Production of Modified Nucleosides in a Continuous Enzyme Membrane Reactor
Abstract
1. Introduction
2. Results and Discussion
2.1. Setup of the EMR
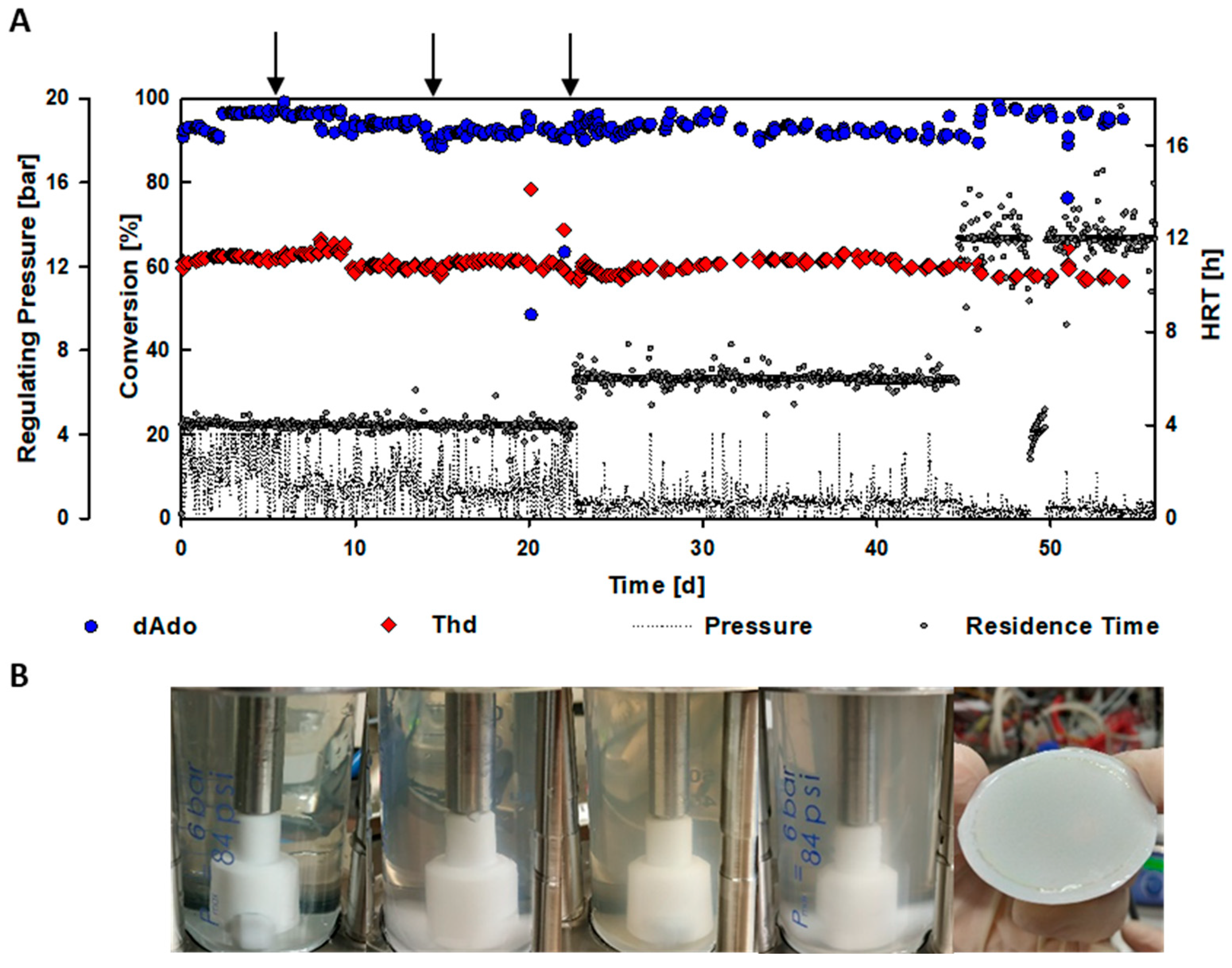
2.2. Validation of the Functionality of the EMR Using 2′-Deoxyadenosine
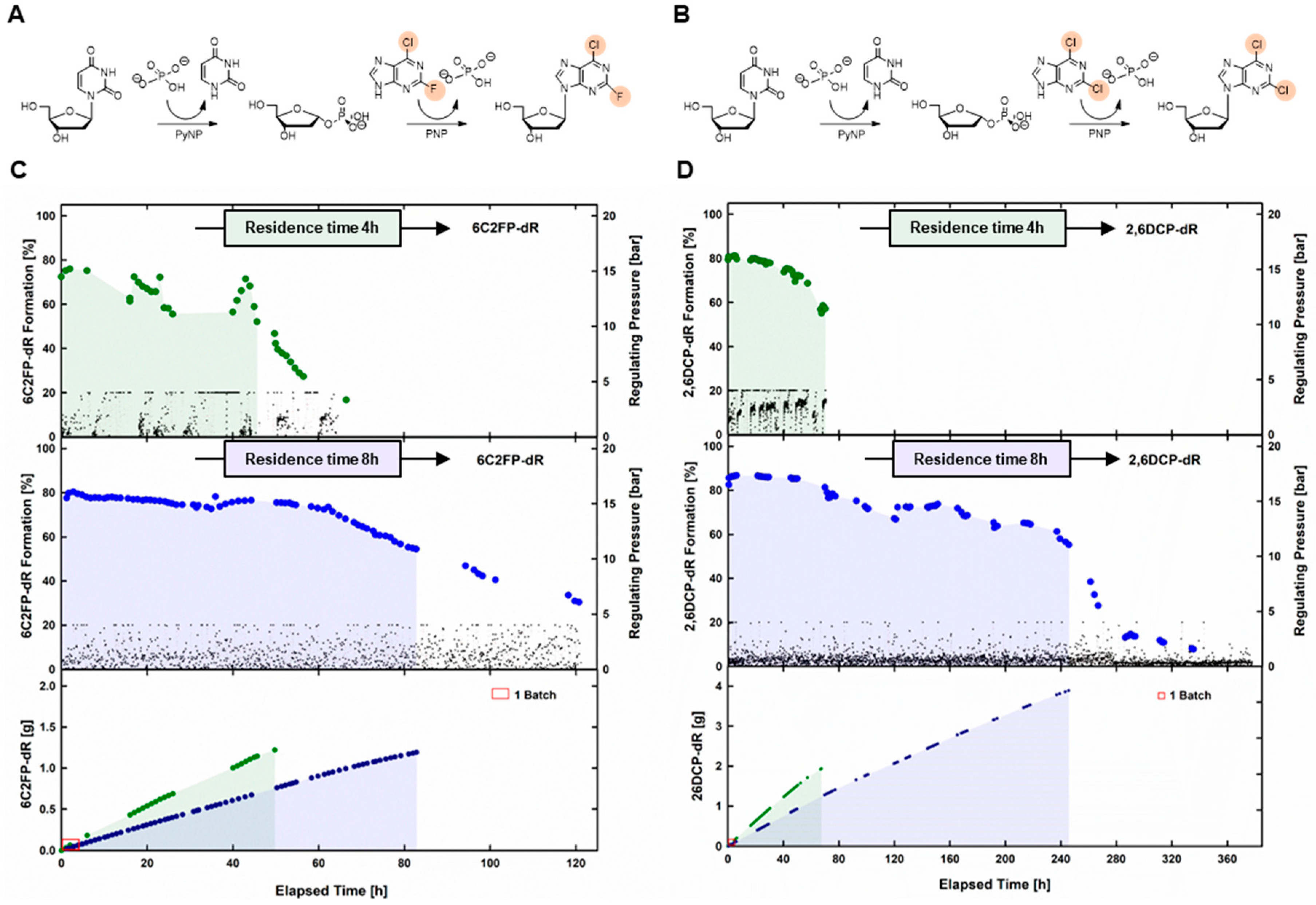
2.3. Synthesis of Dihalogenated Nucleoside by Nucleoside Phosphorylases
3. Materials and Methods
3.1. Reactor Configurations
3.2. Control Design
3.3. Chemicals
3.4. Synthesis of Natural and Modified Nucleosides in Batch Reactions
3.5. Operational Procedure to Produce Natural and Modified Nucleosides in Enzyme Membrane Reactors
3.6. Measurement of Enzyme Activity
3.7. Determination of Nucleosides and Nucleobases by HPLC
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
References
- Yehia, H.; Kamel, S.; Paulick, K.; Neubauer, P.; Wagner, A. Substrate spectra of nucleoside phosphorylases and their potential in the production of pharmaceutically active compounds. Curr. Pharm. Des. 2017, 23, 13–35. [Google Scholar] [CrossRef] [PubMed]
- Schultz, D.C.; Johnson, R.M.; Ayyanathan, K.; Miller, J.; Whig, K.; Kamalia, B.; Dittmar, M.; Weston, S.; Hammond, H.L.; Dillen, C.; et al. Pyrimidine inhibitors synergize with nucleoside analogues to block SARS-CoV-2. Nature 2022, 604, 134–140. [Google Scholar] [CrossRef]
- Scarabel, L.; Guardascione, M.; Dal Bo, M.; Toffoli, G. Pharmacological strategies to prevent SARS-CoV-2 infection and treat the early phases of COVID-19. Int. J. Infect. Dis. 2021, 104, 441–451. [Google Scholar] [CrossRef] [PubMed]
- Ashour, N.A.; Abo Elmaaty, A.; Sarhan, A.A.; Elkaeed, E.B.; Moussa, A.M.; Erfan, I.A.; Al-Karmalawy, A.A. A Systematic Review of the Global Intervention for SARS-CoV-2 Combating: From Drugs Repurposing to Molnupiravir Approval. Drug Des. Dev. Ther. 2022, 16, 685–715. [Google Scholar] [CrossRef] [PubMed]
- Mikhailopulo, I.A.; Miroshnikov, A.I. New Trends in Nucleoside Biotechnology. Acta Nat. 2010, 2, 36–58. [Google Scholar] [CrossRef]
- Mikhailopulo, I.A.; Miroshnikov, A.I. Some recent findings in the biotechnology of biologically important nucleosides. In Biochemistry and Biotechnology for Modern Medicine; Komisarenko, S., Ed.; Publishing House Moskalenko O.M.: Kiev, Ukraine, 2013; pp. 328–353. [Google Scholar]
- Ashcroft, C.P.; Dessi, Y.; Entwistle, D.A.; Hesmondhalgh, L.C.; Longstaff, A.; Smith, J.D. Route selection and process development of a multikilogram route to the inhaled A 2a agonist UK-432,097. Org. Process Res. Dev. 2012, 16, 470–483. [Google Scholar] [CrossRef]
- Nguyen-Trung, N.Q.; Botta, O.; Terenzi, S.; Strazewski, P. A Practical Route to 3‘-Amino-3‘-deoxyadenosine Derivatives and Puromycin Analogues. J. Org. Chem. 2003, 68, 2038–2041. [Google Scholar] [CrossRef]
- Divakar, K.J.; Sawant, C.M.; Mulla, Y.A.; Zemse, D.V.; Sitabkhan, S.M.; Ross, B.S.; Sanghvi, Y.S. Commercial-scale synthesis of protected 2′-deoxycytidine and cytidine nucleosides. Nucleosides Nucleotides Nucleic Acids 2003, 22, 1321–1325. [Google Scholar] [CrossRef]
- Mackman, R.L. Anti-HIV nucleoside phosphonate GS-9148 and its prodrug GS-9131: Scale up of a 2′-F modified cyclic nucleoside phosphonate and synthesis of selected amidate prodrugs. Curr. Protoc. Nucleic Acid Chem. 2014, 56, 14.10.1–14.10.21. [Google Scholar] [CrossRef]
- Kaspar, F.; Stone MR, L.; Neubauer, P.; Kurreck, A. Route efficiency assessment and review of the synthesis of β-nucleosides: Via N -glycosylation of nucleobases. Green Chem. 2021, 23, 37–50. [Google Scholar] [CrossRef]
- Del Arco, J.; Fernández-Lucas, J. Purine and pyrimidine salvage pathway in thermophiles: A valuable source of biocatalysts for the industrial production of nucleic acid derivatives. Appl. Microbiol. Biotechnol. 2018, 102, 7805–7820. [Google Scholar] [CrossRef] [PubMed]
- Zhou, X.; Szeker, K.; Jiao, L.Y.; Oestreich, M.; Mikhailopulo, I.A.; Neubauer, P. Synthesis of 2,6-dihalogenated purine nucleosides by thermostable nucleoside phosphorylases. Adv. Synth. Catal. 2015, 357, 1237–1244. [Google Scholar] [CrossRef]
- Kamel, S.; Thiele, I.; Neubauer, P.; Wagner, A. Thermophilic nucleoside phosphorylases: Their properties, characteristics and applications. Biochim. Biophys. Acta Proteins Proteom. 2020, 1868, 140304. [Google Scholar] [CrossRef] [PubMed]
- Zhou, X.; Mikhailopulo, I.A.; Cruz Bournazou, M.N.; Neubauer, P. Immobilization of thermostable nucleoside phosphorylases on MagReSyn® epoxide microspheres and their application for the synthesis of 2,6-dihalogenated purine nucleosides. J. Mol. Catal. B Enzym. 2015, 115, 119–127. [Google Scholar] [CrossRef]
- Okuma, H.; Watanabe, E. Flow system for fish freshness determination based on double multi-enzyme reactor electrodes. Biosens. Bioelectron. 2002, 17, 367–372. [Google Scholar] [CrossRef]
- Carsol, M.A.; Mascini, M. Development of a system with enzyme reactors for the determination of fish freshness. Talanta 1998, 47, 335–342. [Google Scholar] [CrossRef]
- Calleri, E.; Ubiali, D.; Serra, I.; Temporini, C.; Cattaneo, G.; Speranza, G.; Morelli, C.F.; Massolini, G. Immobilized purine nucleoside phosphorylase from Aeromonas hydrophila as an on-line enzyme reactor for biocatalytic applications. J. Chromatogr. B 2014, 968, 79–86. [Google Scholar] [CrossRef]
- Hori, N.; Watanabe, M.; Sunagawa, K.; Uehara, K.; Mikami, Y. Production of 5-methyluridine by immobilized thermostable purine nucleoside phosphorylase and pyrimidine nucleoside phosphorylase from Bacillus stearothermophilus JTS 859. J. Biotechnol. 1991, 17, 121–131. [Google Scholar] [CrossRef]
- Calleri, E.; Cattaneo, G.; Rabuffetti, M.; Serra, I.; Bavaro, T.; Massolini, G.; Speranza, G.; Ubiali, D. Flow-Synthesis of Nucleosides Catalyzed by an Immobilized Purine Nucleoside Phosphorylase from Aeromonas hydrophila: Integrated Systems of Reaction Control and Product Purification. Adv. Synth. Catal. 2015, 357, 2520–2528. [Google Scholar] [CrossRef]
- Cowan, D.A.; Fernandez-Lafuente, R. Enhancing the functional properties of thermophilic enzymes by chemical modification and immobilization. Enzym. Microb. Technol. 2011, 49, 326–346. [Google Scholar] [CrossRef]
- Rinaldi, F.; Fernández-Lucas, J.; de la Fuente, D.; Zheng, C.; Bavaro, T.; Peters, B.; Massolini, G.; Annunziata, F.; Conti, P.; de la Mata, I.; et al. Immobilized enzyme reactors based on nucleoside phosphorylases and 2′-deoxyribosyltransferase for the in-flow synthesis of pharmaceutically relevant nucleoside analogues. Bioresour. Technol. 2020, 307, 123258. [Google Scholar] [CrossRef] [PubMed]
- Tamborini, L.; Previtali, C.; Annunziata, F.; Bavaro, T.; Terreni, M.; Calleri, E.; Rinaldi, F.; Pinto, A.; Speranza, G.; Ubiali, D.; et al. An Enzymatic Flow-Based Preparative Route to Vidarabine. Molecules 2020, 25, 1223. [Google Scholar] [CrossRef] [PubMed]
- Benítez-Mateos, A.I.; Paradisi, F. Sustainable Flow-Synthesis of (Bulky) Nucleoside Drugs by a Novel and Highly Stable Nucleoside Phosphorylase Immobilized on Reusable Supports. ChemSusChem 2022, 15, e202102030. [Google Scholar] [CrossRef] [PubMed]
- Visser, D.F.; Hennessy, F.; Rashamuse, J.; Pletschke, B.; Brady, D. Stabilization of Escherichia coli uridine phosphorylase by evolution and immobilization. J. Mol. Catal. B Enzym. 2011, 68, 279–285. [Google Scholar] [CrossRef]
- Kim, S.; Jiménez-González, C.; Dale, B.E. Enzymes for pharmaceutical applications-a cradle-to-gate life cycle assessment. Int. J. Life Cycle Assess. 2009, 14, 392–400. [Google Scholar] [CrossRef]
- Sitanggang, A.B.; Drews, A.; Kraume, M. Continuous synthesis of lactulose in an enzymatic membrane reactor reduces lactulose secondary hydrolysis. Bioresour. Technol. 2014, 167, 108–115. [Google Scholar] [CrossRef]
- Cao, T.; Pázmándi, M.; Galambos, I.; Kovács, Z. Continuous production of galacto-oligosaccharides by an enzyme membrane reactor utilizing free enzymes. Membranes 2020, 10, 203. [Google Scholar] [CrossRef]
- Rios, G.M.; Belleville, M.P.; Paolucci, D.; Sanchez, J. Progress in enzymatic membrane reactors—A review. J. Membr. Sci. 2004, 242, 189–196. [Google Scholar] [CrossRef]
- Wöltinger, J.; Karau, A.; Leuchtenberger, W.; Drauz, K. Membrane reactors at Degussa. Adv. Biochem. Eng. Biotechnol. 2005, 92, 289–316. [Google Scholar] [CrossRef]
- Sitanggang, A.B.; Drews, A.; Kraume, M. Development of a continuous membrane reactor process for enzyme-catalyzed lactulose synthesis. Biochem. Eng. J. 2016, 109, 65–80. [Google Scholar] [CrossRef]
- Birkholz, M.; Mai, A.; Wenger, C.; Meliani, C.; Scholz, R. Technology modules from micro- and nano-electronics for the life sciences. WIREs Nanomed. Nanobiotechnology 2016, 8, 355–377. [Google Scholar] [CrossRef] [PubMed]
- Zhou, X.; Szeker, K.; Janocha, B.; Böhme, T.; Albrecht, D.; Mikhailopulo, I.A.; Neubauer, P. Recombinant purine nucleoside phosphorylases from thermophiles: Preparation, properties and activity towards purine and pyrimidine nucleosides. FEBS J. 2013, 280, 1475–1490. [Google Scholar] [CrossRef] [PubMed]
- Yuryev, R.; Strompen, S.; Liese, A. Coupled chemo(enzymatic) reactions in continuous flow. Beilstein J. Org. Chem. 2011, 7, 1449–1467. [Google Scholar] [CrossRef] [PubMed]
- Almendros, M.; Gago JV, S.; Carlos, J.B. Thermus thermophilus strains active in purine nucleoside synthesis. Molecules 2009, 14, 1279–1287. [Google Scholar] [CrossRef] [PubMed]
- Padua, R.; Geiger, J.D.; Dambock, S.; Nagy, J.I. 2′-Deoxycoformycin Inhibition of Adenosine Deaminase in Rat Brain: In Vivo and In Vitro Analysis of Specificity, Potency, and Enzyme Recovery. J. Neurochem. 1990, 54, 1169–1178. [Google Scholar] [CrossRef] [PubMed]
- Bzowska, A.; Kulikowska, E.; Shugar, D. Purine nucleoside phosphorylases: Properties, functions, and clinical aspects. Pharmacol. Ther. 2000, 88, 349–425. [Google Scholar] [CrossRef] [PubMed]
- Yehia, H.; Westarp, S.; Röhrs, V.; Kaspar, F.; Giessmann, R.T.; Klare HF, T.; Paulick, K.; Neubauer, P.; Wagner, A. Efficient Biocatalytic Synthesis of Dihalogenated Thermodynamic Calculations. Molecules 2020, 25, 934. [Google Scholar] [CrossRef]
- Anand, R.; Kaminski, P.A.; Ealick, S.E. Structures of Purine 2′-Deoxyribosyltransferase, Substrate Complexes, and the Ribosylated Enzyme Intermediate at 2.0 Å Resolution. Biochemistry 2004, 43, 2384–2393. [Google Scholar] [CrossRef]
- Bonate, P.L.; Arthaud, L.; Cantrell, W.R.; Stephenson, K.; Secrist, J.A.; Weitman, S. Discovery and development of clofarabine: A nucleoside analogue for treating cancer. Nat. Rev. Drug Discov. 2006, 5, 855–863. [Google Scholar] [CrossRef]
- Avramis, V.I.; Plunkett, W. 2-Fluoro-ATP: A toxic metabolite of 9-β-D-arabinosyl-2-fluoroadenine. Biochem. Biophys. Res. Commun. 1983, 113, 35–43. [Google Scholar] [CrossRef]
- Skipper, H.E.; Montgomery, J.A.; Thomson, J.R.; Schabel FM, J. Structure-activity relationships and cross-resistance observed on evaluation of a series of purine analogs against experimental neoplasms. Cancer Res. 1959, 19, 425–437. [Google Scholar] [PubMed]
- Bennett, L.L.; Vail, M.H.; Chumley, S.; Montgomery, J.A. Activity of adenosine analogs against a cell culture, line resistant to 2-fluoroadenine. Biochem. Pharmacol. 1966, 15, 1719–1728. [Google Scholar] [CrossRef]
- de Giuseppe, P.O.; Martins, N.H.; Meza, A.N.; dos Santos, C.R.; Pereira HD, M.; Murakami, M.T. Insights into Phosphate Cooperativity and Influence of Substrate Modifications on Binding and Catalysis of Hexameric Purine Nucleoside Phosphorylases. PLoS ONE 2012, 7, e44282. [Google Scholar] [CrossRef] [PubMed]
- Liu, P.; Sharon, A.; Chu, C.K. Fluorinated nucleosides: Synthesis and biological implication. J. Fluor. Chem. 2008, 129, 743–766. [Google Scholar] [CrossRef] [PubMed]


| Enzymes | Product | Productivity [g L−1 h−1] | Specific Productivity [mg U−1 h−1] | Product Per Enzymatic Unit [mg U−1] | Reaction System | Reference |
|---|---|---|---|---|---|---|
| PyNP 02/PNP 02 | dAdo | 2.49 | 0.22 | 1.32 | Batch | This study |
| PyNP 02/PNP 02 | dAdo | 2.5 | 0.22 | 290.4 * | EMR | This study |
| PyNP 02/PNP 02 | 2,6DCP-dR | 1.6 | 0.14 | 0.84 | Batch | This study |
| PyNP 02/PNP 02 | 6C2FP-dR | 1.4 | 0.12 | 0.72 | Batch | This study |
| PyNP 02/PNP 02 | 2,6DCP-dR | 1.5 1.6 | 0.13 0.14 | 7.8 (HRT 4 h) 33.6 (HRT 8 h/16 h) | EMR | This study |
| PyNP 02/PNP 02 | 6C2FP-dR | 1.3 1.3 | 0.11 0.11 | 5.5 (HRT 4 h) 6.49 (HRT 8 h) | EMR | This study |
| TtPyNP a/GtPNP b | 2,6DCP-R | 1.5 | - | - | Batch, immob. enzyme | [13] |
| TtPyNP/GtPNP | 6C2FP-R | 2.0 | - | - | Batch, immob. enzyme | [13] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Thiele, I.; Yehia, H.; Krausch, N.; Birkholz, M.; Cruz Bournazou, M.N.; Sitanggang, A.B.; Kraume, M.; Neubauer, P.; Kurreck, A. Production of Modified Nucleosides in a Continuous Enzyme Membrane Reactor. Int. J. Mol. Sci. 2023, 24, 6081. https://doi.org/10.3390/ijms24076081
Thiele I, Yehia H, Krausch N, Birkholz M, Cruz Bournazou MN, Sitanggang AB, Kraume M, Neubauer P, Kurreck A. Production of Modified Nucleosides in a Continuous Enzyme Membrane Reactor. International Journal of Molecular Sciences. 2023; 24(7):6081. https://doi.org/10.3390/ijms24076081
Chicago/Turabian StyleThiele, Isabel, Heba Yehia, Niels Krausch, Mario Birkholz, Mariano Nicolas Cruz Bournazou, Azis Boing Sitanggang, Matthias Kraume, Peter Neubauer, and Anke Kurreck. 2023. "Production of Modified Nucleosides in a Continuous Enzyme Membrane Reactor" International Journal of Molecular Sciences 24, no. 7: 6081. https://doi.org/10.3390/ijms24076081
APA StyleThiele, I., Yehia, H., Krausch, N., Birkholz, M., Cruz Bournazou, M. N., Sitanggang, A. B., Kraume, M., Neubauer, P., & Kurreck, A. (2023). Production of Modified Nucleosides in a Continuous Enzyme Membrane Reactor. International Journal of Molecular Sciences, 24(7), 6081. https://doi.org/10.3390/ijms24076081










