Metso, A.J.; Kinnunen, P.K. Sphingomyelinase Activity Associated with Human Plasma Low Density Lipoprotein. J. Biol. Chem. 2000, 275, 16484–16489. [Google Scholar] [CrossRef]
- Rivas-Urbina, A.; Rull, A.; Ordóñez-Llanos, J.; Sánchez–Quesada, J.L. Electronegative LDL: An Active Player in Atherogenesis or a ByProduct of Atherosclerosis? Curr. Med. Chem. 2019, 26, 1665–1679. [Google Scholar] [CrossRef]
- Sánchez-Quesada, J.L.; Villegas, S.; Ordóñez-Llanos, J. Electronegative low-density lipoprotein. A link between apolipoprotein B misfolding, lipoprotein aggregation and proteoglycan binding. Curr. Opin. Lipidol. 2012, 23, 479–486. [Google Scholar] [CrossRef]
- Brunelli, R.; Spirito, M.; Mei, G.; Papi, M.; Perrone, G.; Stefanutti, C.; Parasassi, T. Misfolding of Apoprotein B-100, LDL Aggregation and 17-β -estradiol in Atherogenesis. Curr. Med. Chem. 2014, 21, 2276–2283. [Google Scholar] [CrossRef]
- Parasassi, T.; De Spirito, M.; Mei, G.; Brunelli, R.; Greco, G.; Lenzi, L.; Maulucci, G.; Nicolai, E.; Papi, M.; Arcovito, G.; et al. Low density lipoprotein misfolding and amyloidogenesis. FASEB J. 2008, 22, 2350–2356. [Google Scholar] [CrossRef]
- Canals, D.; Perry, D.M.; Jenkins, R.W.; Hannun, Y.A. Drug targeting of sphingolipid metabolism: Sphingomyelinases and ceramidases. J. Cereb. Blood Flow Metab. 2011, 163, 694–712. [Google Scholar] [CrossRef] [PubMed]
- Tani, M.; Igarashi, Y.; Ito, M. Involvement of Neutral Ceramidase in Ceramide Metabolism at the Plasma Membrane and in Extracellular Milieu. J. Biol. Chem. 2005, 280, 36592–36600. [Google Scholar] [CrossRef] [PubMed]
- Bielawska, A.; Greenberg, M.S.; Perry, D.; Jayadev, S.; Shayman, J.A.; McKay, C.; Hannun, Y.A. (1S,2R)-D-erythro-2-(N-Myristoylamino)-1-phenyl-1-propanol as an Inhibitor of Ceramidase. J. Biol. Chem. 1996, 271, 12646–12654. [Google Scholar] [CrossRef] [PubMed]
- Tani, M.; Okino, N.; Mitsutake, S.; Ito, M. Specific and sensitive assay for alkaline and neutral ceramidases involving C12-NBD-ceramide. J. Biochem. 1999, 125, 746–749. [Google Scholar] [CrossRef] [PubMed]
- Bancells, C.; Benítez, S.; Ordóñez-Llanos, J.; Öörni, K.; Kovanen, P.T.; Milne, R.W.; Sánchez-Quesada, J.L. Immunochemical Analysis of the Electronegative LDL Subfraction Shows That Abnormal N-terminal Apolipoprotein B Conformation Is Involved in Increased Binding to Proteoglycans. J. Biol. Chem. 2011, 286, 1125–1133. [Google Scholar] [CrossRef]
- Ostermann, A.I.; Müller, M.; Willenberg, I.; Schebb, N.H. Determining the fatty acid composition in plasma and tissues as fatty acid methyl esters using gas chromatography—A comparison of different derivatization and extraction procedures. Prostaglandins Leukot. Essent. Fat. Acids 2014, 91, 235–241. [Google Scholar] [CrossRef] [PubMed]
- Chiu, H.-H.; Tsai, S.-J.; Tseng, Y.J.; Wu, M.-S.; Liao, W.-C.; Huang, C.-S.; Kuo, C.-H. An efficient and robust fatty acid profiling method for plasma metabolomic studies by gas chromatography–mass spectrometry. Clin. Chim. Acta 2015, 451 Pt B, 183–190. [Google Scholar] [CrossRef]
- Karlsson, H.; Mörtstedt, H.; Lindqvist, H.; Tagesson, C.; Lindahl, M. Protein profiling of low-density lipoprotein from obese subjects. Proteom.–Clin. Appl. 2009, 3, 663–671. [Google Scholar] [CrossRef]

| PAF-AH Activity (mmol/min/mL) | SMase Activity (mU/mg apoB) | |
|---|---|---|
| LDL(−) 24 h | 1.25 ± 0.35 | 205.85 ± 57.77 |
| LDL(−) 24 h plus MAPP | 1.29 ± 0.30 | 215.92 ± 34.48 |
| Number of PSMs | % of Total PSMs | Number of PSMs | % of Total PSMs | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Accession | Description | LDL(−)_1 | LDL(−)_2 | LDL(−)_3 | Mean LDL(−) | SD LDL(−) | LDL(+)_1 | LDL(+)_2 | LDL(+)_3 | Mean LDL(+) | SD LDL(+) |
| ALBU_HUMAN | Albumin OS = Homo sapiens OX = 9606 GN = ALB PE = 1 SV = 2 | 6 | 0 | 3 | 0.45 | 0.42 | 1 | 19 | 2 | 1.05 | 1.50 |
| APOA_HUMAN | Apolipoprotein(a) OS = Homo sapiens OX = 9606 GN = LPA PE = 1 SV = 1 | 61 | 13 | 63 | 6.95 | 4.65 | 0 | 0 | 2 | 0.09 | 0.15 |
| APOA1_HUMAN | Apolipoprotein A-I OS = Homo sapiens OX = 9606 GN = APOA1 PE = 1 SV = 1 | 59 | 9 | 32 | 4.94 | 3.53 | 16 | 14 | 37 | 2.92 | 1.68 |
| APOA2_HUMAN | Apolipoprotein A-II OS = Homo sapiens OX = 9606 GN = APOA2 PE = 1 SV = 1 | 21 | 6 | 7 | 1.63 | 1.13 | 7 | 5 | 16 | 1.22 | 0.77 |
| APOB_HUMAN | Apolipoprotein B-100 OS = Homo sapiens OX = 9606 GN = APOB PE = 1 SV = 2 | 460 | 661 | 373 | 71.71 | 14.23 | 801 | 575 | 645 | 87.32 | 5.04 |
| APOC1_HUMAN | Apolipoprotein C-I OS = Homo sapiens OX = 9606 GN = APOC1 PE = 1 SV = 1 | 4 | 0 | 2 | 0.30 | 0.28 | 2 | 5 | 4 | 0.50 | 0.25 |
| APOC2_HUMAN | Apolipoprotein C-II OS = Homo sapiens OX = 9606 GN = APOC2 PE = 1 SV = 1 | 7 | 0 | 6 | 0.66 | 0.57 | 0 | 4 | 1 | 0.24 | 0.31 |
| APOC3_HUMAN | Apolipoprotein C-III OS = Homo sapiens OX = 9606 GN = APOC3 PE = 1 SV = 1 | 8 | 3 | 7 | 0.90 | 0.43 | 5 | 6 | 4 | 0.66 | 0.19 |
| APOC4_HUMAN | Apolipoprotein C-IV OS = Homo sapiens OX = 9606 GN = APOC4 PE = 1 SV = 1 | 1 | 0 | 2 | 0.16 | 0.17 | 0 | 0 | 0 | ||
| APOD_HUMAN | Apolipoprotein D OS = Homo sapiens OX = 9606 GN = APOD PE = 1 SV = 1 | 13 | 11 | 13 | 1.82 | 0.36 | 7 | 8 | 15 | 1.32 | 0.59 |
| APOE_HUMAN | Apolipoprotein E OS = Homo sapiens OX = 9606 GN = APOE PE = 1 SV = 1 | 45 | 32 | 46 | 6.10 | 1.76 | 14 | 28 | 20 | 2.78 | 1.24 |
| APOF_HUMAN | Apolipoprotein F OS = Homo sapiens OX = 9606 GN = APOF PE = 1 SV = 2 | 3 | 11 | 7 | 1.02 | 0.54 | 0 | 0 | 3 | 0.13 | 0.23 |
| APOL1_HUMAN | Apolipoprotein L1 OS = Homo sapiens OX = 9606 GN = APOL1 PE = 1 SV = 5 | 2 | 0 | 2 | 0.21 | 0.18 | 2 | 1 | 3 | 0.26 | 0.13 |
| APOM_HUMAN | Apolipoprotein M OS = Homo sapiens OX = 9606 GN = APOM PE = 1 SV = 2 | 4 | 0 | 1 | 0.24 | 0.29 | 1 | 1 | 4 | 0.26 | 0.23 |
| CLUS_HUMAN | Clusterin OS = Homo sapiens OX = 9606 GN = CLU PE = 1 SV = 1 | 4 | 0 | 8 | 0.64 | 0.68 | 0 | 0 | 0 | ||
| CO4A_HUMAN | Complement C4-A OS = Homo sapiens OX = 9606 GN = C4A PE = 1 SV = 1 | 2 | 2 | 1 | 0.24 | 0.06 | 0 | 0 | 0 | ||
| FIBA_HUMAN | Fibrinogen alpha chain OS = Homo sapiens OX = 9606 GN = FGA PE = 1 SV = 2 | 2 | 0 | 0 | 0.09 | 0.16 | 0 | 4 | 1 | 0.24 | 0.31 |
| PON1_HUMAN | Serum paraoxonase/arylesterase 1 OS = Homo sapiens OX = 9606 GN = PON1 PE = 1 SV = 3 | 6 | 0 | 5 | 0.56 | 0.48 | 0 | 0 | 0 | 0.00 | 0.00 |
| SAA1_HUMAN | Serum amyloid A-1 protein OS = Homo sapiens OX = 9606 GN = SAA1 PE = 1 SV = 1 | 3 | 0 | 2 | 0.25 | 0.22 | 0 | 2 | 0 | 0.10 | 0.17 |
| SAA4_HUMAN | Serum amyloid A-4 protein OS = Homo sapiens OX = 9606 GN = SAA4 PE = 1 SV = 1 | 8 | 2 | 10 | 1.02 | 0.72 | 4 | 11 | 5 | 0.91 | 0.61 |
| SPRL1_HUMAN | SPARC-like protein 1 OS = Homo sapiens OX = 9606 GN = SPARCL1 PE = 1 SV = 2 | 0 | 0 | 1 | 0.06 | 0.10 | 0 | 0 | 0 | ||
| VTNC_HUMAN | Vitronectin OS = Homo sapiens OX = 9606 GN = VTN PE = 1 SV = 1 | 0 | 0 | 1 | 0.06 | 0.10 | 0 | 0 | 0 | ||
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Puig, N.; Rives, J.; Estruch, M.; Aguilera-Simon, A.; Rotllan, N.; Camacho, M.; Colomé, N.; Canals, F.; Sánchez-Quesada, J.L.; Benitez, S. Presence of Ceramidase Activity in Electronegative LDL. Int. J. Mol. Sci. 2023, 24, 165. https://doi.org/10.3390/ijms24010165
Puig N, Rives J, Estruch M, Aguilera-Simon A, Rotllan N, Camacho M, Colomé N, Canals F, Sánchez-Quesada JL, Benitez S. Presence of Ceramidase Activity in Electronegative LDL. International Journal of Molecular Sciences. 2023; 24(1):165. https://doi.org/10.3390/ijms24010165
Chicago/Turabian StylePuig, Núria, Jose Rives, Montserrat Estruch, Ana Aguilera-Simon, Noemi Rotllan, Mercedes Camacho, Núria Colomé, Francesc Canals, José Luis Sánchez-Quesada, and Sonia Benitez. 2023. "Presence of Ceramidase Activity in Electronegative LDL" International Journal of Molecular Sciences 24, no. 1: 165. https://doi.org/10.3390/ijms24010165
APA StylePuig, N., Rives, J., Estruch, M., Aguilera-Simon, A., Rotllan, N., Camacho, M., Colomé, N., Canals, F., Sánchez-Quesada, J. L., & Benitez, S. (2023). Presence of Ceramidase Activity in Electronegative LDL. International Journal of Molecular Sciences, 24(1), 165. https://doi.org/10.3390/ijms24010165









