Insight into CAZymes of Alicyclobacillus mali FL18: Characterization of a New Multifunctional GH9 Enzyme
Abstract
1. Introduction
2. Results and Discussion
2.1. Analysis of CAZymes in A. mali FL18
Mining of Enzymes for Cellulose Degradation
2.2. Sequence Analysis of AmCel9
2.3. Recombinant AmCel9: Expression, Purification, and Molecular Weight Analyses
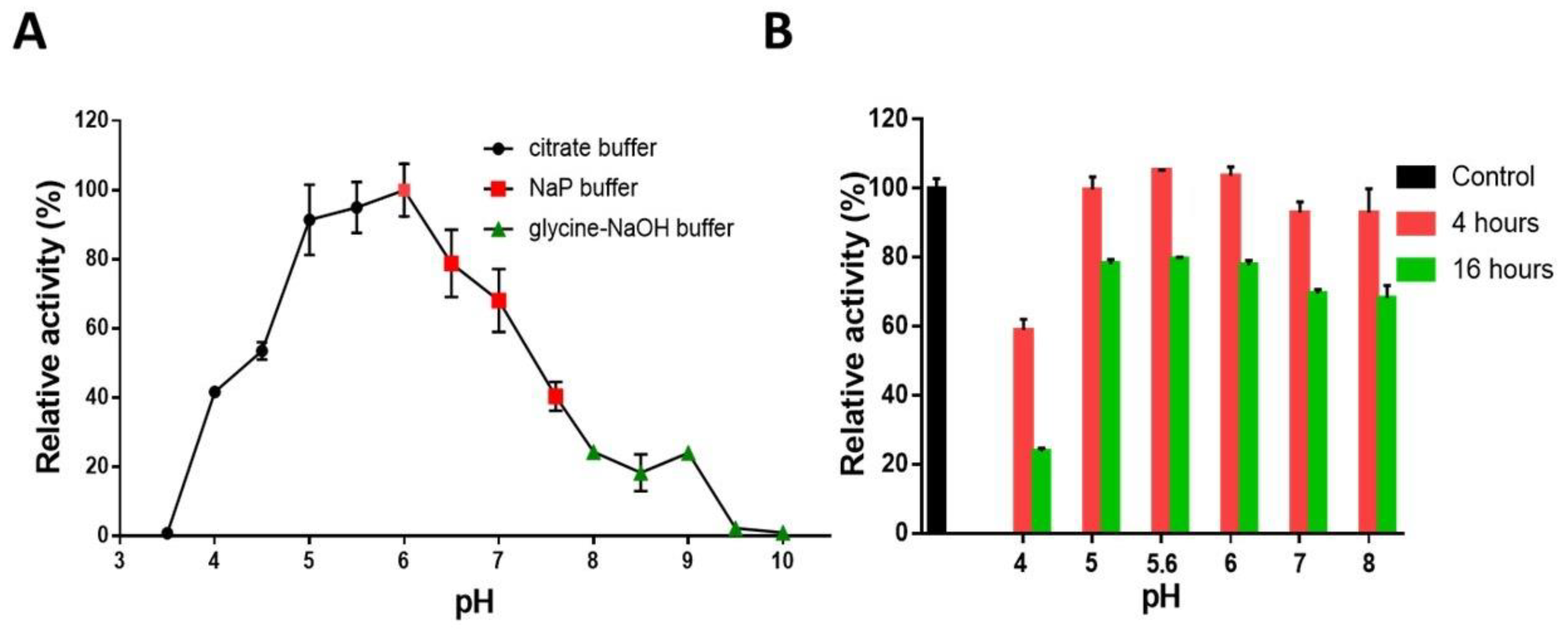
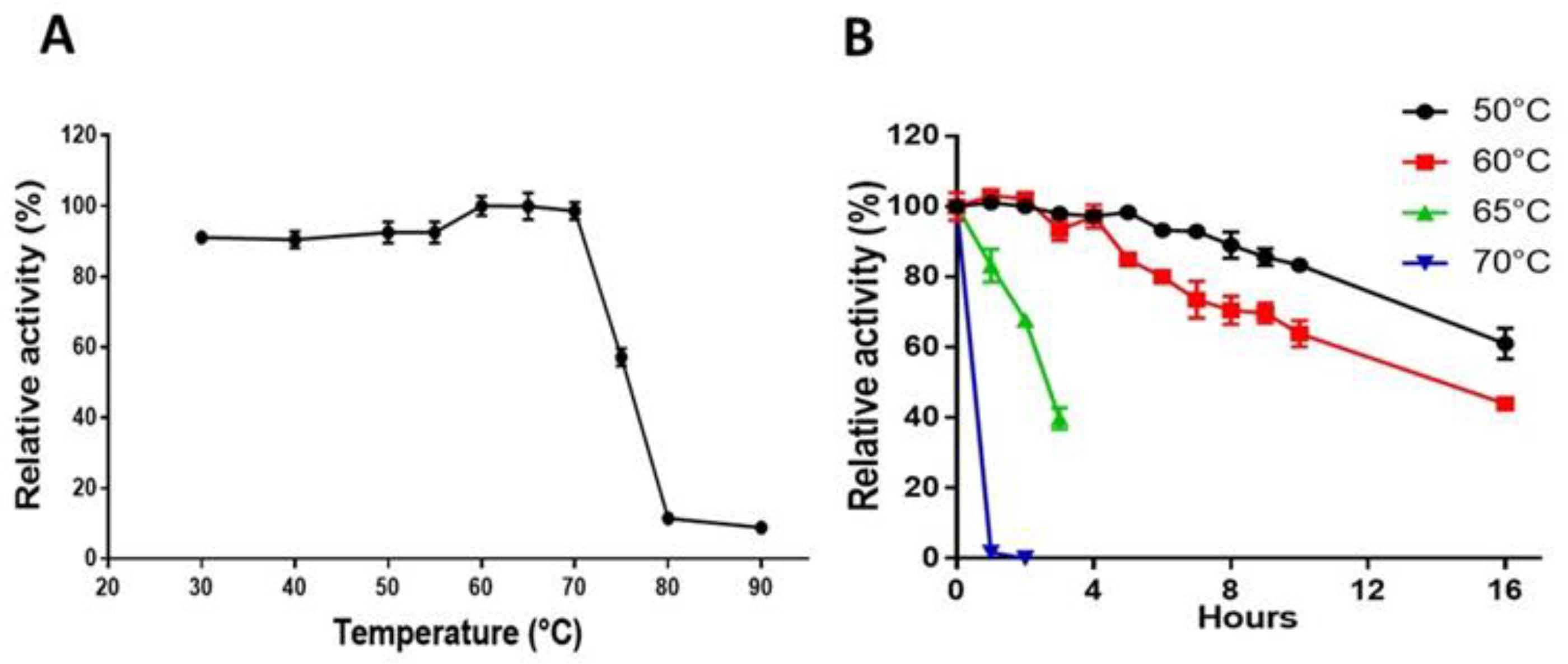
2.4. pH and Temperature Profile and Stability Properties of AmCel9
2.5. Substrate Specificity and Catalytic Properties
3. Materials and Methods
3.1. Functional Annotation of A. mali FL18 CAZymes
3.2. Reagents and Substrates
3.3. AmCel9 In Silico Analyses
3.4. Expression and Purification of Recombinant AmCel9
3.5. Determination of AmCel9 Native Molecular Weight
3.6. Enzyme Assay, Temperature and pH Profile
3.7. Substrate Specificity and Kinetic Studies
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Aich, S.; Datta, S. Efficient Utilization of Lignocellulosic Biomass: Hydrolysis Methods for Biorefineries. In Biorefineries: A Step Towards Renewable and Clean Energy; Verma, P., Ed.; Springer: Singapore, 2020; pp. 273–295. ISBN 978-981-15-9593-6. [Google Scholar]
- Gallo, G.; Puopolo, R.; Carbonaro, M.; Maresca, E.; Fiorentino, G. Extremophiles, a Nifty Tool to Face Environmental Pollution: From Exploitation of Metabolism to Genome Engineering. Int. J. Environ. Res. Public Health 2021, 18, 5228. [Google Scholar] [CrossRef] [PubMed]
- Piras, C.C.; Fernández-Prieto, S.; de Borggraeve, W.M. Ball Milling: A Green Technology for the Preparation and Functionalisation of Nanocellulose Derivatives. Nanoscale Adv. 2019, 1, 937–947. [Google Scholar] [CrossRef] [PubMed]
- Glasgow, E.; vander Meulen, K.; Kuch, N.; Fox, B.G. Multifunctional Cellulases Are Potent, Versatile Tools for a Renewable Bioeconomy. Curr. Opin. Biotechnol. 2021, 67, 141–148. [Google Scholar] [CrossRef] [PubMed]
- Salzano, F.; Aulitto, M.; Fiorentino, G.; Pedone, E.; Contursi, P.; Limauro, D. Alicyclobacillus mali FL18 as a Novel Source of Glycosyl Hydrolases: Characterization of a New Thermophilic β-Xylosidase Tolerant to Monosaccharides. Int. J. Mol. Sci. 2022, 23, 14310. [Google Scholar] [CrossRef] [PubMed]
- Drula, E.; Garron, M.-L.; Dogan, S.; Lombard, V.; Henrissat, B.; Terrapon, N. The Carbohydrate-Active Enzyme Database: Functions and Literature. Nucleic Acids Res. 2022, 50, D571–D577. [Google Scholar] [CrossRef]
- Andlar, M.; Rezić, T.; Marđetko, N.; Kracher, D.; Ludwig, R.; Šantek, B. Lignocellulose Degradation: An Overview of Fungi and Fungal Enzymes Involved in Lignocellulose Degradation. Eng. Life Sci. 2018, 18, 768–778. [Google Scholar] [CrossRef]
- Konar, A.; Aich, S.; Katakojwala, R.; Datta, S.; Mohan, S.V. A Processive GH9 Family Endoglucanase of Bacillus licheniformis and the Role of Its Carbohydrate-Binding Domain. Appl. Microbiol. Biotechnol. 2022, 106, 6059–6075. [Google Scholar] [CrossRef]
- Ajeje, S.B.; Hu, Y.; Song, G.; Peter, S.B.; Afful, R.G.; Sun, F.; Asadollahi, M.A.; Amiri, H.; Abdulkhani, A.; Sun, H. Thermostable Cellulases/Xylanases from Thermophilic and Hyperthermophilic Microorganisms: Current Perspective. Front. Bioeng. Biotechnol. 2021, 9, 794304. [Google Scholar] [CrossRef]
- Botturi, A.; Battista, F.; Andreolli, M.; Faccenda, F.; Fusco, S.; Bolzonella, D.; Lampis, S.; Frison, N. Polyhydroxyalkanoated-Rich Microbial Cells from Bio-Based Volatile Fatty Acids as Potential Ingredient for Aquaculture Feed. Energies 2021, 14, 38. [Google Scholar] [CrossRef]
- Wang, R.; Lorantfy, B.; Fusco, S.; Olsson, L.; Franzén, C.J. Analysis of Methods for Quantifying Yeast Cell Concentration in Complex Lignocellulosic Fermentation Processes. Sci. Rep. 2021, 11, 11293. [Google Scholar] [CrossRef]
- Fusco, F.A.; Ronca, R.; Fiorentino, G.; Pedone, E.; Contursi, P.; Bartolucci, S.; Limauro, D. Biochemical Characterization of a Thermostable Endomannanase/Endoglucanase from Dictyoglomus Turgidum. Extremophiles 2018, 22, 131–140. [Google Scholar] [CrossRef] [PubMed]
- Antonucci, I.; Gallo, G.; Limauro, D.; Contursi, P.; Ribeiro, A.L.; Blesa, A.; Berenguer, J.; Bartolucci, S.; Fiorentino, G. Characterization of a Promiscuous Cadmium and Arsenic Resistance Mechanism in Thermus Thermophilus HB27 and Potential Application of a Novel Bioreporter System. Microb Cell Fact. 2018, 17, 78. [Google Scholar] [CrossRef] [PubMed]
- Koga, Y. Thermal Adaptation of the Archaeal and Bacterial Lipid Membranes. Archaea 2012, 2012, 789652. [Google Scholar] [CrossRef] [PubMed]
- Morana, A.; Esposito, A.; Maurelli, L.; Ruggiero, G.; Ionata, E.; Rossi, M.; la Cara, F. A Novel Thermoacidophilic Cellulase from Alicyclobacillus acidocaldarius. Protein Pept. Lett. 2008, 15, 1017–1021. [Google Scholar] [CrossRef] [PubMed]
- Bai, Y.; Wang, J.; Zhang, Z.; Shi, P.; Luo, H.; Huang, H.; Luo, C.; Yao, B. Expression of an Extremely Acidic β-1,4-Glucanase from Thermoacidophilic Alicyclobacillus sp. A4 in Pichia Pastoris Is Improved by Truncating the Gene Sequence. Microb Cell Fact. 2010, 9, 33. [Google Scholar] [CrossRef] [PubMed]
- Aulitto, M.; Gallo, G.; Puopolo, R.; Mormone, A.; Limauro, D.; Contursi, P.; Piochi, M.; Bartolucci, S.; Fiorentino, G. Genomic Insight of Alicyclobacillus mali FL18 Isolated from an Arsenic-Rich Hot Spring. Front. Microbiol. 2021, 12, 669. [Google Scholar] [CrossRef] [PubMed]
- Aulitto, M.; Fusco, S.; Limauro, D.; Fiorentino, G.; Bartolucci, S.; Contursi, P. Galactomannan Degradation by Thermophilic Enzymes: A Hot Topic for Biotechnological Applications. World J. Microbiol. Biotechnol. 2019, 35, 32. [Google Scholar] [CrossRef]
- Aulitto, M.; Fusco, F.A.; Fiorentino, G.; Bartolucci, S.; Contursi, P.; Limauro, D. A Thermophilic Enzymatic Cocktail for Galactomannans Degradation. Enzyme Microb Technol. 2018, 111, 7–11. [Google Scholar] [CrossRef]
- Ing, N.; Deng, K.; Chen, Y.; Aulitto, M.; Gin, J.W.; Pham, T.L.M.; Petzold, C.J.; Singer, S.W.; Bowen, B.; Sale, K.L.; et al. A Multiplexed Nanostructure-Initiator Mass Spectrometry (NIMS) Assay for Simultaneously Detecting Glycosyl Hydrolase and Lignin Modifying Enzyme Activities. Sci. Rep. 2021, 11, 11803. [Google Scholar] [CrossRef]
- Dadwal, A.; Sharma, S.; Satyanarayana, T. Thermostable Cellulose Saccharifying Microbial Enzymes: Characteristics, Recent Advances and Biotechnological Applications. Int. J. Biol. Macromol. 2021, 188, 226–244. [Google Scholar] [CrossRef]
- Turner, P.; Mamo, G.; Karlsson, E.N. Potential and Utilization of Thermophiles and Thermostable Enzymes in Biorefining. Microb Cell Fact. 2007, 6, 9. [Google Scholar] [CrossRef] [PubMed]
- Politi, J.; Spadavecchia, J.; Fiorentino, G.; Antonucci, I.; Casale, S.; de Stefano, L. Interaction of Thermus Thermophilus ArsC Enzyme and Gold Nanoparticles Naked-Eye Assays Speciation between As(III) and As(V). Nanotechnology 2015, 26, 435703. [Google Scholar] [CrossRef] [PubMed]
- Aulitto, M.; Tom, L.M.; Ceja-Navarro, J.A.; Simmons, B.A.; Singer, S.W. Whole-Genome Sequence of Brevibacillus borstelensis SDM, Isolated from a Sorghum-Adapted Microbial Community. Microbiol. Resour. Announc. 2020, 9, 15–16. [Google Scholar] [CrossRef] [PubMed]
- Tom, L.M.; Aulitto, M.; Wu, Y.W.; Deng, K.; Gao, Y.; Xiao, N.; Rodriguez, B.G.; Louime, C.; Northen, T.R.; Eudes, A.; et al. Low—Abundance Populations Distinguish Microbiome Performance in Plant Cell Wall Deconstruction. Microbiome 2022, 10, 183. [Google Scholar] [CrossRef] [PubMed]
- Aulitto, M.; Strazzulli, A.; Sansone, F.; Cozzolino, F.; Monti, M.; Moracci, M.; Fiorentino, G.; Limauro, D.; Bartolucci, S.; Contursi, P. Prebiotic Properties of Bacillus coagulans MA-13: Production of Galactoside Hydrolyzing Enzymes and Characterization of the Transglycosylation Properties of a GH42 β-Galactosidase. Microb Cell Fact. 2021, 20, 71. [Google Scholar] [CrossRef]
- Aulitto, M.; Fusco, S.; Franzén, C.J.; Strazzulli, A.; Moracci, M.; Bartolucci, S.; Contursi, P. Draft Genome Sequence of Bacillus coagulans Ma-13, a Thermophilic Lactic Acid Producer from Lignocellulose. Microbiol. Resour. Announc. 2019, 8, e00341-19. [Google Scholar] [CrossRef]
- Fiorentino, G.; Cannio, R.; Rossi, M.; Bartolucci, S. Decreasing the Stability and Changing the Substrate Specificity of the Bacillus stearothermophilus Alcohol Dehydrogenase by Single Amino Acid Replacements. Protein Eng. Des. Sel. 1998, 11, 925–930. [Google Scholar] [CrossRef]
- Si, M.; Liu, D.; Liu, M.; Yan, X.; Gao, C.; Chai, L.; Shi, Y. Complementary Effect of Combined Bacterial-Chemical Pretreatment to Promote Enzymatic Digestibility of Lignocellulose Biomass. Bioresour. Technol. 2019, 272, 275–280. [Google Scholar] [CrossRef]
- Viikari, L.; Alapuranen, M.; Puranen, T.; Vehmaanperä, J.; Siika-Aho, M. Thermostable Enzymes in Lignocellulose Hydrolysis. Adv. Biochem. Eng. Biotechnol. 2007, 108, 121–145. [Google Scholar] [CrossRef]
- Zuliani, L.; Serpico, A.; de Simone, M.; Frison, N.; Fusco, S. Biorefinery Gets Hot: Thermophilic Enzymes and Microorganisms for Second-Generation Bioethanol Production. Processes 2021, 9, 1583. [Google Scholar] [CrossRef]
- Huang, L.; Zhang, H.; Wu, P.; Entwistle, S.; Li, X.; Yohe, T.; Yi, H.; Yang, Z.; Yin, Y. DbCAN-Seq: A Database of Carbohydrate-Active Enzyme (CAZyme) Sequence and Annotation. Nucleic Acids Res. 2018, 46, D516–D521. [Google Scholar] [CrossRef] [PubMed]
- Aulitto, M.; Martinez-Alvarez, L.; Fiorentino, G.; Limauro, D.; Peng, X.; Contursi, P. A Comparative Analysis of Weizmannia coagulans Genomes Unravels the Genetic Potential for Biotechnological Applications. Int. J. Mol. Sci. 2022, 23, 3135. [Google Scholar] [CrossRef] [PubMed]
- Tong, X.; Qi, Z.; Zheng, D.; Pei, J.; Li, Q.; Zhao, L. High-Level Expression of a Novel Multifunctional GH3 Family β-Xylosidase/α-Arabinosidase/β-Glucosidase from Dictyoglomus Turgidum in Escherichia coli. Bioorg. Chem. 2021, 111. [Google Scholar] [CrossRef] [PubMed]
- Wu, Y.; Mao, G.; Fan, H.; Song, A.; Zhang, Y.H.P.; Chen, H. Biochemical Properties of GH94 Cellodextrin Phosphorylase THA_1941 from a Thermophilic Eubacterium Thermosipho Africanus TCF52B with Cellobiose Phosphorylase Activity. Sci. Rep. 2017, 7, 4849. [Google Scholar] [CrossRef] [PubMed]
- Eckert, K.; Schneider, E. A Thermoacidophilic Endoglucanase (CelB) from Alicyclobacillus acidocaldarius Displays High Sequence Similarity to Arabinofuranosidases Belonging to Family 51 of Glycoside Hydrolases. Eur. J. Biochem. 2003, 270, 3593–3602. [Google Scholar] [CrossRef] [PubMed]
- Eckert, K.; Zielinski, F.; lo Leggio, L.; Schneider, E. Gene Cloning, Sequencing, and Characterization of a Family 9 Endoglucanase (CelA) with an Unusual Pattern of Activity from the Thermoacidophile Alicyclobacillus acidocaldarius ATCC27009. Appl. Microbiol. Biotechnol. 2002, 60, 428–436. [Google Scholar] [CrossRef]
- Yeoman, C.J.; Fields, C.J.; Lepercq, P.; Ruiz, P.; Forano, E.; White, B.A.; Mosoni, P. In Vivo Competitions between Fibrobacter succinogenes, Ruminococcus flavefaciens, and Ruminoccus albus in a Gnotobiotic Sheep Model Revealed by Multi-Omic Analyses. mBio 2021, 12, e03533-20. [Google Scholar] [CrossRef]
- Gilad, R.; Rabinovich, L.; Yaron, S.; Bayer, E.A.; Lamed, R.; Gilbert, H.J.; Shoham, Y. CelI, a Noncellulosomal Family 9 Enzyme from Clostridium thermocellum, Is a Processive Endoglucanase That Degrades Crystalline Cellulose. J. Bacteriol. 2003, 185, 391–398. [Google Scholar] [CrossRef]
- Aymé, L.; Hébert, A.; Henrissat, B.; Lombard, V.; Franche, N.; Perret, S.; Jourdier, E.; Heiss-Blanquet, S. Characterization of Three Bacterial Glycoside Hydrolase Family 9 Endoglucanases with Different Modular Architectures Isolated from a Compost Metagenome. Biochim. Biophys. Acta (BBA) Gen. Subj. 2021, 1865, 129848. [Google Scholar] [CrossRef]
- Wu, L.; Davies, G.J. Structure of the GH9 Glucosidase/Glucosaminidase from Vibrio Cholerae. Acta Crystallogr. Sect. F 2018, 74, 512–523. [Google Scholar] [CrossRef]
- Liu, H.; Pereira, J.H.; Adams, P.D.; Sapra, R.; Simmons, B.A.; Sale, K.L. Molecular Simulations Provide New Insights into the Role of the Accessory Immunoglobulin-like Domain of Cel9A. FEBS Lett. 2010, 584, 3431–3435. [Google Scholar] [CrossRef] [PubMed]
- Phakeenuya, V.; Ratanakhanokchai, K.; Kosugi, A.; Tachaapaikoon, C. A Novel Multifunctional GH9 Enzyme from Paenibacillus curdlanolyticus B-6 Exhibiting Endo/Exo Functions of Cellulase, Mannanase and Xylanase Activities. Appl. Microbiol. Biotechnol. 2020, 104, 2079–2096. [Google Scholar] [CrossRef] [PubMed]
- Schröder, C.; Burkhardt, C.; Busch, P.; Schirrmacher, G.; Claren, J.; Antranikian, G. Characterization of a Theme C Glycoside Hydrolase Family 9 Endo-β-Glucanase from a Biogas Reactor Metagenome. Protein J. 2018, 37, 454–460. [Google Scholar] [CrossRef] [PubMed]
- Pereira, J.H.; Sapra, R.; Volponi, J.V.; Kozina, C.L.; Simmons, B.; Adams, P.D. Structure of Endoglucanase Cel9A from the Thermoacidophilic Alicyclobacillus acidocaldarius. Acta Crystallogr. D Biol. Crystallogr. 2009, 65, 744–750. [Google Scholar] [CrossRef]
- Kesavulu, M.M.; Tsai, J.Y.; Lee, H.L.; Liang, P.H.; Hsiao, C.D. Structure of the Catalytic Domain of the Clostridium thermocellum Cellulase CelT. Acta Crystallogr. D Biol. Crystallogr. 2012, 68, 310–320. [Google Scholar] [CrossRef]
- Bakker, E.P. The Role of Alkali-Cation Transport in Energy Coupling of Neutrophilic and Acidophilic Bacteria: An Assessment of Methods and Concepts. FEMS Microbiol. Rev. 1990, 6, 319–334. [Google Scholar] [CrossRef]
- Bai, Y.; Wang, J.; Zhang, Z.; Shi, P.; Luo, H.; Huang, H.; Luo, C.; Yao, B. A Novel Family 9 β-1,3(4)-Glucanase from Thermoacidophilic Alicyclobacillus sp. A4 with Potential Applications in the Brewing Industry. Appl. Microbiol. Biotechnol. 2010, 87, 251–259. [Google Scholar] [CrossRef]
- Akram, F.; Khan, M.A.; Hussain, Z.; Nawaz, A.; Iqbal, K.; Shah, A.J. CenC, a Multidomain Thermostable GH9 Processive Endoglucanase from Clostridium thermocellum: Cloning, Characterization and Saccharification Studies. World J. Microbiol. Biotechnol. 2015, 31, 1699–1710. [Google Scholar] [CrossRef]
- de Araújo, E.A.; de Oliveira Neto, M.; Polikarpov, I. Biochemical Characterization and Low-Resolution SAXS Structure of Two-Domain Endoglucanase BlCel9 from Bacillus licheniformis. Appl. Microbiol. Biotechnol. 2019, 103, 1275–1287. [Google Scholar] [CrossRef]
- Celestino, K.R.S.; Cunha, R.B.; Felix, C.R. Characterization of a β-Glucanase Produced by Rhizopus microsporus Var. microsporus, and Its Potential for Application in the Brewing Industry. BMC Biochem. 2006, 7, 23. [Google Scholar] [CrossRef]
- Schubot, F.D.; Kataeva, I.A.; Chang, J.; Shah, A.K.; Ljungdahl, L.G.; Rose, J.P.; Wang, B.C. Structural Basis for the Exocellulase Activity of the Cellobiohydrolase CbhA from Clostridium thermocellum. Biochemistry 2004, 43, 1163–1170. [Google Scholar] [CrossRef] [PubMed]
- Kataeva, I.; Li, X.-L.; Chen, H.; Choi, S.-K.; Ljungdahl, L.G. Cloning and Sequence Analysis of a New Cellulase Gene Encoding CelK, a Major Cellulosome Component of Clostridium thermocellum: Evidence for Gene Duplication and Recombination. J. Bacteriol. 1999, 181, 5288–5295. [Google Scholar] [CrossRef] [PubMed]
- Sakon, J.; Irwin, D.; Wilson, D.B.; Karplus, P.A. Structure and Mechanism of Endo/Exocellulase E4 from Thermomonospora fusca. Nat. Struct. Biol. 1997, 4, 810–818. [Google Scholar] [CrossRef] [PubMed]
- Ravachol, J.; Borne, R.; Tardif, C.; de Philip, P.; Fierobe, H.P. Characterization of All Family-9 Glycoside Hydrolases Synthesized by the Cellulosome-Producing Bacterium Clostridium cellulolyticum. J. Biol. Chem. 2014, 289, 7335–7348. [Google Scholar] [CrossRef] [PubMed]
- Gao, S.; You, C.; Renneckar, S.; Bao, J.; Zhang, Y.-H.P. New Insights into Enzymatic Hydrolysis of Heterogeneous Cellulose by Using Carbohydrate-Binding Module 3 Containing GFP and Carbohydrate-Binding Module 17 Containing CFP. Biotechnol. Biofuels 2014, 7, 24. [Google Scholar] [CrossRef]
- Mirdita, M.; Schütze, K.; Moriwaki, Y.; Heo, L.; Ovchinnikov, S.; Steinegger, M. ColabFold: Making Protein Folding Accessible to All. Nat. Methods 2022, 19, 679–682. [Google Scholar] [CrossRef]
- Pirone, L.; Pitzer, J.E.; D’Abrosca, G.; Fattorusso, R.; Malgieri, G.; Pedone, E.M.; Pedone, P.V.; Roop, R.M.; Baglivo, I. Identifying the Region Responsible for Brucella abortus MucR Higher-Order Oligomer Formation and Examining Its Role in Gene Regulation. Sci. Rep. 2018, 8, 17238. [Google Scholar] [CrossRef]
- Kim, J.J.; Kwon, Y.K.; Kim, J.H.; Heo, S.J.; Lee, Y.; Lee, S.J.; Shim, W.B.; Jung, W.K.; Hyun, J.H.; Kwon, K.K.; et al. Effective Microwell Plate-Based Screening Method for Microbes Producing Cellulase and Xylanase and Its Application. J. Microbiol. Biotechnol. 2014, 24, 1559–1565. [Google Scholar] [CrossRef]
- Miller, G.L. Use of Dinitrosalicylic Acid Reagent for Determination of Reducing Sugar. Anal. Chem. 1959, 31, 426–428. [Google Scholar] [CrossRef]
- Ogonda, L.A.; Saumonneau, A.; Dion, M.; Muge, E.K.; Wamalwa, B.M.; Mulaa, F.J.; Tellier, C. Characterization and Engineering of Two New GH9 and GH48 Cellulases from a Bacillus pumilus Isolated from Lake Bogoria. Biotechnol. Lett. 2021, 43, 691–700. [Google Scholar] [CrossRef]
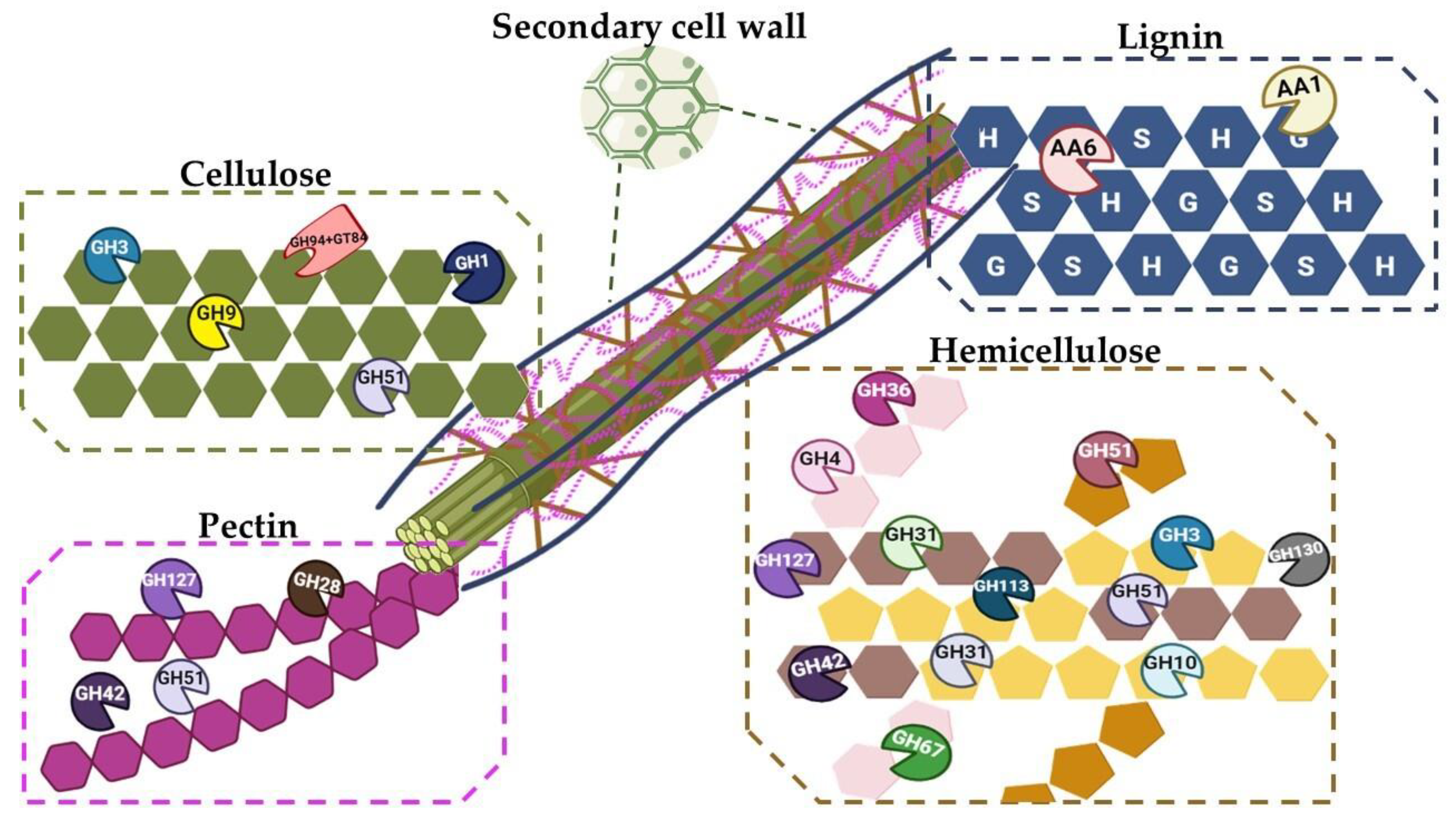
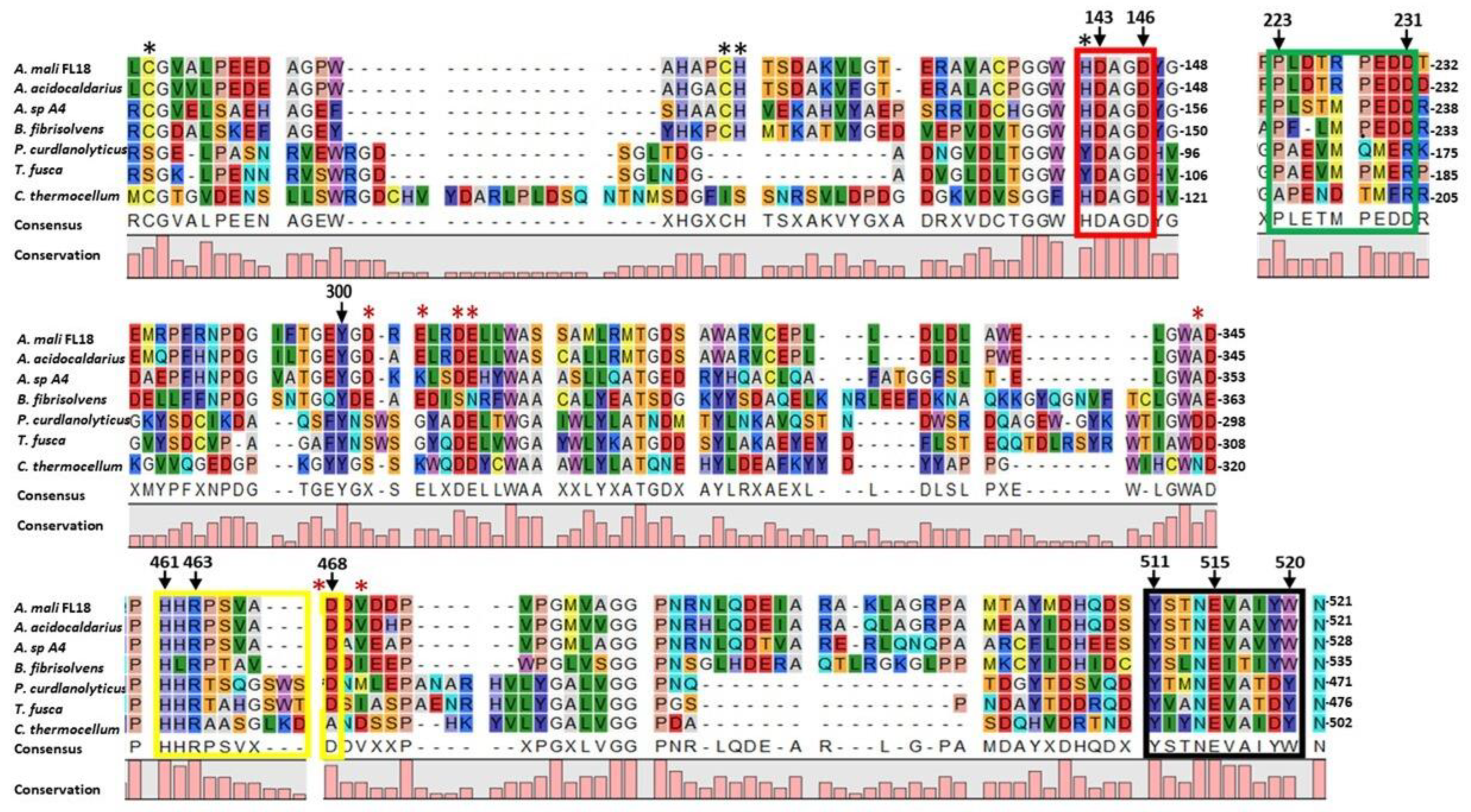
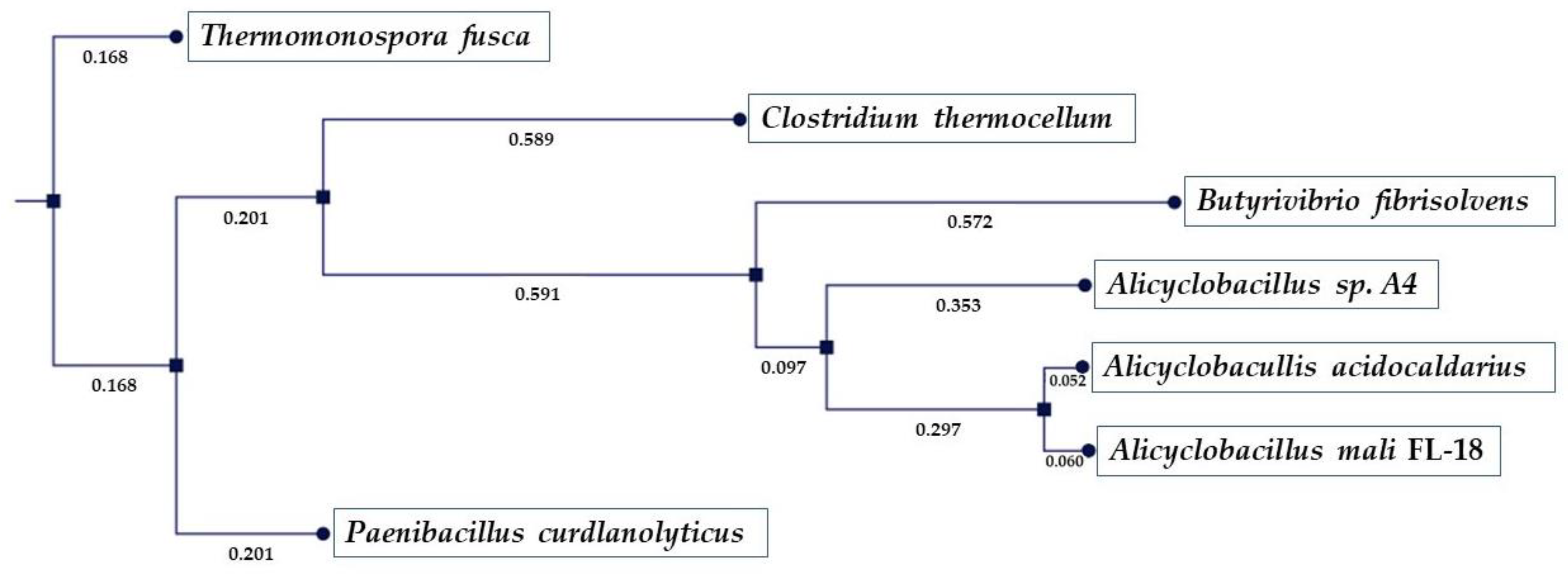
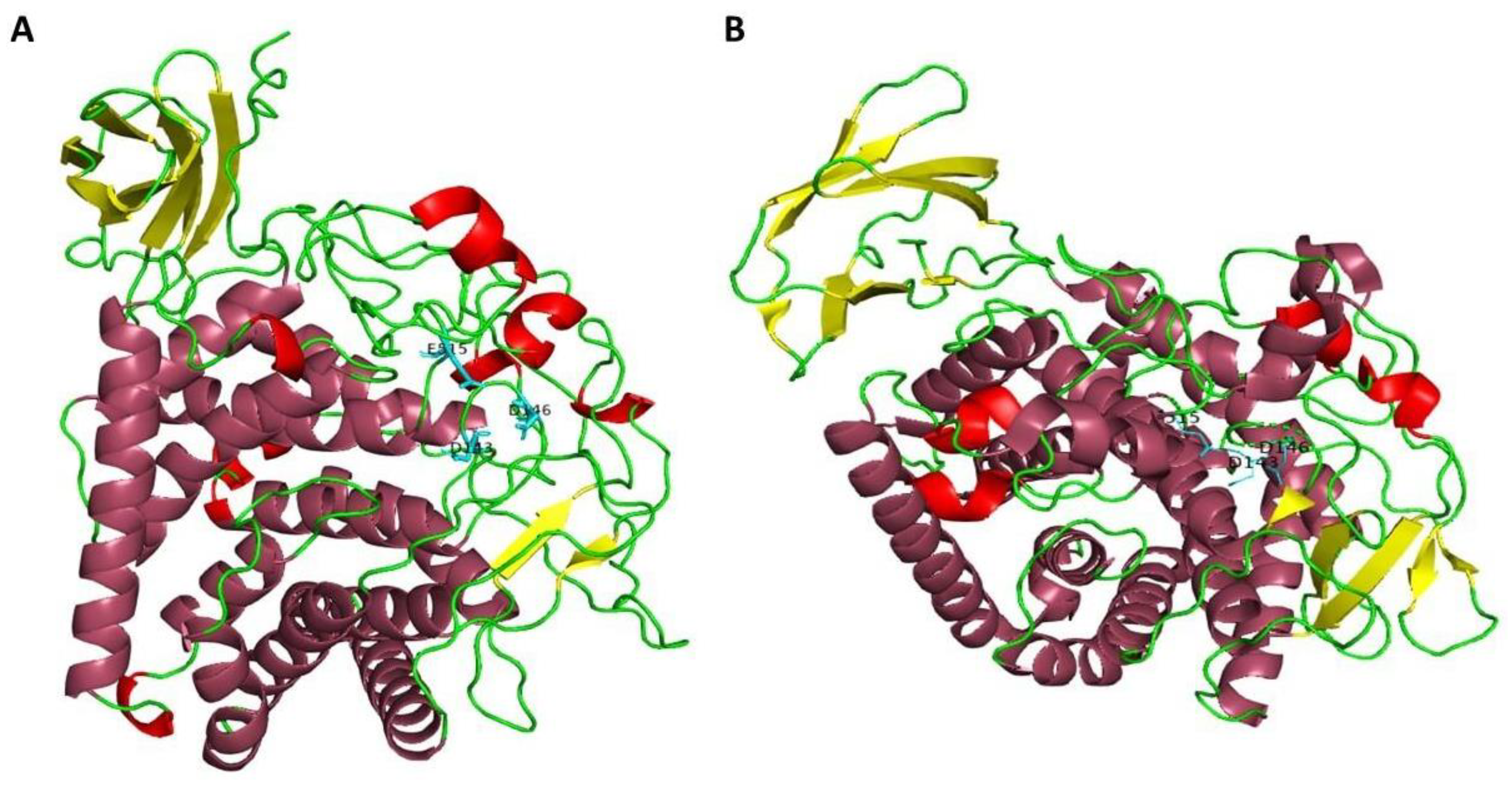
| Accession Number | E.C. Number | NCBI Annotation | CAZy Family |
|---|---|---|---|
| Cellulose | |||
| MBF8378818.1 | 3.2.1.21|3.2.1.-|3.2.1.38|3.2.1.23|3.2.1.74 | β-glucosidase | GH1 |
| MBF8377998.1 | 3.2.1.4|3.2.1.6|3.2.1.151 | glycoside hydrolase family 9 protein | GH9 |
| MBF8377873.1 | 3.2.1.4|3.2.1.8 | α-l-arabinofuranosidase | GH51 |
| MBF8378553.1 | 2.4.1.333|2.4.1.-|- | carbohydrate-binding protein | GH94 + GT84 |
| Hemicellulose | |||
| MBF8378422.1 | 3.2.1.37|3.2.1.55|3.2.1.21 | glycoside hydrolase family 3 C-terminal domain-containing protein | GH3 |
| MBF8376310.1 | 3.2.1.22 | α-glucosidase/α-galactosidase | GH4 |
| MBF8379065.1 | 3.2.1.8 | endo-1,4-β-xylanase | GH10 |
| MBF8377308.1 | 3.2.1.20|3.2.1.10 | α-glucosidase | GH31 |
| MBF8377285.1 | 3.2.1.177 | α-xylosidase | GH31 |
| MBF8378558.1 | 3.2.1.22 | α-galactosidase | GH36 |
| MBF8377281.1 | 3.2.1.23 | β-galactosidase | GH42 |
| MBF8377873.1 | 3.2.1.4|3.2.1.8 | α-l-arabinofuranosidase | GH51 |
| MBF8377278.1 | 3.2.1.55 | α-N-arabinofuranosidase | GH51 |
| MBF8377854.1 | 3.2.1.139|3.2.1.131 | α-glucuronidase | GH67 |
| MBF8378555.1 | 3.2.1.78|3.2.1.25 | 1,4-β-xylanase | GH113 |
| MBF8376699.1 | 3.2.1.185 | glycoside hydrolase family 127 protein | GH127 |
| MBF8377082.1 | 2.4.1.339 | glycoside hydrolase family 130 protein | GH130 |
| MBF8376362.1 | - | polysaccharide deacetylase family sporulation protein PdaB | CE4 |
| MBF8376532.1 | - | polysaccharide deacetylase family protein | CE4 |
| MBF8377192.1 | - | polysaccharide deacetylase family protein | CE4 |
| MBF8377262.1 | - | polysaccharide deacetylase family protein | CE4 |
| MBF8377649.1 | - | polysaccharide deacetylase family protein | CE4 |
| MBF8378334.1 | - | polysaccharide deacetylase family protein | CE4 |
| MBF8377707.1 | - | N-acetylglucosamine-6-phosphate deacetylase | CE9 |
| MBF8376962.1 | - | PIG-L family deacetylase | CE14 |
| MBF8377409.1 | 3.5.1.- | PIG-L family deacetylase | CE14 |
| MBF8377724.1 | - | bacillithiol biosynthesis deacetylase BshB2 | CE14 |
| Pectin | |||
| MBF8377980.1 | - | glycoside hydrolase family 28 protein | GH28 |
| MBF8377281.1 | 3.2.1.23 | β-galactosidase | GH42 |
| MBF8377278.1 | 3.2.1.55 | α-N-arabinofuranosidase | GH51 |
| MBF8376699.1 | 3.2.1.185 | glycoside hydrolase family 127 protein | GH127 |
| Lignin | |||
| MBF8377978.1 | 1.10.3.2 | multicopper oxidase domain-containing protein | AA1 |
| MBF8377540.1 | - | NAD(P)H:quinone oxidoreductase type IV | AA6 |
| Substrate | KM (mg mL−1) | kcat (s−1) | kcat/KM (mL mg−1 s−1) |
|---|---|---|---|
| CMC | 10.9 | 847.2 | 77.7 |
| Lichenan | 7.5 | 1626.5 | 216.9 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Carbonaro, M.; Aulitto, M.; Gallo, G.; Contursi, P.; Limauro, D.; Fiorentino, G. Insight into CAZymes of Alicyclobacillus mali FL18: Characterization of a New Multifunctional GH9 Enzyme. Int. J. Mol. Sci. 2023, 24, 243. https://doi.org/10.3390/ijms24010243
Carbonaro M, Aulitto M, Gallo G, Contursi P, Limauro D, Fiorentino G. Insight into CAZymes of Alicyclobacillus mali FL18: Characterization of a New Multifunctional GH9 Enzyme. International Journal of Molecular Sciences. 2023; 24(1):243. https://doi.org/10.3390/ijms24010243
Chicago/Turabian StyleCarbonaro, Miriam, Martina Aulitto, Giovanni Gallo, Patrizia Contursi, Danila Limauro, and Gabriella Fiorentino. 2023. "Insight into CAZymes of Alicyclobacillus mali FL18: Characterization of a New Multifunctional GH9 Enzyme" International Journal of Molecular Sciences 24, no. 1: 243. https://doi.org/10.3390/ijms24010243
APA StyleCarbonaro, M., Aulitto, M., Gallo, G., Contursi, P., Limauro, D., & Fiorentino, G. (2023). Insight into CAZymes of Alicyclobacillus mali FL18: Characterization of a New Multifunctional GH9 Enzyme. International Journal of Molecular Sciences, 24(1), 243. https://doi.org/10.3390/ijms24010243









