CRISPR/Cas9-Induced Inactivation of the Autism-Risk Gene setd5 Leads to Social Impairments in Zebrafish
Abstract
:1. Introduction
2. Results
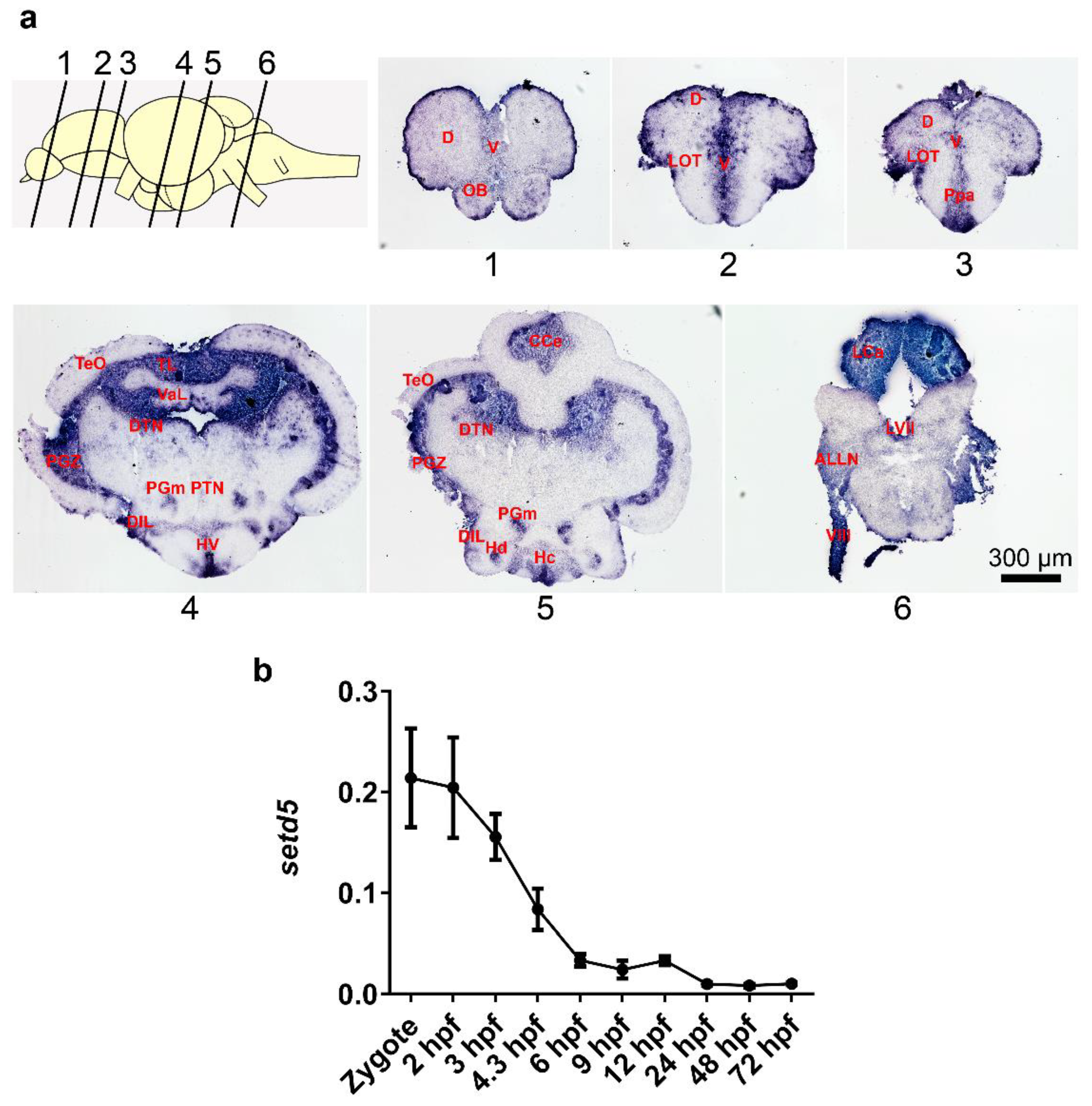
2.1. setd5 Is Expressed in Zebrafish Adult Brain and during Early Embryo Development
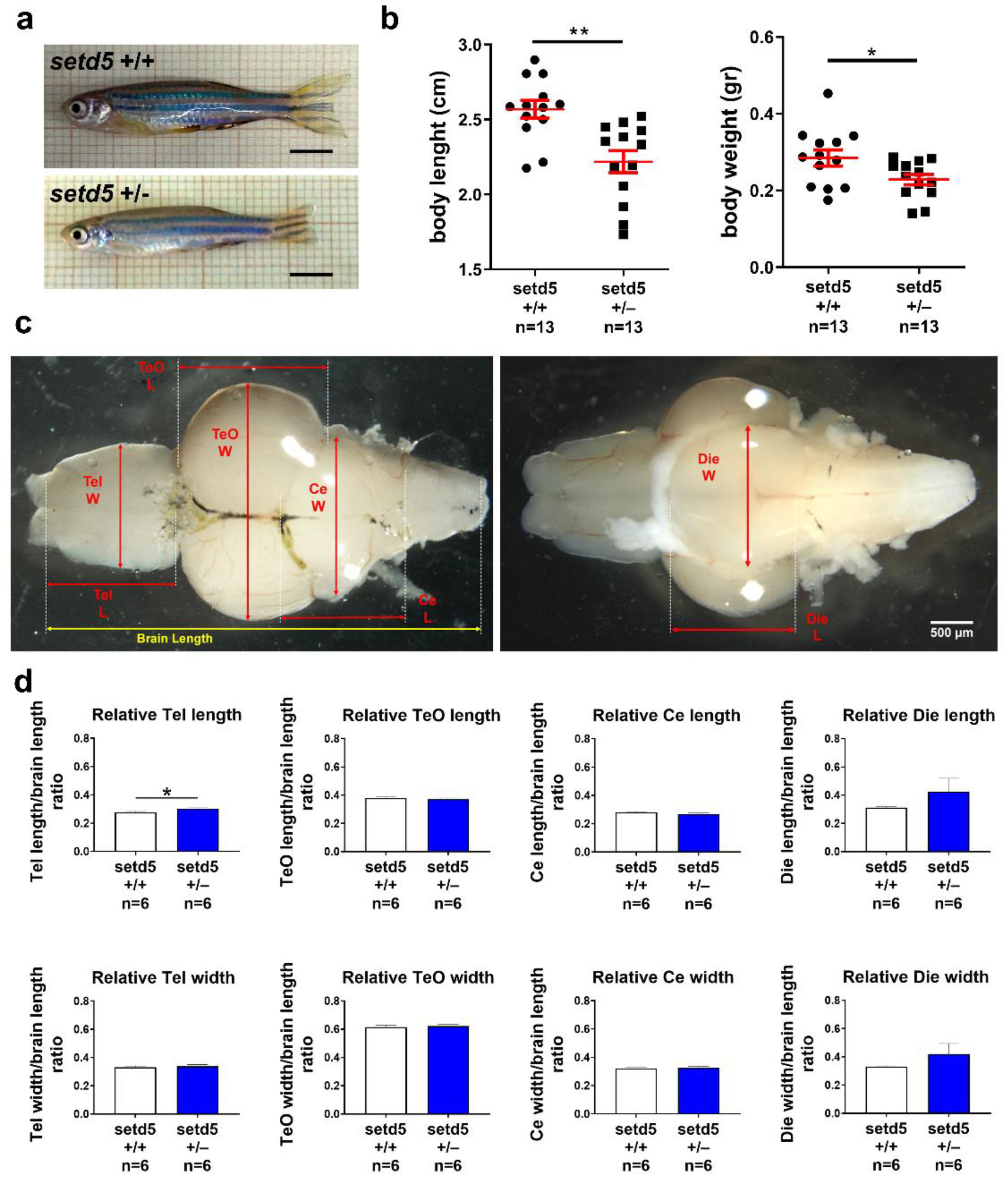
2.2. setd5 Knock-Out Causes a Growth Delay in Zebrafish
2.3. setd5 Mutant Embryos Show Growth Delay, Microphthalmia and Deficits in Locomotor Behavior
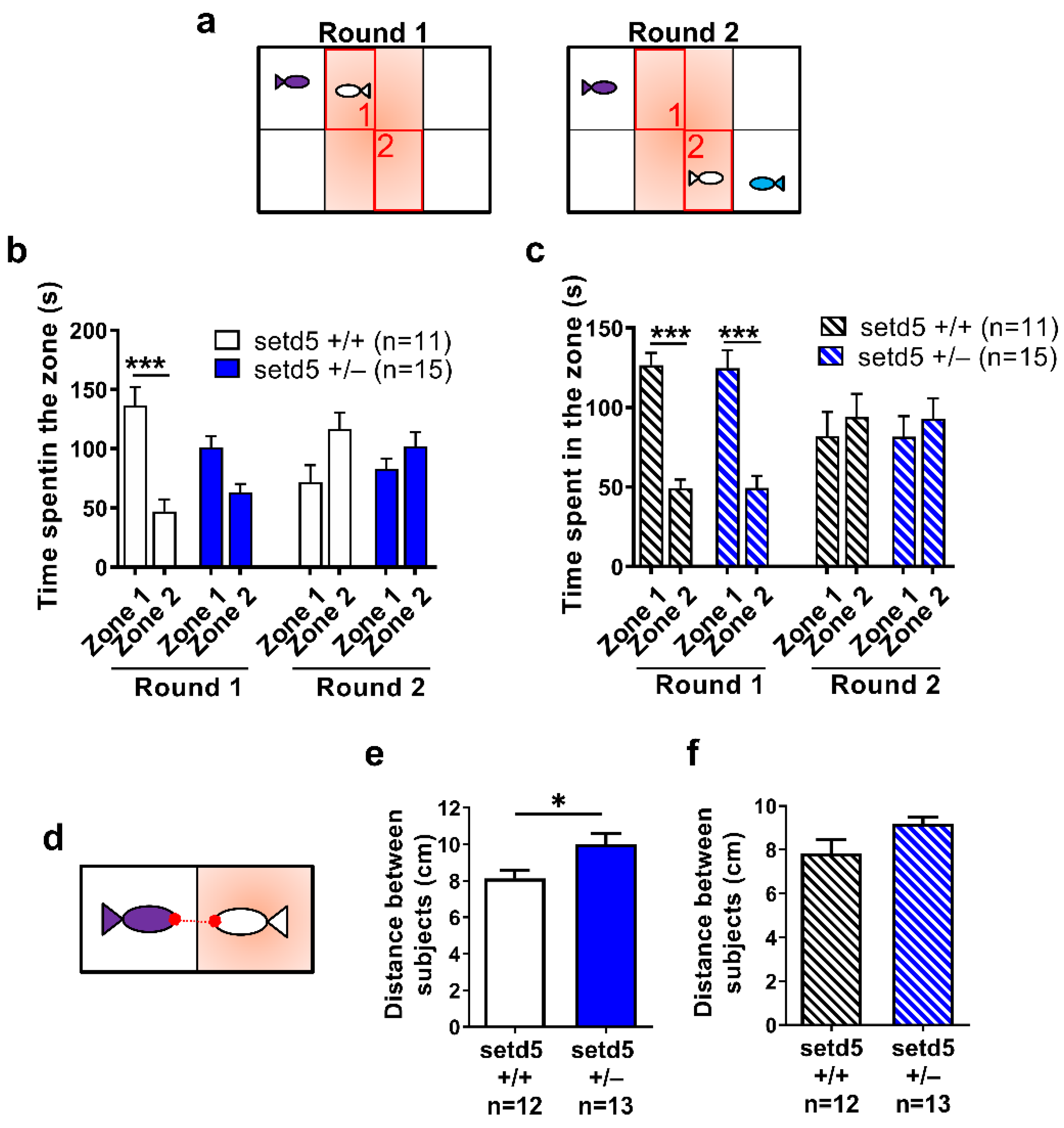
2.4. setd5 Mutant Adults Show a Tight Shoal
2.5. setd5 Mutant Adults Display Perturbed Social Interaction, Ameliorated by the Antipsychotic Drug Risperidone
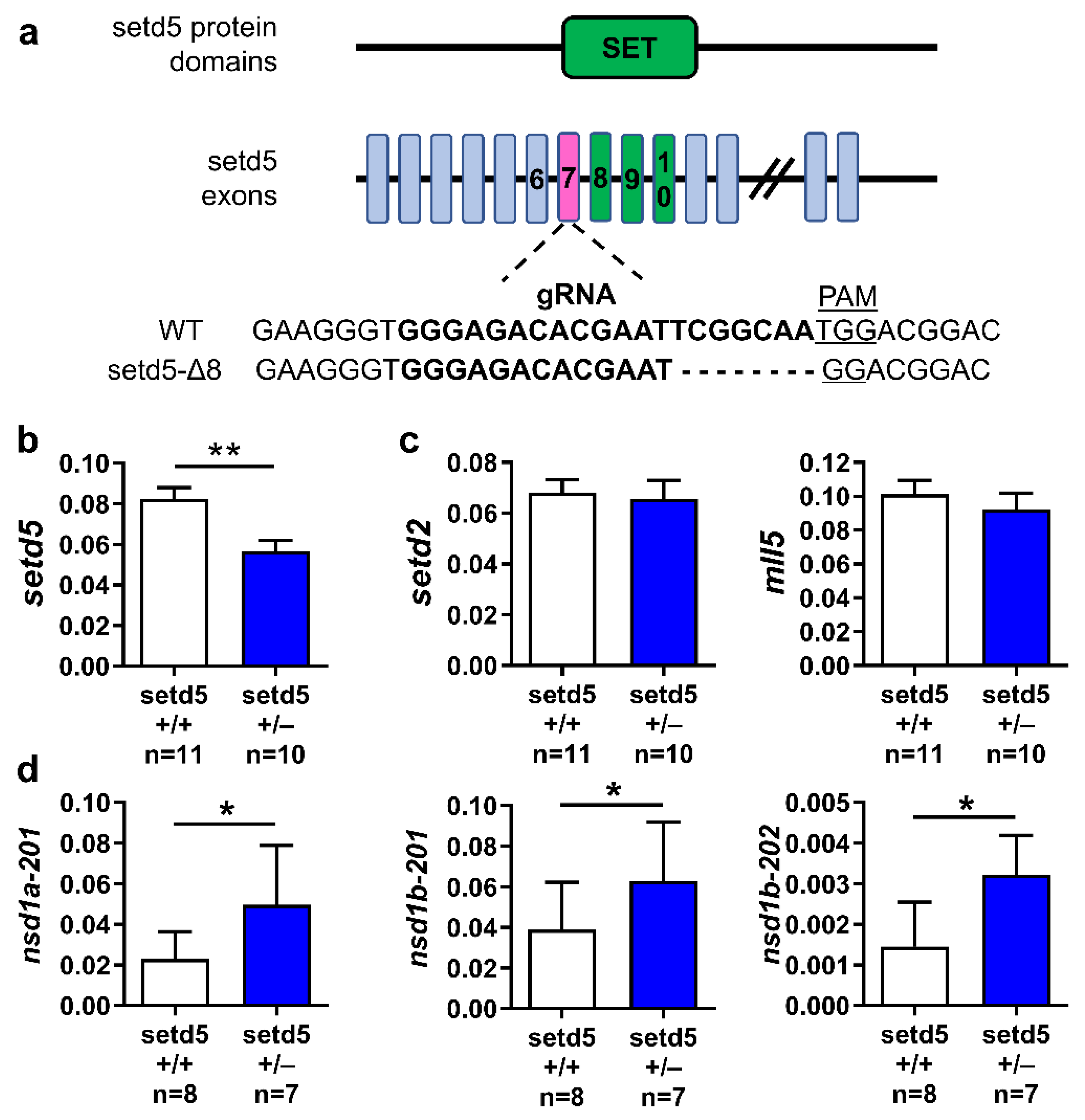
2.6. setd5 Mutation Affects the mRNA Expression Levels of Proteins Involved in Neurotransmission
3. Discussion
4. Materials and Methods
4.1. Zebrafish Care
4.2. Generation of setd5 Mutant Zebrafish
4.3. Genotyping
4.4. Morphological Analysis
4.5. Adult Brain Dissection
4.6. Extraction of Total RNA and RT-qPCR
4.7. In Situ Hybridization on Frozen Tissue Sections
4.8. Immunofluorescence and Quantitative Analysis
4.9. Behavioral Analyses
4.10. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Lai, M.C.; Lombardo, M.V.; Baron-Cohen, S. Autism. Lancet 2014, 383, 896–910. [Google Scholar] [CrossRef] [PubMed]
- Satterstrom, F.K.; Kosmicki, J.A.; Wang, J.; Breen, M.S.; De Rubeis, S.; An, J.Y.; Peng, M.; Collins, R.; Grove, J.; Klei, L.; et al. Large-Scale Exome Sequencing Study Implicates Both Developmental and Functional Changes in the Neurobiology of Autism. Cell 2020, 180, 568–584.e23. [Google Scholar] [CrossRef]
- Basilico, B.; Morandell, J.; Novarino, G. Molecular mechanisms for targeted ASD treatments. Curr. Opin. Genet. Dev. 2020, 65, 126–137. [Google Scholar] [CrossRef] [PubMed]
- Mossink, B.; Negwer, M.; Schubert, D.; Nadif Kasri, N. The emerging role of chromatin remodelers in neurodevelopmental disorders: A developmental perspective. Cell. Mol. Life Sci. 2021, 78, 2517–2563. [Google Scholar] [CrossRef] [PubMed]
- Study, D.D.D. Prevalence and architecture of de novo mutations in developmental disorders. Nature 2017, 542, 433–438. [Google Scholar] [CrossRef]
- Fernandes, I.R.; Cruz, A.C.P.; Ferrasa, A.; Phan, D.; Herai, R.H.; Muotri, A.R. Genetic variations on SETD5 underlying autistic conditions. Dev. Neurobiol. 2018, 78, 500–518. [Google Scholar] [CrossRef]
- Green, C.; Willoughby, J.; Balasubramanian, M.; Study, D. De novo SETD5 loss-of-function variant as a cause for intellectual disability in a 10-year old boy with an aberrant blind ending bronchus. Am. J. Med. Genet. A 2017, 173, 3165–3171. [Google Scholar] [CrossRef] [Green Version]
- Grozeva, D.; Carss, K.; Spasic-Boskovic, O.; Parker, M.J.; Archer, H.; Firth, H.V.; Park, S.M.; Canham, N.; Holder, S.E.; Wilson, M.; et al. De novo loss-of-function mutations in SETD5, encoding a methyltransferase in a 3p25 microdeletion syndrome critical region, cause intellectual disability. Am. J. Hum. Genet. 2014, 94, 618–624. [Google Scholar] [CrossRef] [Green Version]
- Kuechler, A.; Zink, A.M.; Wieland, T.; Lüdecke, H.J.; Cremer, K.; Salviati, L.; Magini, P.; Najafi, K.; Zweier, C.; Czeschik, J.C.; et al. Loss-of-function variants of SETD5 cause intellectual disability and the core phenotype of microdeletion 3p25.3 syndrome. Eur. J. Hum. Genet. 2015, 23, 753–760. [Google Scholar] [CrossRef]
- Pinto, D.; Delaby, E.; Merico, D.; Barbosa, M.; Merikangas, A.; Klei, L.; Thiruvahindrapuram, B.; Xu, X.; Ziman, R.; Wang, Z.; et al. Convergence of genes and cellular pathways dysregulated in autism spectrum disorders. Am. J. Hum. Genet. 2014, 94, 677–694. [Google Scholar] [CrossRef]
- Stur, E.; Soares, L.A.; Louro, I.D. SETD5 gene variant associated with mild intellectual disability—A case report. Genet. Mol. Res. 2017, 16, 1–6. [Google Scholar] [CrossRef] [PubMed]
- Szczałuba, K.; Brzezinska, M.; Kot, J.; Rydzanicz, M.; Walczak, A.; Stawiński, P.; Werner, B.; Płoski, R. SETD5 loss-of-function mutation as a likely cause of a familial syndromic intellectual disability with variable phenotypic expression. Am. J. Med. Genet. A 2016, 170, 2322–2327. [Google Scholar] [CrossRef] [PubMed]
- Powis, Z.; Farwell Hagman, K.D.; Mroske, C.; McWalter, K.; Cohen, J.S.; Colombo, R.; Serretti, A.; Fatemi, A.; David, K.L.; Reynolds, J.; et al. Expansion and further delineation of the SETD5 phenotype leading to global developmental delay, variable dysmorphic features, and reduced penetrance. Clin. Genet. 2017, 93, 752–761. [Google Scholar] [CrossRef] [PubMed]
- Pinheiro, I.; Margueron, R.; Shukeir, N.; Eisold, M.; Fritzsch, C.; Richter, F.M.; Mittler, G.; Genoud, C.; Goyama, S.; Kurokawa, M.; et al. Prdm3 and Prdm16 are H3K9me1 methyltransferases required for mammalian heterochromatin integrity. Cell 2012, 150, 948–960. [Google Scholar] [CrossRef] [Green Version]
- Sessa, A.; Fagnocchi, L.; Mastrototaro, G.; Massimino, L.; Zaghi, M.; Indrigo, M.; Cattaneo, S.; Martini, D.; Gabellini, C.; Pucci, C.; et al. SETD5 Regulates Chromatin Methylation State and Preserves Global Transcriptional Fidelity during Brain Development and Neuronal Wiring. Neuron 2019, 104, 271–289. [Google Scholar] [CrossRef]
- Deliu, E.; Arecco, N.; Morandell, J.; Dotter, C.P.; Contreras, X.; Girardot, C.; Käsper, E.L.; Kozlova, A.; Kishi, K.; Chiaradia, I.; et al. Haploinsufficiency of the intellectual disability gene SETD5 disturbs developmental gene expression and cognition. Nat. Neurosci. 2018, 21, 1717–1727. [Google Scholar] [CrossRef]
- Osipovich, A.B.; Gangula, R.; Vianna, P.G.; Magnuson, M.A. Setd5 is essential for mammalian development and the co-transcriptional regulation of histone acetylation. Development 2016, 143, 4595–4607. [Google Scholar] [CrossRef] [Green Version]
- Yu, S.E.; Kim, M.S.; Park, S.H.; Yoo, B.C.; Kim, K.H.; Jang, Y.K. SET domain-containing protein 5 is required for expression of primordial germ cell specification-associated genes in murine embryonic stem cells. Cell Biochem. Funct. 2017, 35, 247–253. [Google Scholar] [CrossRef]
- Wang, Z.; Hausmann, S.; Lyu, R.; Li, T.M.; Lofgren, S.M.; Flores, N.M.; Fuentes, M.E.; Caporicci, M.; Yang, Z.; Meiners, M.J.; et al. SETD5-Coordinated Chromatin Reprogramming Regulates Adaptive Resistance to Targeted Pancreatic Cancer Therapy. Cancer Cell 2020, 37, 834–849.e13. [Google Scholar] [CrossRef]
- Moore, S.M.; Seidman, J.S.; Ellegood, J.; Gao, R.; Savchenko, A.; Troutman, T.D.; Abe, Y.; Stender, J.; Lee, D.; Wang, S.; et al. Setd5 haploinsufficiency alters neuronal network connectivity and leads to autistic-like behaviors in mice. Transl. Psychiatry 2019, 9, 24. [Google Scholar] [CrossRef]
- Rea, V.; Van Raay, T.J. Using Zebrafish to Model Autism Spectrum Disorder: A Comparison of ASD Risk Genes Between Zebrafish and Their Mammalian Counterparts. Front. Mol. Neurosci. 2020, 13, 575575. [Google Scholar] [CrossRef] [PubMed]
- Choi, T.Y.; Choi, T.I.; Lee, Y.R.; Choe, S.K.; Kim, C.H. Zebrafish as an animal model for biomedical research. Exp. Mol. Med. 2021, 53, 310–317. [Google Scholar] [CrossRef] [PubMed]
- Salanga, C.M.; Salanga, M.C. Genotype to Phenotype: CRISPR Gene Editing Reveals Genetic Compensation as a Mechanism for Phenotypic Disjunction of Morphants and Mutants. Int. J. Mol. Sci. 2021, 22, 3472. [Google Scholar] [CrossRef] [PubMed]
- Zaghi, M.; Broccoli, V.; Sessa, A. H3K36 Methylation in Neural Development and Associated Diseases. Front. Genet. 2019, 10, 1291. [Google Scholar] [CrossRef] [PubMed]
- Dalla Vecchia, E.; Di Donato, V.; Young, A.M.J.; Del Bene, F.; Norton, W.H.J. Reelin Signaling Controls the Preference for Social Novelty in Zebrafish. Front. Behav. Neurosci. 2019, 13, 214. [Google Scholar] [CrossRef]
- Norton, W.H.J.; Manceau, L.; Reichmann, F. The Visually Mediated Social Preference Test: A Novel Technique to Measure Social Behavior and Behavioral Disturbances in Zebrafish. Methods. Mol. Biol. 2019, 2011, 121–132. [Google Scholar] [CrossRef]
- de Bartolomeis, A.; Latte, G.; Tomasetti, C.; Iasevoli, F. Glutamatergic postsynaptic density protein dysfunctions in synaptic plasticity and dendritic spines morphology: Relevance to schizophrenia and other behavioral disorders pathophysiology, and implications for novel therapeutic approaches. Mol. Neurobiol. 2014, 49, 484–511. [Google Scholar] [CrossRef]
- Atas-Ozcan, H.; Brault, V.; Duchon, A.; Herault, Y. from Gene Function in Development and Physiology to Dosage Correction across Life Span in Down Syndrome. Genes 2021, 12, 1833. [Google Scholar] [CrossRef]
- Teles, M.C.; Almeida, O.; Lopes, J.S.; Oliveira, R.F. Social interactions elicit rapid shifts in functional connectivity in the social decision-making network of zebrafish. Proc. Biol. Sci. 2015, 282, 20151099. [Google Scholar] [CrossRef]
- Kobayashi, Y.; Tohyama, J.; Kato, M.; Akasaka, N.; Magara, S.; Kawashima, H.; Ohashi, T.; Shiraishi, H.; Nakashima, M.; Saitsu, H.; et al. High prevalence of genetic alterations in early-onset epileptic encephalopathies associated with infantile movement disorders. Brain. Dev. 2016, 38, 285–292. [Google Scholar] [CrossRef]
- Iwagawa, T.; Kawabata, R.; Fukushima, M.; Kuribayashi, H.; Watanabe, S. Setd5, but not Setd2, is indispensable for retinal cell survival and proliferation. FEBS Lett. 2022. [Google Scholar] [CrossRef] [PubMed]
- Pietri, T.; Roman, A.C.; Guyon, N.; Romano, S.A.; Washbourne, P.; Moens, C.B.; de Polavieja, G.G.; Sumbre, G. The first mecp2-null zebrafish model shows altered motor behaviors. Front. Neural Circuits 2013, 7, 118. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, C.X.; Li, C.Y.; Hu, C.C.; Wang, Y.; Lin, J.; Jiang, Y.H.; Li, Q.; Xu, X. CRISPR/Cas9-induced shank3b mutant zebrafish display autism-like behaviors. Mol. Autism. 2018, 9, 23. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Stewart, A.M.; Nguyen, M.; Wong, K.; Poudel, M.K.; Kalueff, A.V. Developing zebrafish models of autism spectrum disorder (ASD). Prog. Neuropsychopharmacol. Biol. Psychiatry 2014, 50, 27–36. [Google Scholar] [CrossRef] [PubMed]
- Kim, O.H.; Cho, H.J.; Han, E.; Hong, T.I.; Ariyasiri, K.; Choi, J.H.; Hwang, K.S.; Jeong, Y.M.; Yang, S.Y.; Yu, K.; et al. Zebrafish knockout of Down syndrome gene. Mol. Autism. 2017, 8, 50. [Google Scholar] [CrossRef] [PubMed]
- Tang, W.; Davidson, J.D.; Zhang, G.; Conen, K.E.; Fang, J.; Serluca, F.; Li, J.; Xiong, X.; Coble, M.; Tsai, T.; et al. Genetic Control of Collective Behavior in Zebrafish. iScience 2020, 23, 100942. [Google Scholar] [CrossRef] [Green Version]
- Greco, B.; Managò, F.; Tucci, V.; Kao, H.T.; Valtorta, F.; Benfenati, F. Autism-related behavioral abnormalities in synapsin knockout mice. Behav. Brain. Res. 2013, 251, 65–74. [Google Scholar] [CrossRef]
- Lovinger, D.M. Communication networks in the brain: Neurons, receptors, neurotransmitters, and alcohol. Alcohol. Res. Health 2008, 31, 196–214. [Google Scholar]
- Clifton, N.E.; Trent, S.; Thomas, K.L.; Hall, J. Regulation and Function of Activity-Dependent Homer in Synaptic Plasticity. Mol. Neuropsychiatry 2019, 5, 147–161. [Google Scholar] [CrossRef]
- Aranda, S.; Laguna, A.; de la Luna, S. DYRK family of protein kinases: Evolutionary relationships, biochemical properties, and functional roles. FASEB J. 2011, 25, 449–462. [Google Scholar] [CrossRef]
- Hayashi, M.K.; Tang, C.; Verpelli, C.; Narayanan, R.; Stearns, M.H.; Xu, R.M.; Li, H.; Sala, C.; Hayashi, Y. The postsynaptic density proteins Homer and Shank form a polymeric network structure. Cell 2009, 137, 159–171. [Google Scholar] [CrossRef] [PubMed]
- Michetti, C.; Caruso, A.; Pagani, M.; Sabbioni, M.; Medrihan, L.; David, G.; Galbusera, A.; Morini, M.; Gozzi, A.; Benfenati, F.; et al. The Knockout of Synapsin II in Mice Impairs Social Behavior and Functional Connectivity Generating an ASD-like Phenotype. Cereb. Cortex 2017, 27, 5014–5023. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yoon, S.; Piguel, N.H.; Khalatyan, N.; Dionisio, L.E.; Savas, J.N.; Penzes, P. Homer1 promotes dendritic spine growth through ankyrin-G and its loss reshapes the synaptic proteome. Mol. Psychiatry 2021, 26, 1775–1789. [Google Scholar] [CrossRef] [PubMed]
- Bourgeron, T. From the genetic architecture to synaptic plasticity in autism spectrum disorder. Nat. Rev. Neurosci. 2015, 16, 551–563. [Google Scholar] [CrossRef]
- Dente, L.; Gestri, G.; Tsang, M.; Kudoh, T.; Wilson, S.W.; Dawid, I.B.; Andreazzoli, M. Cloning and developmental expression of zebrafish pdzrn3. Int. J. Dev. Biol. 2011, 55, 989–993. [Google Scholar] [CrossRef] [Green Version]
- Jao, L.E.; Wente, S.R.; Chen, W. Efficient multiplex biallelic zebrafish genome editing using a CRISPR nuclease system. Proc. Natl. Acad. Sci. USA 2013, 110, 13904–13909. [Google Scholar] [CrossRef] [Green Version]
- Brinkman, E.K.; Chen, T.; Amendola, M.; van Steensel, B. Easy quantitative assessment of genome editing by sequence trace decomposition. Nucleic Acids Res. 2014, 42, e168. [Google Scholar] [CrossRef]
- Dehairs, J.; Talebi, A.; Cherifi, Y.; Swinnen, J.V. CRISP-ID: Decoding CRISPR mediated indels by Sanger sequencing. Sci. Rep. 2016, 6, 28973. [Google Scholar] [CrossRef] [Green Version]
- Hill, J.T.; Demarest, B.L.; Bisgrove, B.W.; Su, Y.C.; Smith, M.; Yost, H.J. Poly peak parser: Method and software for identification of unknown indels using sanger sequencing of polymerase chain reaction products. Dev. Dyn. 2014, 243, 1632–1636. [Google Scholar] [CrossRef] [Green Version]
- Martini, D.; Pucci, C.; Gabellini, C.; Pellegrino, M.; Andreazzoli, M. Exposure to the natural alkaloid Berberine affects cardiovascular system morphogenesis and functionality during zebrafish development. Sci. Rep. 2020, 10, 17358. [Google Scholar] [CrossRef]
- Smith, A.; Zhang, J.; Guay, D.; Quint, E.; Johnson, A.; Akimenko, M.A. Gene expression analysis on sections of zebrafish regenerating fins reveals limitations in the whole-mount in situ hybridization method. Dev. Dyn. 2008, 237, 417–425. [Google Scholar] [CrossRef] [PubMed]
- Stern, H.M.; Murphey, R.D.; Shepard, J.L.; Amatruda, J.F.; Straub, C.T.; Pfaff, K.L.; Weber, G.; Tallarico, J.A.; King, R.W.; Zon, L.I. Small molecules that delay S phase suppress a zebrafish bmyb mutant. Nat. Chem. Biol. 2005, 1, 366–370. [Google Scholar] [CrossRef] [PubMed]
- Pineda, R.H.; Svoboda, K.R.; Wright, M.A.; Taylor, A.D.; Novak, A.E.; Gamse, J.T.; Eisen, J.S.; Ribera, A.B. Knockdown of Nav1.6a Na+ channels affects zebrafish motoneuron development. Development 2006, 133, 3827–3836. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lange, M.; Norton, W.; Coolen, M.; Chaminade, M.; Merker, S.; Proft, F.; Schmitt, A.; Vernier, P.; Lesch, K.P.; Bally-Cuif, L. The ADHD-susceptibility gene lphn3.1 modulates dopaminergic neuron formation and locomotor activity during zebrafish development. Mol. Psychiatry 2012, 17, 946–954. [Google Scholar] [CrossRef] [PubMed]



Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gabellini, C.; Pucci, C.; De Cesari, C.; Martini, D.; Di Lauro, C.; Digregorio, M.; Norton, W.; Zippo, A.; Sessa, A.; Broccoli, V.; et al. CRISPR/Cas9-Induced Inactivation of the Autism-Risk Gene setd5 Leads to Social Impairments in Zebrafish. Int. J. Mol. Sci. 2023, 24, 167. https://doi.org/10.3390/ijms24010167
Gabellini C, Pucci C, De Cesari C, Martini D, Di Lauro C, Digregorio M, Norton W, Zippo A, Sessa A, Broccoli V, et al. CRISPR/Cas9-Induced Inactivation of the Autism-Risk Gene setd5 Leads to Social Impairments in Zebrafish. International Journal of Molecular Sciences. 2023; 24(1):167. https://doi.org/10.3390/ijms24010167
Chicago/Turabian StyleGabellini, Chiara, Cecilia Pucci, Chiara De Cesari, Davide Martini, Caterina Di Lauro, Matteo Digregorio, William Norton, Alessio Zippo, Alessandro Sessa, Vania Broccoli, and et al. 2023. "CRISPR/Cas9-Induced Inactivation of the Autism-Risk Gene setd5 Leads to Social Impairments in Zebrafish" International Journal of Molecular Sciences 24, no. 1: 167. https://doi.org/10.3390/ijms24010167
APA StyleGabellini, C., Pucci, C., De Cesari, C., Martini, D., Di Lauro, C., Digregorio, M., Norton, W., Zippo, A., Sessa, A., Broccoli, V., & Andreazzoli, M. (2023). CRISPR/Cas9-Induced Inactivation of the Autism-Risk Gene setd5 Leads to Social Impairments in Zebrafish. International Journal of Molecular Sciences, 24(1), 167. https://doi.org/10.3390/ijms24010167






