Mapping miRNA Research in Schizophrenia: A Scientometric Review
Abstract
:1. Introduction
2. Materials and Methods
2.1. Data Import into CiteSpace
2.2. Document Co-Citation Analysis (DCA)
2.3. DCA Network Evaluation Metrics
3. Results
3.1. Bibliometric Analysis on the Citing References
3.2. Document Co-Citation Analysis
4. Discussion
4.1. Cluster #9: Expanding on Central Dogma
4.2. Cluster #12: Comprehensive Mammalian Non-Coding RNA Database
4.3. Cluster #14: Pioneering Study
4.4. Cluster #7: Identifying Biomarkers of Schizophrenia
4.5. Cluster #8: miRNA and the Brain
4.6. Cluster #0: miRNA and Neurological Disorders
4.7. Cluster #10: Epigenetic Dysregulation
4.8. Cluster #3: Transcriptional Effects of miRNAs
4.9. Cluster #6: Deletion Syndrome
4.10. Cluster #1: miRNA-137 and Schizophrenia
4.11. Cluster #4: Therapeutic Potential
4.12. Cluster #5: Circular RNA
4.13. Cluster #2: Bioinformatics Analysis
4.14. Study Evaluation
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| miRNA | Micro Ribonucleic Acid |
| DCA | Document Co-Citation Analysis |
| WoS | Web of Science |
| PRISMA | Preferred Reporting Items for Systematic Reviews and Meta-Analyses |
| GWAS | Genome-Wide Association Study |
| SCP | Single Country Publications |
| MCP | Multiple Country Publications |
| LLR | Log-Likelihood Ratio |
| GCS | Global Citing Score |
| mRNA | Messenger RNA |
| PFC | Prefrontal Cortex |
| eQTL | Gene Expression Quantitative Trait Loci |
| DGCR8 | DiGeorge Syndrome Critical Region Gene 8 |
| NMDA | N-Methyl-D-Aspartate |
| iPSC | Induced Pluripotent Stem Cells |
References
- Lee, R.C.; Feinbaum, R.L.; Ambros, V. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell 1993, 75, 843–854. [Google Scholar] [CrossRef]
- Pasquinelli, A.E.; Reinhart, B.J.; Slack, F.; Martindale, M.Q.; Kuroda, M.I.; Maller, B.; Hayward, D.C.; Ball, E.E.; Degnan, B.; Müller, P.; et al. Conservation of the sequence and temporal expression of let-7 heterochronic regulatory RNA. Nature 2000, 408, 86–89. [Google Scholar] [CrossRef] [PubMed]
- Hammond, S.M. An overview of microRNAs. Adv. Drug Deliv. Rev. 2015, 87, 3–14. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sullivan, P.F.; Kendler, K.S.; Neale, M.C. Schizophrenia as a complex trait: Evidence from a meta-analysis of twin studies. Arch. Gen. Psychiatry 2003, 60, 1187–1192. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Beveridge, N.J.; Cairns, M.J. MicroRNA dysregulation in schizophrenia. Neurobiol. Dis. 2012, 46, 263–271. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cao, X.; Yeo, G.; Muotri, A.R.; Kuwabara, T.; Gage, F.H. Noncoding RNAs in the mammalian central nervous system. Annu. Rev. Neurosci. 2006, 29, 77–103. [Google Scholar] [CrossRef] [Green Version]
- Perkins, D.O.; Jeffries, C.D.; Jarskog, L.F.; Thomson, J.M.; Woods, K.; Newman, M.A.; Parker, J.S.; Jin, J.; Hammond, S.M. microRNA expression in the prefrontal cortex of individuals with schizophrenia and schizoaffective disorder. Genome Biol. 2007, 8, 1–11. [Google Scholar] [CrossRef] [Green Version]
- Siegert, S.; Seo, J.; Kwon, E.J.; Rudenko, A.; Cho, S.; Wang, W.; Flood, Z.; Martorell, A.J.; Ericsson, M.; Mungenast, A.E.; et al. The schizophrenia risk gene product miR-137 alters presynaptic plasticity. Nat. Neurosci. 2015, 18, 1008–1016. [Google Scholar] [CrossRef] [Green Version]
- Pantelis, C.; Papadimitriou, G.N.; Papiol, S.; Parkhomenko, E.; Pato, M.T.; Paunio, T.; Pejovic-Milovancevic, M.; Perkins, D.O.; Pietiläinen, O.; Pimm, J. Biological insights from 108 schizophrenia-associated genetic loci. Nature 2014, 511, 421–427. [Google Scholar]
- Liu, S.; Zhang, F.; Wang, X.; Shugart, Y.Y.; Zhao, Y.; Li, X.; Liu, Z.; Sun, N.; Yang, C.; Zhang, K.; et al. Diagnostic value of blood-derived microRNAs for schizophrenia: Results of a meta-analysis and validation. Sci. Rep. 2017, 7, 15328. [Google Scholar] [CrossRef] [Green Version]
- Jahan, N.; Naveed, S.; Zeshan, M.; Tahir, M.A. How to conduct a systematic review: A narrative literature review. Cureus 2016, 8, e864. [Google Scholar] [CrossRef] [PubMed]
- Mulchenko, Z. Measurement of Science. Study of the Development of Science as an Information Process; USAF Foreign Technology Division Translation AD735634; Defense Technical Information Center: Fort Belvoir, VA, USA, 1969. [Google Scholar]
- Börner, K.; Chen, C.; Boyack, K.W. Visualizing knowledge domains. Annu. Rev. Inf. Sci. Technol. 2003, 37, 179–255. [Google Scholar] [CrossRef]
- Junior, O.F. The Legacy of David Bohm in Physics—An Essay in Scientometry. In David Bohm; Springer: Cham, Switzerland, 2019; pp. 223–240. [Google Scholar]
- Falagas, M.E.; Pitsouni, E.I.; Malietzis, G.A.; Pappas, G. Comparison of PubMed, Scopus, web of science, and Google scholar: strengths and weaknesses. FASEB J. 2008, 22, 338–342. [Google Scholar] [CrossRef]
- Cataldo, I.; Lieu, A.A.; Carollo, A.; Bornstein, M.H.; Gabrieli, G.; Lee, A.; Esposito, G. From the cradle to the web: The growth of “sharenting”—A scientometric perspective. Hum. Behav. Emerg. Technol. 2022, 2022, 5607422. [Google Scholar] [CrossRef]
- Aria, M.; Cuccurullo, C. bibliometrix: An R-tool for comprehensive science mapping analysis. J. Inf. 2017, 11, 959–975. [Google Scholar] [CrossRef]
- Chen, C. The citespace manual. Coll. Comput. Inform. 2014, 1, 1–84. [Google Scholar]
- Lim, M.; Carollo, A.; Dimitriou, D.; Esposito, G. Recent Developments in Autism Genetic Research: A Scientometric Review from 2018 to 2022. Genes 2022, 13, 1646. [Google Scholar] [CrossRef]
- Chen, C. CiteSpace: A Practical Guide for Mapping Scientific Literature; Nova Science Publishers: Hauppauge, NY, USA, 2016. [Google Scholar]
- Small, H. Co-citation context analysis and the structure of paradigms. J. Doc. 1980, 36, 183–196. [Google Scholar] [CrossRef]
- Chen, C.; Ibekwe-SanJuan, F.; Hou, J. The structure and dynamics of cocitation clusters: A multiple-perspective cocitation analysis. J. Am. Soc. Inf. Sci. Technol. 2010, 61, 1386–1409. [Google Scholar] [CrossRef] [Green Version]
- Egghe, L. An improvement of the h-index: The g-index. ISSI Newsl. 2006, 2, 8–9. [Google Scholar]
- Bornmann, L.; Daniel, H.D. What do we know about the h index? J. Am. Soc. Inf. Sci. Technol. 2007, 58, 1381–1385. [Google Scholar] [CrossRef]
- Alonso, S.; Cabrerizo, F.J.; Herrera-Viedma, E.; Herrera, F. h-Index: A review focused in its variants, computation and standardization for different scientific fields. J. Inf. 2009, 3, 273–289. [Google Scholar] [CrossRef] [Green Version]
- Newman, M.E. Modularity and community structure in networks. Proc. Natl. Acad. Sci. USA 2006, 103, 8577–8582. [Google Scholar] [CrossRef] [PubMed]
- Rousseeuw, P.J. Silhouettes: A graphical aid to the interpretation and validation of cluster analysis. J. Comput. Appl. Math. 1987, 20, 53–65. [Google Scholar] [CrossRef] [Green Version]
- Aryadoust, V.; Ang, B.H. Exploring the frontiers of eye tracking research in language studies: A novel co-citation scientometric review. Comput. Assist. Lang. Learn. 2019, 34, 898–933. [Google Scholar] [CrossRef] [Green Version]
- Freeman, L.C. A set of measures of centrality based on betweenness. Sociometry 1977, 40, 35–41. [Google Scholar] [CrossRef]
- Kleinberg, J. Bursty and hierarchical structure in streams. Data Min. Knowl. Discov. 2003, 7, 373–397. [Google Scholar] [CrossRef]
- Chen, C. Science mapping: A systematic review of the literature. J. Data Inf. Sci. 2017, 2, 1–40. [Google Scholar] [CrossRef] [Green Version]
- The Schizophrenia Psychiatric Genome-Wide Association Study (GWAS) Consortium. Genome-wide association study identifies five new schizophrenia loci. Nat. Genet. 2011, 43, 969–976. [Google Scholar] [CrossRef]
- Ingelman-Sundberg, M.; Sim, S.C.; Gomez, A.; Rodriguez-Antona, C. Influence of cytochrome P450 polymorphisms on drug therapies: pharmacogenetic, pharmacoepigenetic and clinical aspects. Pharmacol. Ther. 2007, 116, 496–526. [Google Scholar] [CrossRef]
- Chen, C. CiteSpace II: Detecting and visualizing emerging trends and transient patterns in scientific literature. J. Am. Soc. Inf. Sci. Technol. 2006, 57, 359–377. [Google Scholar] [CrossRef] [Green Version]
- Carollo, A.; Lim, M.; Aryadoust, V.; Esposito, G. Interpersonal Synchrony in the Context of Caregiver-Child Interactions: A Document Co-citation Analysis. Front. Psychol. 2021, 12, 2977. [Google Scholar] [CrossRef] [PubMed]
- Gaggero, G.; Bonassi, A.; Dellantonio, S.; Pastore, L.; Aryadoust, V.; Esposito, G. A scientometric review of alexithymia: Mapping thematic and disciplinary shifts in half a century of research. Front. Psychiatry 2020, 11, 1405. [Google Scholar] [CrossRef] [PubMed]
- Carollo, A.; Balagtas, J.P.M.; Neoh, M.J.Y.; Esposito, G. A scientometric approach to review the role of the medial preoptic area (MPOA) in parental behavior. Brain Sci. 2021, 11, 393. [Google Scholar] [CrossRef] [PubMed]
- Bartel, D.P. MicroRNAs: Genomics, biogenesis, mechanism, and function. Cell 2004, 116, 281–297. [Google Scholar] [CrossRef] [Green Version]
- Filipowicz, W.; Bhattacharyya, S.N.; Sonenberg, N. Mechanisms of post-transcriptional regulation by microRNAs: Are the answers in sight? Nat. Rev. Genet. 2008, 9, 102–114. [Google Scholar] [CrossRef]
- Abelson, J.F.; Kwan, K.Y.; O’Roak, B.J.; Baek, D.Y.; Stillman, A.A.; Morgan, T.M.; Mathews, C.A.; Pauls, D.L.; Rasin, M.R.; Gunel, M.; et al. Sequence variants in SLITRK1 are associated with Tourette’s syndrome. Science 2005, 310, 317–320. [Google Scholar] [CrossRef] [Green Version]
- Ripke, S.; O’dushlaine, C.; Chambert, K.; Moran, J.L.; Kähler, A.K.; Akterin, S.; Bergen, S.E.; Collins, A.L.; Crowley, J.J.; Fromer, M.; et al. Genome-wide association analysis identifies 13 new risk loci for schizophrenia. Nat. Genet. 2013, 45, 1150–1159. [Google Scholar] [CrossRef]
- Schratt, G.M.; Tuebing, F.; Nigh, E.A.; Kane, C.G.; Sabatini, M.E.; Kiebler, M.; Greenberg, M.E. A brain-specific microRNA regulates dendritic spine development. Nature 2006, 439, 283–289. [Google Scholar] [CrossRef]
- Wright, C.; Turner, J.A.; Calhoun, V.D.; Perrone-Bizzozero, N. Potential impact of miR-137 and its targets in schizophrenia. Front. Genet. 2013, 4, 58. [Google Scholar] [CrossRef] [Green Version]
- Lewis, B.P.; Burge, C.B.; Bartel, D.P. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell 2005, 120, 15–20. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Barry, G.; Briggs, J.; Vanichkina, D.; Poth, E.; Beveridge, N.; Ratnu, V.; Nayler, S.; Nones, K.; Hu, J.; Bredy, T.; et al. The long non-coding RNA Gomafu is acutely regulated in response to neuronal activation and involved in schizophrenia-associated alternative splicing. Mol. Psychiatry 2014, 19, 486–494. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shi, W.; Du, J.; Qi, Y.; Liang, G.; Wang, T.; Li, S.; Xie, S.; Zeshan, B.; Xiao, Z. Aberrant expression of serum miRNAs in schizophrenia. J. Psychiatr. Res. 2012, 46, 198–204. [Google Scholar] [CrossRef] [PubMed]
- Hansen, T.; Olsen, L.; Lindow, M.; Jakobsen, K.D.; Ullum, H.; Jonsson, E.; Andreassen, O.A.; Djurovic, S.; Melle, I.; Agartz, I.; et al. Brain expressed microRNAs implicated in schizophrenia etiology. PLoS ONE 2007, 2, e873. [Google Scholar] [CrossRef] [PubMed]
- Sun, G.; Ye, P.; Murai, K.; Lang, M.F.; Li, S.; Zhang, H.; Li, W.; Fu, C.; Yin, J.; Wang, A.; et al. miR-137 forms a regulatory loop with nuclear receptor TLX and LSD1 in neural stem cells. Nat. Commun. 2011, 2, 1–10. [Google Scholar] [CrossRef] [Green Version]
- Kim, J.; Inoue, K.; Ishii, J.; Vanti, W.B.; Voronov, S.V.; Murchison, E.; Hannon, G.; Abeliovich, A. A MicroRNA feedback circuit in midbrain dopamine neurons. Science 2007, 317, 1220–1224. [Google Scholar] [CrossRef] [Green Version]
- Gardiner, E.; Beveridge, N.; Wu, J.; Carr, V.; Scott, R.; Tooney, P.; Cairns, M. Imprinted DLK1-DIO3 region of 14q32 defines a schizophrenia-associated miRNA signature in peripheral blood mononuclear cells. Mol. Psychiatry 2012, 17, 827–840. [Google Scholar] [CrossRef] [Green Version]
- Zhou, R.; Yuan, P.; Wang, Y.; Hunsberger, J.G.; Elkahloun, A.; Wei, Y.; Damschroder-Williams, P.; Du, J.; Chen, G.; Manji, H.K. Evidence for selective microRNAs and their effectors as common long-term targets for the actions of mood stabilizers. Neuropsychopharmacology 2009, 34, 1395–1405. [Google Scholar] [CrossRef] [Green Version]
- Guan, F.; Zhang, B.; Yan, T.; Li, L.; Liu, F.; Li, T.; Feng, Z.; Zhang, B.; Liu, X.; Li, S. MIR137 gene and target gene CACNA1C of miR-137 contribute to schizophrenia susceptibility in Han Chinese. Schizophr. Res. 2014, 152, 97–104. [Google Scholar] [CrossRef]
- Perkins, D.O.; Jeffries, C.; Sullivan, P. Expanding the ‘central dogma’: The regulatory role of nonprotein coding genes and implications for the genetic liability to schizophrenia. Mol. Psychiatry 2005, 10, 69–78. [Google Scholar] [CrossRef]
- Miller, B.H.; Wahlestedt, C. MicroRNA dysregulation in psychiatric disease. Brain Res. 2010, 1338, 89–99. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Turner, J.D.; Alt, S.R.; Cao, L.; Vernocchi, S.; Trifonova, S.; Battello, N.; Muller, C.P. Transcriptional control of the glucocorticoid receptor: CpG islands, epigenetics and more. Biochem. Pharmacol. 2010, 80, 1860–1868. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pang, K.C.; Stephen, S.; Engström, P.G.; Tajul-Arifin, K.; Chen, W.; Wahlestedt, C.; Lenhard, B.; Hayashizaki, Y.; Mattick, J.S. RNAdb—A comprehensive mammalian noncoding RNA database. Nucleic Acids Res. 2005, 33, D125–D130. [Google Scholar] [CrossRef] [PubMed]
- Tiwari, K.; Kananathan, S.; Roberts, M.G.; Meyer, J.P.; Sharif Shohan, M.U.; Xavier, A.; Maire, M.; Zyoud, A.; Men, J.; Ng, S.; et al. Reproducibility in systems biology modelling. Mol. Syst. Biol. 2021, 17, e9982. [Google Scholar] [CrossRef]
- Harrison, P.J.; Weinberger, D.R. Schizophrenia genes, gene expression, and neuropathology: On the matter of their convergence. Mol. Psychiatry 2005, 10, 40–68. [Google Scholar] [CrossRef] [Green Version]
- Straub, R.E.; Weinberger, D.R. Schizophrenia genes-famine to feast. Biol. Psychiatry 2006, 60, 81–83. [Google Scholar] [CrossRef]
- Tabares-Seisdedos, R.; Rubenstein, J. Chromosome 8p as a potential hub for developmental neuropsychiatric disorders: Implications for schizophrenia, autism and cancer. Mol. Psychiatry 2009, 14, 563–589. [Google Scholar] [CrossRef]
- Lai, C.Y.; Yu, S.L.; Hsieh, M.H.; Chen, C.H.; Chen, H.Y.; Wen, C.C.; Huang, Y.H.; Hsiao, P.C.; Hsiao, C.K.; Liu, C.M.; et al. MicroRNA expression aberration as potential peripheral blood biomarkers for schizophrenia. PLoS ONE 2011, 6, e21635. [Google Scholar] [CrossRef] [Green Version]
- Woelk, C.H.; Singhania, A.; Pérez-Santiago, J.; Glatt, S.J.; Tsuang, M.T. The utility of gene expression in blood cells for diagnosing neuropsychiatric disorders. Int. Rev. Neurobiol. 2011, 101, 41–63. [Google Scholar]
- Vawter, M.P.; Mamdani, F.; Macciardi, F. An integrative functional genomics approach for discovering biomarkers in schizophrenia. Briefings Funct. Genom. 2011, 10, 387–399. [Google Scholar] [CrossRef] [Green Version]
- Glatt, S.J.; Everall, I.P.; Kremen, W.S.; Corbeil, J.; Šášik, R.; Khanlou, N.; Han, M.; Liew, C.C.; Tsuang, M.T. Comparative gene expression analysis of blood and brain provides concurrent validation of SELENBP1 up-regulation in schizophrenia. Proc. Natl. Acad. Sci. USA 2005, 102, 15533–15538. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kuzman, M.R.; Medved, V.; Terzic, J.; Krainc, D. Genome-wide expression analysis of peripheral blood identifies candidate biomarkers for schizophrenia. J. Psychiatr. Res. 2009, 43, 1073–1077. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Perkins, D.O.; Jeffries, C.D. miRNA and Schizophrenia. In Current Perspectives in microRNAs (miRNA); Springer: Cham, Switzerland, 2008; pp. 267–281. [Google Scholar]
- Omahen, D.A. MicroRNA and diseases of the nervous system. Neurosurgery 2011, 69, 440–454. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Caputo, V.; Sinibaldi, L.; Fiorentino, A.; Parisi, C.; Catalanotto, C.; Pasini, A.; Cogoni, C.; Pizzuti, A. Brain derived neurotrophic factor (BDNF) expression is regulated by microRNAs miR-26a and miR-26b allele-specific binding. PLoS ONE 2011, 6, e28656. [Google Scholar] [CrossRef] [PubMed]
- Im, H.I.; Kenny, P.J. MicroRNAs in neuronal function and dysfunction. Trends Neurosci. 2012, 35, 325–334. [Google Scholar] [CrossRef] [Green Version]
- Xu, B.; Hsu, P.K.; Karayiorgou, M.; Gogos, J.A. MicroRNA dysregulation in neuropsychiatric disorders and cognitive dysfunction. Neurobiol. Dis. 2012, 46, 291–301. [Google Scholar] [CrossRef] [Green Version]
- Sun, E.; Shi, Y. MicroRNAs: Small molecules with big roles in neurodevelopment and diseases. Exp. Neurol. 2015, 268, 46–53. [Google Scholar] [CrossRef] [Green Version]
- Wright, C.; Ross, D.A.; Weinberger, D.R. Small RNAs may answer big questions in mental illness. Biol. Psychiatry 2018, 83, e1–e3. [Google Scholar] [CrossRef] [Green Version]
- Beveridge, N.J.; Tooney, P.A.; Carroll, A.P.; Gardiner, E.; Bowden, N.; Scott, R.J.; Tran, N.; Dedova, I.; Cairns, M.J. Dysregulation of miRNA 181b in the temporal cortex in schizophrenia. Hum. Mol. Genet. 2008, 17, 1156–1168. [Google Scholar] [CrossRef] [Green Version]
- Stark, K.L.; Xu, B.; Bagchi, A.; Lai, W.S.; Liu, H.; Hsu, R.; Wan, X.; Pavlidis, P.; Mills, A.A.; Karayiorgou, M.; et al. Altered brain microRNA biogenesis contributes to phenotypic deficits in a 22q11-deletion mouse model. Nat. Genet. 2008, 40, 751–760. [Google Scholar] [CrossRef]
- Zelena, D. Co-regulation and epigenetic dysregulation in schizophrenia and bipolar disorder. In Patho-Epigenetics of Disease; Springer: Cham, Switzerland, 2012; pp. 281–347. [Google Scholar]
- Gavin, D.P.; Akbarian, S. Epigenetic and post-transcriptional dysregulation of gene expression in schizophrenia and related disease. Neurobiol. Dis. 2012, 46, 255–262. [Google Scholar] [CrossRef] [PubMed]
- Day, J.J.; Sweatt, J.D. Epigenetic treatments for cognitive impairments. Neuropsychopharmacology 2012, 37, 247–260. [Google Scholar] [CrossRef] [PubMed]
- Feinstein, A.; Goldberg, T.E.; Nowlin, B.; Weinberger, D.R. Types and characteristics of remote memory impairment in schizophrenia. Schizophr. Res. 1998, 30, 155–163. [Google Scholar] [CrossRef] [PubMed]
- Luoni, A.; Riva, M.A. MicroRNAs and psychiatric disorders: From aetiology to treatment. Pharmacol. Ther. 2016, 167, 13–27. [Google Scholar] [CrossRef]
- Merico, D.; Costain, G.; Butcher, N.J.; Warnica, W.; Ogura, L.; Alfred, S.E.; Brzustowicz, L.M.; Bassett, A.S. MicroRNA dysregulation, gene networks, and risk for schizophrenia in 22q11. 2 deletion syndrome. Front. Neurol. 2014, 5, 238. [Google Scholar] [CrossRef] [Green Version]
- Kolshus, E.; Dalton, V.; Ryan, K.; McLoughlin, D. When less is more–microRNAs and psychiatric disorders. Acta Psychiatr. Scand. 2014, 129, 241–256. [Google Scholar] [CrossRef] [PubMed]
- Giridharan, V.; Thandavarayan, R.; Fries, G.; Walss-Bass, C.; Barichello, T.; Justice, N.; Reddy, M.; Quevedo, J. Newer insights into the role of miRNA a tiny genetic tool in psychiatric disorders: Focus on post-traumatic stress disorder. Transl. Psychiatry 2016, 6, e954. [Google Scholar] [CrossRef]
- Millan, M.J. An epigenetic framework for neurodevelopmental disorders: From pathogenesis to potential therapy. Neuropharmacology 2013, 68, 2–82. [Google Scholar]
- Geaghan, M.; Cairns, M.J. MicroRNA and posttranscriptional dysregulation in psychiatry. Biol. Psychiatry 2015, 78, 231–239. [Google Scholar] [CrossRef] [Green Version]
- Mellios, N.; Huang, H.S.; Baker, S.P.; Galdzicka, M.; Ginns, E.; Akbarian, S. Molecular determinants of dysregulated GABAergic gene expression in the prefrontal cortex of subjects with schizophrenia. Biol. Psychiatry 2009, 65, 1006–1014. [Google Scholar] [CrossRef]
- Banigan, M.G.; Kao, P.F.; Kozubek, J.A.; Winslow, A.R.; Medina, J.; Costa, J.; Schmitt, A.; Schneider, A.; Cabral, H.; Cagsal-Getkin, O.; et al. Differential expression of exosomal microRNAs in prefrontal cortices of schizophrenia and bipolar disorder patients. PLoS ONE 2013, 8, e48814. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Smalheiser, N.R.; Lugli, G.; Zhang, H.; Rizavi, H.; Cook, E.H.; Dwivedi, Y. Expression of microRNAs and other small RNAs in prefrontal cortex in schizophrenia, bipolar disorder and depressed subjects. PLoS ONE 2014, 9, e86469. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Merico, D.; Costain, G.; Butcher, N.; Warnica, W.; Ogura, L.; Alfred, S.E.; Brzustowicz, L.; Bassett, A.S. MicroRNA target genes and risk for schizophrenia in 22q11. 2 deletion syndrome. Front. Neurol. 2014, 5, 221. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Réthelyi, J.M.; Benkovits, J.; Bitter, I. Genes and environments in schizophrenia: The different pieces of a manifold puzzle. Neurosci. Biobehav. Rev. 2013, 37, 2424–2437. [Google Scholar] [CrossRef]
- Merico, D.; Zarrei, M.; Costain, G.; Ogura, L.; Alipanahi, B.; Gazzellone, M.J.; Butcher, N.J.; Thiruvahindrapuram, B.; Nalpathamkalam, T.; Chow, E.W.; et al. Whole-genome sequencing suggests schizophrenia risk mechanisms in humans with 22q11. 2 deletion syndrome. G3 Genes Genomes Genet. 2015, 5, 2453–2461. [Google Scholar]
- Forstner, A.J.; Degenhardt, F.; Schratt, G.; Nöthen, M.M. MicroRNAs as the cause of schizophrenia in 22q11. 2 deletion carriers, and possible implications for idiopathic disease: A mini-review. Front. Mol. Neurosci. 2013, 6, 47. [Google Scholar] [CrossRef] [PubMed]
- Karayiorgou, M.; Simon, T.J.; Gogos, J.A. 22q11. 2 microdeletions: Linking DNA structural variation to brain dysfunction and schizophrenia. Nat. Rev. Neurosci. 2010, 11, 402–416. [Google Scholar] [CrossRef]
- Earls, L.R.; Fricke, R.G.; Yu, J.; Berry, R.B.; Baldwin, L.T.; Zakharenko, S.S. Age-dependent microRNA control of synaptic plasticity in 22q11 deletion syndrome and schizophrenia. J. Neurosci. 2012, 32, 14132–14144. [Google Scholar] [CrossRef] [Green Version]
- Hoffmann, A.; Ziller, M.; Spengler, D. Childhood-onset schizophrenia: Insights from induced pluripotent stem cells. Int. J. Mol. Sci. 2018, 19, 3829. [Google Scholar] [CrossRef] [Green Version]
- Ahmad, R.; Sportelli, V.; Ziller, M.; Spengler, D.; Hoffmann, A. Tracing early neurodevelopment in schizophrenia with induced pluripotent stem cells. Cells 2018, 7, 140. [Google Scholar] [CrossRef] [Green Version]
- Liu, Y.N.; Lu, S.Y.; Yao, J. Application of induced pluripotent stem cells to understand neurobiological basis of bipolar disorder and schizophrenia. Psychiatry Clin. Neurosci. 2017, 71, 579–599. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Eyles, D.W. How do established developmental risk-factors for schizophrenia change the way the brain develops? Transl. Psychiatry 2021, 11, 1–15. [Google Scholar] [CrossRef] [PubMed]
- Gibbons, A.; Udawela, M.; Dean, B. Non-coding RNA as novel players in the pathophysiology of schizophrenia. Non-Coding RNA 2018, 4, 11. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cao, T.; Zhen, X.C. Dysregulation of mi RNA and its potential therapeutic application in schizophrenia. CNS Neurosci. Ther. 2018, 24, 586–597. [Google Scholar] [CrossRef] [Green Version]
- Wright, C.; Gupta, C.; Chen, J.; Patel, V.; Calhoun, V.D.; Ehrlich, S.; Wang, L.; Bustillo, J.; Perrone-Bizzozero, N.; Turner, J. Polymorphisms in MIR137HG and microRNA-137-regulated genes influence gray matter structure in schizophrenia. Transl. Psychiatry 2016, 6, e724. [Google Scholar] [CrossRef] [Green Version]
- Smrt, R.D.; Szulwach, K.E.; Pfeiffer, R.L.; Li, X.; Guo, W.; Pathania, M.; Teng, Z.Q.; Luo, Y.; Peng, J.; Bordey, A.; et al. MicroRNA miR-137 regulates neuronal maturation by targeting ubiquitin ligase mind bomb-1. Stem Cells 2010, 28, 1060–1070. [Google Scholar] [CrossRef]
- Alural, B.; Genc, S.; Haggarty, S.J. Diagnostic and therapeutic potential of microRNAs in neuropsychiatric disorders: Past, present, and future. Prog. Neuro-Psychopharmacol. Biol. Psychiatry 2017, 73, 87–103. [Google Scholar] [CrossRef] [Green Version]
- Lin, C.C.; Huang, T.L. Epigenetic biomarkers in neuropsychiatric disorders. In Neuropsychiatric Disorders and Epigenetics; Elsevier: Amsterdam, The Netherlands, 2017; pp. 35–66. [Google Scholar]
- Gruzdev, S.; Yakovlev, A.; Druzhkova, T.; Guekht, A.; Gulyaeva, N. The missing link: How exosomes and miRNAs can help in bridging psychiatry and molecular biology in the context of depression, bipolar disorder and schizophrenia. Cell. Mol. Neurobiol. 2019, 39, 729–750. [Google Scholar] [CrossRef]
- Swathy, B.; Banerjee, M. Understanding epigenetics of schizophrenia in the backdrop of its antipsychotic drug therapy. Epigenomics 2017, 9, 721–736. [Google Scholar] [CrossRef] [Green Version]
- Deraredj Nadim, W.; Simion, V.; Bénédetti, H.; Pichon, C.; Baril, P.; Morisset-Lopez, S. MicroRNAs in neurocognitive dysfunctions: New molecular targets for pharmacological treatments? Curr. Neuropharmacol. 2017, 15, 260–275. [Google Scholar] [CrossRef] [Green Version]
- Kocerha, J.; Faghihi, M.A.; Lopez-Toledano, M.A.; Huang, J.; Ramsey, A.J.; Caron, M.G.; Sales, N.; Willoughby, D.; Elmen, J.; Hansen, H.F.; et al. MicroRNA-219 modulates NMDA receptor-mediated neurobehavioral dysfunction. Proc. Natl. Acad. Sci. USA 2009, 106, 3507–3512. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Song, H.T.; Sun, X.Y.; Zhang, L.; Zhao, L.; Guo, Z.M.; Fan, H.M.; Zhong, A.F.; Niu, W.; Dai, Y.H.; Zhang, L.Y.; et al. A preliminary analysis of association between the down-regulation of microRNA-181b expression and symptomatology improvement in schizophrenia patients before and after antipsychotic treatment. J. Psychiatr. Res. 2014, 54, 134–140. [Google Scholar] [CrossRef] [PubMed]
- Roy, B.; Yoshino, Y.; Allen, L.; Prall, K.; Schell, G.; Dwivedi, Y. Exploiting circulating MicroRNAs as biomarkers in psychiatric disorders. Mol. Diagn. Ther. 2020, 24, 279–298. [Google Scholar] [CrossRef]
- van Calker, D.; Serchov, T. The “missing heritability”—Problem in psychiatry: Is the interaction of genetics, epigenetics and transposable elements a potential solution? Neurosci. Biobehav. Rev. 2021, 126, 23–42. [Google Scholar] [CrossRef]
- Shi, Y.; Wang, Q.; Song, R.; Kong, Y.; Zhang, Z. Non-coding RNAs in depression: Promising diagnostic and therapeutic biomarkers. EBioMedicine 2021, 71, 103569. [Google Scholar] [CrossRef]
- Smigielski, L.; Jagannath, V.; Rössler, W.; Walitza, S.; Grünblatt, E. Epigenetic mechanisms in schizophrenia and other psychotic disorders: a systematic review of empirical human findings. Mol. Psychiatry 2020, 25, 1718–1748. [Google Scholar] [CrossRef] [PubMed]
- Kuehner, J.N.; Bruggeman, E.C.; Wen, Z.; Yao, B. Epigenetic regulations in neuropsychiatric disorders. Front. Genet. 2019, 10, 268. [Google Scholar] [CrossRef] [Green Version]
- Gürel, Ç.; Kuşçu, G.C.; Yavaşoğlu, A.; Avcı, Ç.B. The clues in solving the mystery of major psychosis: The epigenetic basis of schizophrenia and bipolar disorder. Neurosci. Biobehav. Rev. 2020, 113, 51–61. [Google Scholar] [CrossRef]
- Hansen, T.B.; Jensen, T.I.; Clausen, B.H.; Bramsen, J.B.; Finsen, B.; Damgaard, C.K.; Kjems, J. Natural RNA circles function as efficient microRNA sponges. Nature 2013, 495, 384–388. [Google Scholar] [CrossRef]
- Memczak, S.; Jens, M.; Elefsinioti, A.; Torti, F.; Krueger, J.; Rybak, A.; Maier, L.; Mackowiak, S.D.; Gregersen, L.H.; Munschauer, M.; et al. Circular RNAs are a large class of animal RNAs with regulatory potency. Nature 2013, 495, 333–338. [Google Scholar] [CrossRef]
- Mahmoudi, E.; Fitzsimmons, C.; Geaghan, M.P.; Shannon Weickert, C.; Atkins, J.R.; Wang, X.; Cairns, M.J. Circular RNA biogenesis is decreased in postmortem cortical gray matter in schizophrenia and may alter the bioavailability of associated miRNA. Neuropsychopharmacology 2019, 44, 1043–1054. [Google Scholar] [CrossRef] [PubMed]
- Tsermpini, E.E.; Kalogirou, C.I.; Kyriakopoulos, G.C.; Patrinos, G.P.; Stathopoulos, C. miRNAs as potential diagnostic biomarkers and pharmacogenomic indicators in psychiatric disorders. Pharmacogenomics J. 2022, 22, 211–222. [Google Scholar] [CrossRef] [PubMed]
- Ghafouri-Fard, S.; Eghtedarian, R.; Taheri, M.; Beatrix Brühl, A.; Sadeghi-Bahmani, D.; Brand, S. A review on the expression pattern of non-coding RNAs in patients with schizophrenia: With a special focus on peripheral blood as a source of expression analysis. Front. Psychiatry 2021, 12, 640463. [Google Scholar] [CrossRef] [PubMed]
- Sabaie, H.; Moghaddam, M.M.; Moghaddam, M.M.; Ahangar, N.K.; Asadi, M.R.; Hussen, B.M.; Taheri, M.; Rezazadeh, M. Bioinformatics analysis of long non-coding RNA-associated competing endogenous RNA network in schizophrenia. Sci. Rep. 2021, 11, 24413. [Google Scholar] [CrossRef]
- Sabaie, H.; Mazaheri Moghaddam, M.; Mazaheri Moghaddam, M.; Amirinejad, N.; Asadi, M.R.; Daneshmandpour, Y.; Hussen, B.M.; Taheri, M.; Rezazadeh, M. Long non-coding RNA-associated competing endogenous RNA axes in the olfactory epithelium in schizophrenia: A bioinformatics analysis. Sci. Rep. 2021, 11, 24497. [Google Scholar] [CrossRef]
- Jin, M.; Zhu, X.; Sun, Y.; Li, Z.; Li, X.; Ai, L.; He, Y.; Liu, Y.; Jia, N.; Hu, G.; et al. Identification of Peripheral Blood miRNA Biomarkers in First-Episode Drug-Free Schizophrenia Patients Using Bioinformatics Strategy. Mol. Neurobiol. 2022, 59, 4730–4746. [Google Scholar] [CrossRef]
- Jin, X.; Wah, B.W.; Cheng, X.; Wang, Y. Significance and challenges of big data research. Big Data Res. 2015, 2, 59–64. [Google Scholar] [CrossRef]
- Xie, M.; Li, Z.; Li, X.; Ai, L.; Jin, M.; Jia, N.; Yang, Y.; Li, W.; Xue, F.; Zhang, M.; et al. Identifying crucial biomarkers in peripheral blood of schizophrenia and screening therapeutic agents by comprehensive bioinformatics analysis. J. Psychiatr. Res. 2022, 152, 86–96. [Google Scholar] [CrossRef]
- Haddaway, N.R.; Collins, A.M.; Coughlin, D.; Kirk, S. The role of Google Scholar in evidence reviews and its applicability to grey literature searching. PLoS ONE 2015, 10, e0138237. [Google Scholar] [CrossRef] [Green Version]
- Sutton, A.J. Publication bias. Handb. Res. Synth. Meta-Anal. 2009, 2, 435–452. [Google Scholar]
- Hicks, D.; Wouters, P.; Waltman, L.; De Rijcke, S.; Rafols, I. Bibliometrics: The Leiden Manifesto for research metrics. Nat. News 2015, 520, 429. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Maric, N.; Svrakic, D. Why schizophrenia genetics needs epigenetics: A review. Psychiatr. Danub. 2012, 24, 2–18. [Google Scholar] [PubMed]
- Rajarajan, P.; Akbarian, S. Use of the epigenetic toolbox to contextualize common variants associated with schizophrenia risk. Dialogues Clin. Neurosci. 2022, 21, 407–416. [Google Scholar] [CrossRef] [PubMed]
- Mojarad, B.A.; Engchuan, W.; Trost, B.; Backstrom, I.; Yin, Y.; Thiruvahindrapuram, B.; Pallotto, L.; Mitina, A.; Khan, M.; Pellecchia, G.; et al. Genome-wide tandem repeat expansions contribute to schizophrenia risk. Mol. Psychiatry 2022, 27, 3692–3698. [Google Scholar] [CrossRef]
- Wang, D.; Liu, S.; Warrell, J.; Won, H.; Shi, X.; Navarro, F.C.; Clarke, D.; Gu, M.; Emani, P.; Yang, Y.T.; et al. Comprehensive functional genomic resource and integrative model for the human brain. Science 2018, 362, eaat8464. [Google Scholar] [CrossRef] [Green Version]
- Wang, Q.; Chen, R.; Cheng, F.; Wei, Q.; Ji, Y.; Yang, H.; Zhong, X.; Tao, R.; Wen, Z.; Sutcliffe, J.S.; et al. A Bayesian framework that integrates multi-omics data and gene networks predicts risk genes from schizophrenia GWAS data. Nat. Neurosci. 2019, 22, 691–699. [Google Scholar] [CrossRef]
- Yoshikawa, A.; Kushima, I.; Miyashita, M.; Toriumi, K.; Suzuki, K.; Horiuchi, Y.; Kawaji, H.; Takizawa, S.; Ozaki, N.; Itokawa, M.; et al. Dysregulation of post-transcriptional modification by copy number variable microRNAs in schizophrenia with enhanced glycation stress. Transl. Psychiatry 2021, 11, 1–11. [Google Scholar] [CrossRef]

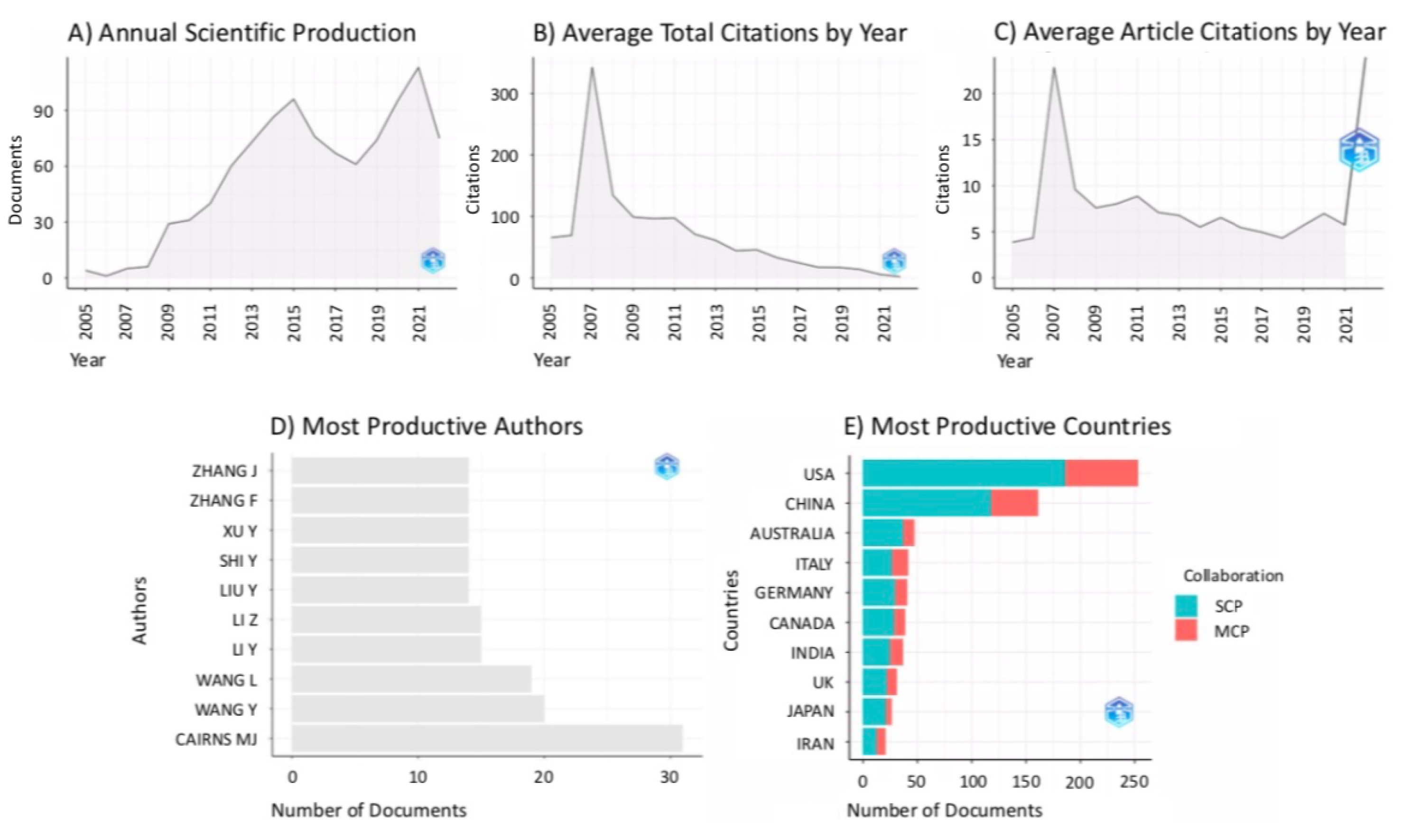
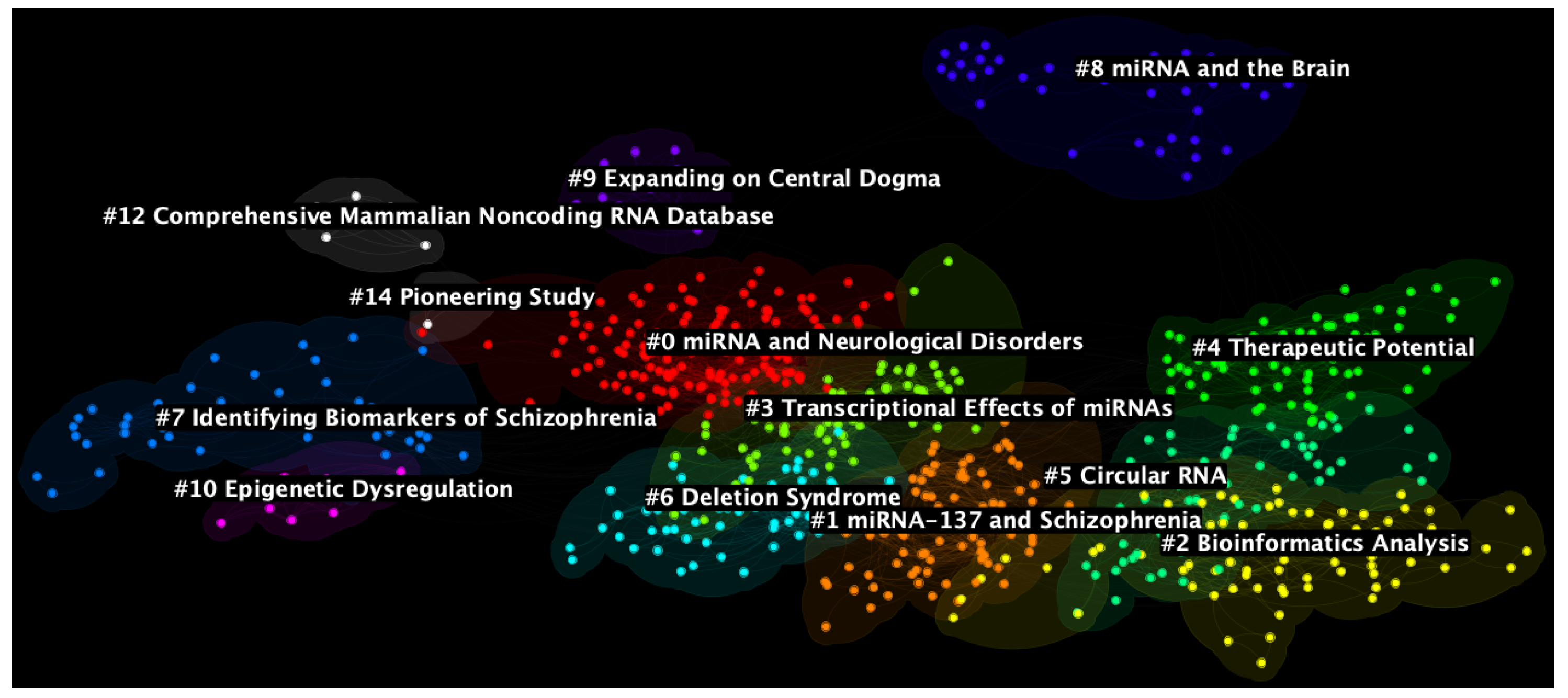
| Cluster ID | Size | Silhouette | Mean Year | LLR Label | Suggested Label |
|---|---|---|---|---|---|
| 0 | 142 | 0.856 | 2010 | mRNA Gene | miRNA and Neurological Disorders |
| 1 | 86 | 0.911 | 2016 | Induced Pluripotent Stem Cell | miRNA-137 and Schizophrenia |
| 2 | 80 | 0.859 | 2020 | Bioinformatics Analysis | Bioinformatics Analysis |
| 3 | 72 | 0.845 | 2013 | Psychiatric Disorder | Transcriptional Effects of miRNAs |
| 4 | 71 | 0.893 | 2016 | Therapeutic Potential | Therapeutic Potential |
| 5 | 63 | 0.898 | 2019 | Exploiting Circulating Micro RNA | Circular RNA |
| 6 | 59 | 0.878 | 2013 | Deletion Syndrome | Deletion Syndrome |
| 7 | 45 | 0.909 | 2008 | Schizophrenia Gene | Identifying Biomarkers of Schizophrenia |
| 8 | 38 | 0.988 | 2008 | Clinical Aspect | miRNA and the Brain |
| 9 | 16 | 0.99 | 2005 | Central Dogma | Expanding on Central Dogma |
| 10 | 10 | 0.995 | 2012 | Epigenetic Dysregulation | Epigenetic Dysregulation |
| 12 | 7 | 0.997 | 2005 | Comprehensive Mammalian Noncoding RNA Database | Comprehensive Mammalian Noncoding RNA Database |
| 14 | 4 | 0.991 | 2007 | Schizophrenia | Pioneering Study |
| Reference | Citation Burstness | Publication Year | Burst Begin | Burst End | Duration | Betweenness Centrality | Sigma |
|---|---|---|---|---|---|---|---|
| Bartel [38] | 9.82 | 2004 | 2009 | 2012 | 3 | 0.08 | 2.09 |
| Pantelis et al. [9] | 9.48 | 2014 | 2016 | 2022 | 6 | 0.02 | 1.24 |
| Perkins et al. [7] | 9.33 | 2007 | 2008 | 2014 | 6 | 0.04 | 1.46 |
| Filipowicz et al. [39] | 8.90 | 2008 | 2009 | 2010 | 1 | 0.01 | 1.07 |
| Abelson et al. [40] | 8.55 | 2005 | 2007 | 2010 | 3 | 0.07 | 1.81 |
| Ripke et al. [41] | 8.53 | 2013 | 2015 | 2019 | 4 | 0.02 | 1.17 |
| Schratt et al. [42] | 8.40 | 2006 | 2007 | 2014 | 7 | 0.03 | 1.29 |
| Siegert et al. [8] | 8.00 | 2015 | 2016 | 2018 | 2 | 0.02 | 1.20 |
| Wright et al. [43] | 7.98 | 2013 | 2014 | 2018 | 4 | 0.01 | 1.05 |
| Lewis et al. [44] | 7.61 | 2005 | 2007 | 2011 | 4 | 0.08 | 1.80 |
| Barry et al. [45] | 7.15 | 2014 | 2018 | 2022 | 4 | 0.02 | 1.15 |
| Shi et al. [46] | 6.72 | 2012 | 2014 | 2016 | 2 | 0.00 | 1.02 |
| Hansen et al. [47] | 6.51 | 2007 | 2009 | 2012 | 3 | 0.03 | 1.19 |
| The Schizophrenia Psychiatric GWAS Consortium [32] | 6.43 | 2011 | 2014 | 2018 | 4 | 0.04 | 1.30 |
| Sun et al. [48] | 6.36 | 2011 | 2014 | 2016 | 2 | 0.00 | 1.00 |
| Kim et al. [49] | 6.21 | 2007 | 2008 | 2013 | 5 | 0.04 | 1.25 |
| Gardiner et al. [50] | 6.14 | 2012 | 2014 | 2017 | 3 | 0.01 | 1.04 |
| Zhou et al. [51] | 6.00 | 2009 | 2010 | 2013 | 3 | 0.02 | 1.12 |
| Guan et al. [52] | 5.98 | 2014 | 2015 | 2018 | 3 | 0.02 | 1.12 |
| Beveridge and Cairns [5] | 5.95 | 2012 | 2016 | 2018 | 2 | 0.00 | 1.01 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lim, M.; Carollo, A.; Neoh, M.J.Y.; Esposito, G. Mapping miRNA Research in Schizophrenia: A Scientometric Review. Int. J. Mol. Sci. 2023, 24, 436. https://doi.org/10.3390/ijms24010436
Lim M, Carollo A, Neoh MJY, Esposito G. Mapping miRNA Research in Schizophrenia: A Scientometric Review. International Journal of Molecular Sciences. 2023; 24(1):436. https://doi.org/10.3390/ijms24010436
Chicago/Turabian StyleLim, Mengyu, Alessandro Carollo, Michelle Jin Yee Neoh, and Gianluca Esposito. 2023. "Mapping miRNA Research in Schizophrenia: A Scientometric Review" International Journal of Molecular Sciences 24, no. 1: 436. https://doi.org/10.3390/ijms24010436
APA StyleLim, M., Carollo, A., Neoh, M. J. Y., & Esposito, G. (2023). Mapping miRNA Research in Schizophrenia: A Scientometric Review. International Journal of Molecular Sciences, 24(1), 436. https://doi.org/10.3390/ijms24010436






