Revealing the Mechanisms for Linalool Antifungal Activity against Fusarium oxysporum and Its Efficient Control of Fusarium Wilt in Tomato Plants
Abstract
:1. Introduction
2. Results and Discussion
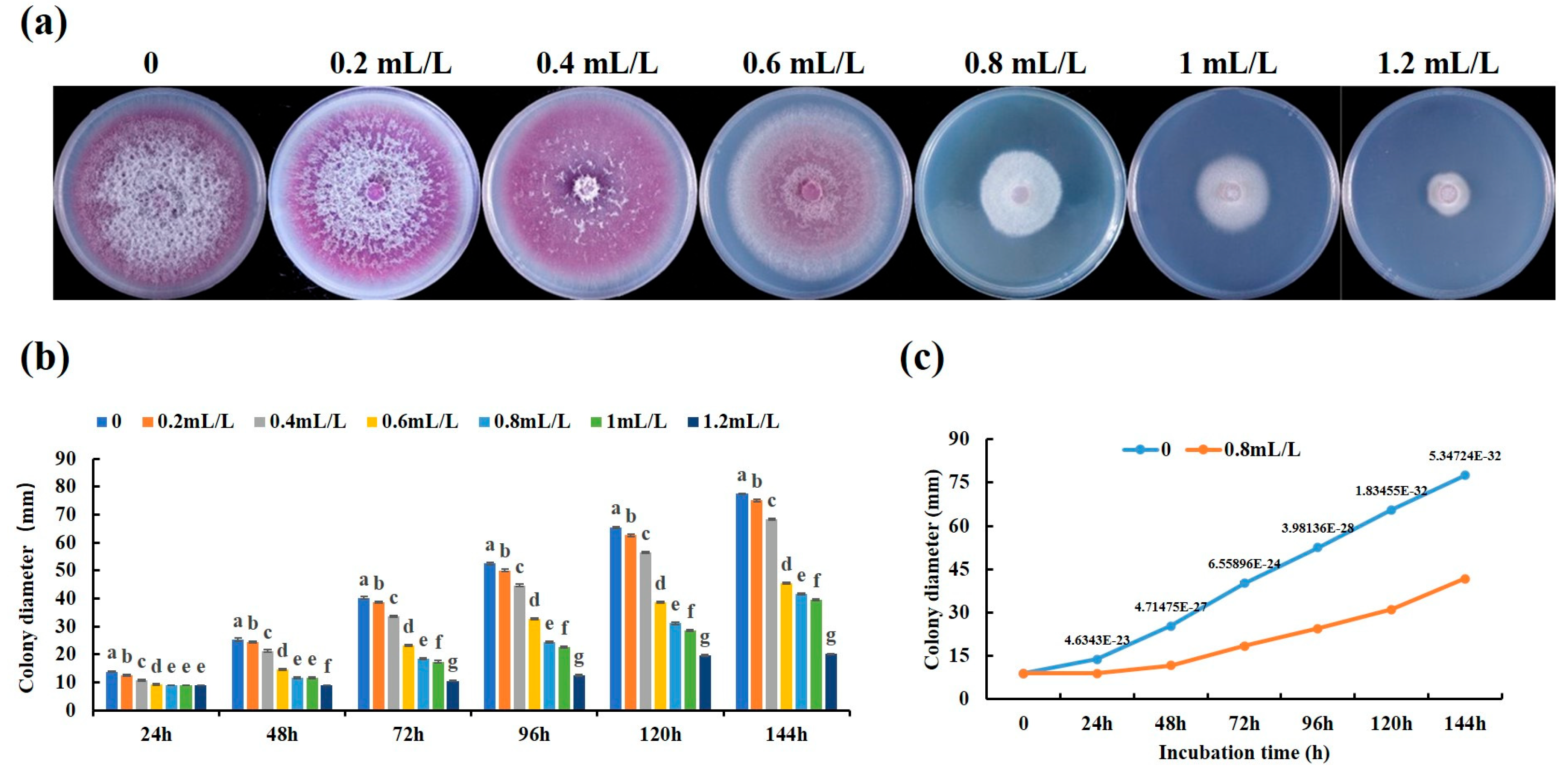
2.1. Linalool Inhibits the Growth of Forl
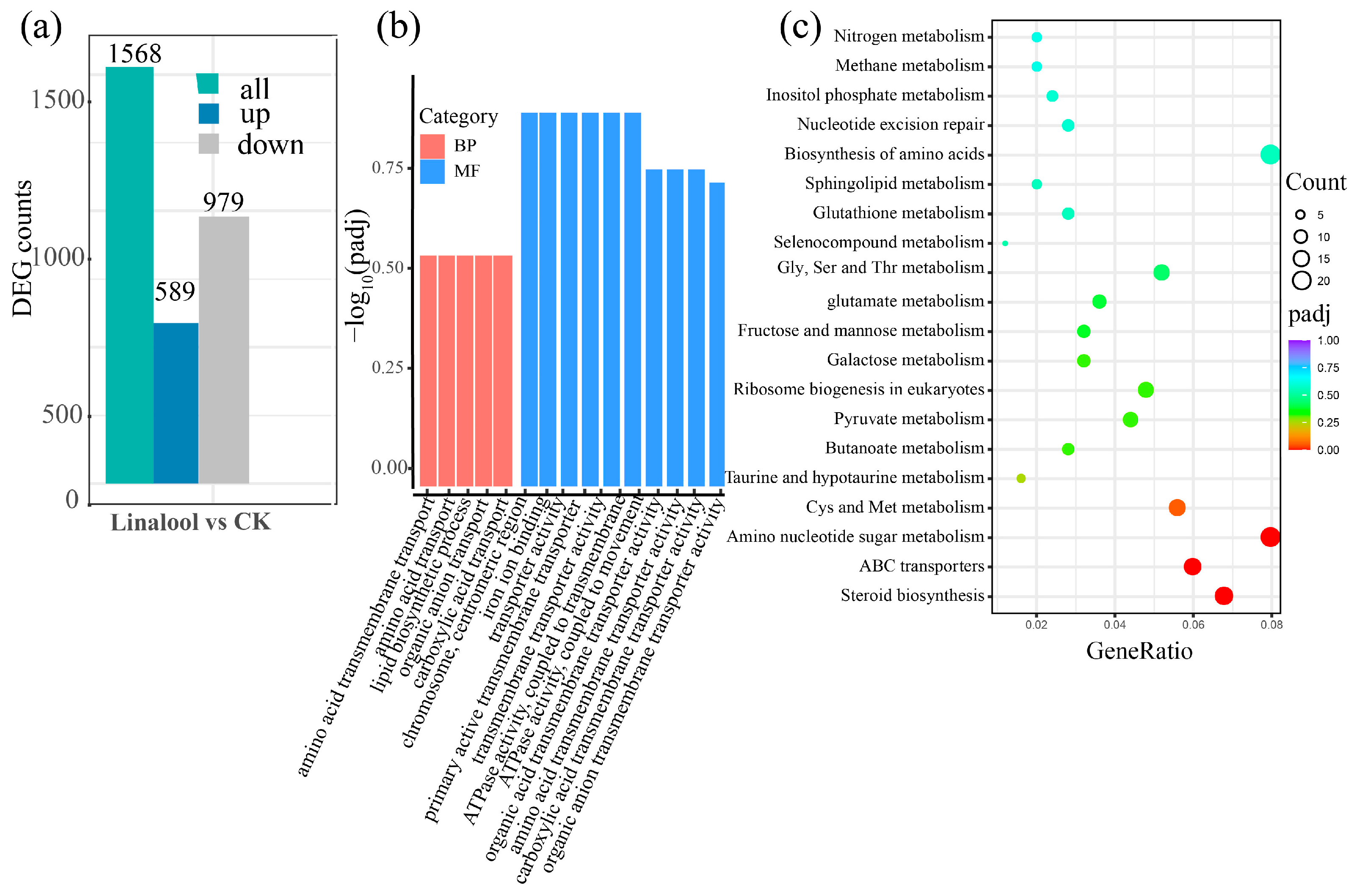
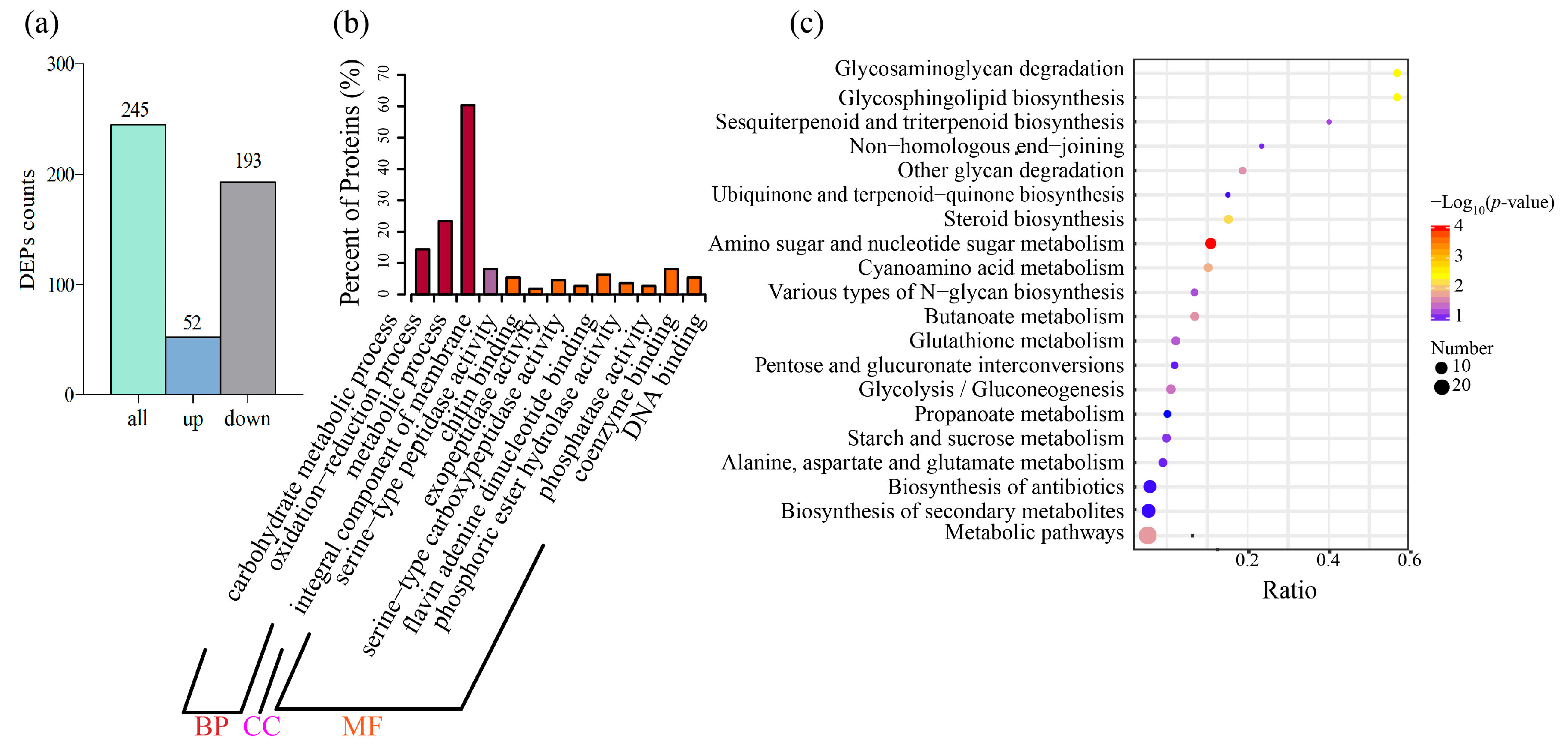
2.2. Transcriptomic and Proteomic Analysis of Forl after the Linalool Treatment
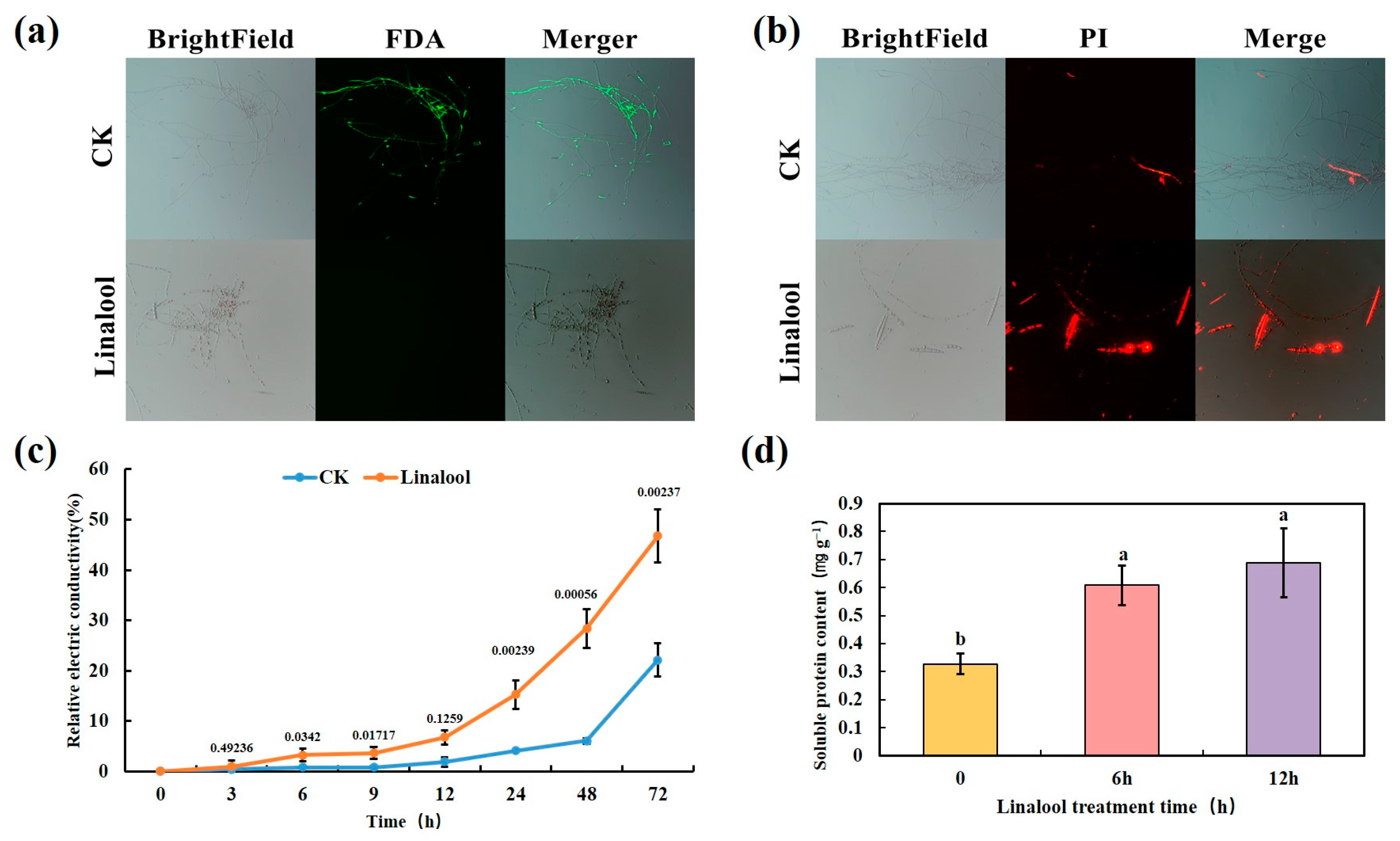
2.3. Linalool Treatment Damages Cell Membrane Integrity
2.4. Linalool Treatment Promotes the Production of Reactive Oxygen Species (ROS) and Inhibits ROS Scavenging in Forl
2.5. Linalool Treatment Affects a Variety of Forl Metabolic Reactions
2.6. Linalool Suppresses the Virulence of Forl and Confers Antifungal Activity against Forl in Tomato Plants
2.7. Effect of Linalool on Growth of Tomato Plants
3. Materials and Methods
3.1. Fungal Strain and Growth Conditions
3.2. Mycelial Growth Inhibition Test
3.3. Transcriptome and RNA-seq and Data Analysis
3.4. Tandem Mass Tag (TMT)-Based Quantitative Proteomic Analysis
3.5. Propidium Iodide (PI) and Fluorescein Diacetate (FDA) Staining of the Forl Mycelia
3.6. Determination of Cell Membrane Permeability
3.7. Determination of Malondialdehyde (MDA) and Soluble Protein Contents
3.8. Determination of Superoxide (O2•−), Hydrogen Peroxide (H2O2), and Glutathione (GSH) Content, and Antioxidant Enzyme Activities
3.9. Determination of Cell Wall Degradation-Related Enzyme Activity
3.10. Effects of Linalool in Controlling Root Rot Disease Caused by Forl
3.10.1. Linalool Application and Forl Inoculation
3.10.2. Disease Severity Assessment
3.10.3. Determination of Chlorophyll and Carotenoid Content
3.11. Plant Total RNA Extraction and Real-Time Quantitative PCR Analysis
3.12. Statistical Analysis
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- McGovern, R. Management of tomato diseases caused by Fusarium oxysporum. Crop. Prot. 2015, 73, 78–92. [Google Scholar] [CrossRef]
- Chitwood-Brown, J.; Vallad, G.E.; Lee, T.G.; Hutton, S.F. Breeding for Resistance to Fusarium Wilt of Tomato: A Review. Genes 2021, 12, 1673. [Google Scholar] [CrossRef] [PubMed]
- Hassan, H.A. Biology and integrated control of tomato wilt caused by Fusarium oxysporum lycopersici: A comprehensive review under the light of recent advancements. J. Bot. Res. 2020, 3, 84–99. [Google Scholar]
- Bosch, F.V.D.; Oliver, R.; Berg, F.V.D.; Paveley, N. Governing Principles Can Guide Fungicide-Resistance Management Tactics. Annu. Rev. Phytopathol. 2014, 52, 175–195. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Abril, M.; Curry, K.J.; Smith, B.J.; Wedge, D.E. Improved microassays used to test natural product-based and conventional fungicides on plant pathogenic fungi. Plant Dis. 2007, 92, 106–112. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cárdenas-Laverde, D.; Barbosa-Cornelio, R.; Coy-Barrera, E. Antifungal Activity against Fusarium oxysporum of Botanical End-Products: An Integration of Chemical Composition and Antifungal Activity Datasets to Identify Antifungal Bioactives. Plants 2021, 10, 2563. [Google Scholar] [CrossRef]
- Seepe, H.A.; Nxumalo, W.; Amoo, S.O. Natural products from medicinal plants against phytopathogenic Fusarium species: Current research endeavours, challenges and prospects. Molecules 2021, 26, 6539. [Google Scholar] [CrossRef]
- Sharifi, R.; Lee, S.-M.; Ryu, C.-M. Microbe-induced plant volatiles. New Phytol. 2017, 220, 684–691. [Google Scholar] [CrossRef] [Green Version]
- Ninkovic, V.; Markovic, D.; Rensing, M. Plant volatiles as cues and signals in plant communication. Plant, Cell Environ. 2020, 44, 1030–1043. [Google Scholar] [CrossRef]
- Boncan, D.A.T.; Tsang, S.S.K.; Li, C.; Lee, I.H.T.; Lam, H.M.; Chan, T.F.; Hui, J.H.L. Terpenes and terpenoids in plants: In-teractions with environment and insects. Int. J. Mol. Sci. 2020, 21, 7382. [Google Scholar] [CrossRef]
- Xu, H.; Turlings, T.C. Plant Volatiles as Mate-Finding Cues for Insects. Trends Plant Sci. 2018, 23, 100–111. [Google Scholar] [CrossRef]
- Picazo-Aragonés, J.; Terrab, A.; Balao, F. Plant Volatile Organic Compounds Evolution: Transcriptional Regulation, Epigenetics and Polyploidy. Int. J. Mol. Sci. 2020, 21, 8956. [Google Scholar] [CrossRef]
- Calo, J.R.; Crandall, P.G.; O’Bryan, C.A.; Ricke, S.C. Essential oils as antimicrobials in food systems—A review. Food Control 2015, 54, 111–119. [Google Scholar] [CrossRef]
- Raguso, R.A. More lessons from linalool: Insights gained from a ubiquitous floral volatile. Curr. Opin. Plant Biol. 2016, 32, 31–36. [Google Scholar] [CrossRef] [Green Version]
- Nagegowda, D.A.; Gupta, P. Advances in biosynthesis, regulation, and metabolic engineering of plant specialized terpenoids. Plant Sci. 2020, 294, 110457. [Google Scholar] [CrossRef]
- Xu, Y.; Tong, Z.; Zhang, X.; Wang, Y.; Fang, W.; Li, L.; Luo, Z. Unveiling the mechanisms for the plant volatile organic com-pound linalool to control gray mold on strawberry fruits. J. Agric. Food Chem. 2019, 67, 9265–9276. [Google Scholar] [CrossRef]
- Yuan, J.S.; Köllner, T.G.; Wiggins, G.; Grant, J.; Degenhardt, J.; Chen, F. Molecular and genomic basis of volatile-mediated indirect defense against insects in rice. Plant J. 2008, 55, 491–503. [Google Scholar] [CrossRef]
- Huang, X.Z.; Xiao, Y.T.; Kollner, T.G.; Jing, W.X.; Kou, J.F.; Chen, J.Y.; Liu, D.F.; Gu, S.H.; Wu, J.X.; Zhang, Y.J.; et al. The terpene synthase gene family in Gossypium hirsutum harbors a linalool synthase GhTPS12 implicated in direct defence responses against herbivores. Plant Cell Environ. 2018, 41, 261–274. [Google Scholar] [CrossRef]
- van Schie, C.C.N.; Haring, M.A.; Schuurink, R.C. Tomato linalool synthase is induced in trichomes by jasmonic acid. Plant Mol. Biol. 2007, 64, 251–263. [Google Scholar] [CrossRef] [Green Version]
- Taniguchi, S.; Hosokawa-Shinonaga, Y.; Tamaoki, D.; Yamada, S.; Akimitsu, K.; Gomi, K. Jasmonate induction of the mono-terpene linalool confers resistance to rice bacterial blight and its biosynthesis is regulated by JAZ protein in rice. Plant Cell Environ. 2014, 37, 451–461. [Google Scholar] [CrossRef]
- Cappellari, L.D.R.; Santoro, M.V.; Schmidt, A.; Gershenzon, J.; Banchio, E. Improving Phenolic Total Content and Monoterpene in Mentha x piperita by Using Salicylic Acid or Methyl Jasmonate Combined with Rhizobacteria Inoculation. Int. J. Mol. Sci. 2019, 21, 50. [Google Scholar] [CrossRef] [PubMed]
- An, Q.; Ren, J.-N.; Li, X.; Fan, G.; Qu, S.-S.; Song, Y.; Li, Y.; Pan, S.-Y. Recent updates on bioactive properties of linalool. Food Funct. 2021, 12, 10370–10389. [Google Scholar] [CrossRef] [PubMed]
- Guo, F.; Chen, Q.; Liang, Q.; Zhang, M.; Chen, W.; Chen, H.; Yun, Y.; Zhong, Q.; Chen, W. Antimicrobial Activity and Proposed Action Mechanism of Linalool Against Pseudomonas fluorescens. Front. Microbiol. 2021, 12. [Google Scholar] [CrossRef] [PubMed]
- Prakash, A.; Vadivel, V.; Rubini, D.; Nithyanand, P. Antibacterial and antibiofilm activities of linalool nanoemulsions against Salmonella Typhimurium. Food Biosci. 2019, 28, 57–65. [Google Scholar] [CrossRef]
- Gao, Z.; Van Nostrand, J.D.; Zhou, J.; Zhong, W.; Chen, K.; Guo, J. Anti-listeria Activities of Linalool and Its Mechanism Revealed by Comparative Transcriptome Analysis. Front. Microbiol. 2019, 10, 2947. [Google Scholar] [CrossRef]
- Shimada, T.; Endo, T.; Fujii, H.; Rodríguez, A.; Peña, L.; Omura, M. Characterization of three linalool synthase genes from Citrus unshiu Marc. and analysis of linalool-mediated resistance against Xanthomonas citri subsp. citri and Penicilium italicum in citrus leaves and fruits. Plant Sci. 2014, 229, 154–166. [Google Scholar] [CrossRef]
- Deepa, N.; Chauhan, S.; Kumari, P.; Rai, A.K.; Tandon, S.; Singh, A. Linalool reduces the virulence of Pseudomonas syringae pv. tomato DC 3000 by modulating the PsyI/PsyR quorum-sensing system. Microb. Pathog. 2022, 173, 105884. [Google Scholar] [CrossRef]
- Shen, Q.; Li, H.; Wang, Q.; Wang, J.; Ge, J.; Yang, X.; Wang, X.; Li, X.; Zhang, Y.; Zhang, R.; et al. Alleviating Effects of Linalool Fumigation on Botrytis cinerea Infections in Postharvest Tomato Fruits. Horticulturae 2022, 8, 1074. [Google Scholar] [CrossRef]
- Jones, K.; Kim, D.W.; Park, J.S.; Khang, C.H. Live-cell fluorescence imaging to investigate the dynamics of plant cell death during infection by the rice blast fungus Magnaporthe oryzae. BMC Plant Biol. 2016, 16, 1–8. [Google Scholar] [CrossRef] [Green Version]
- Bajpai, V.K.; Kang, S.-R.; Xu, H.; Lee, S.-G.; Baek, K.-H.; Kang, S.-C. Potential Roles of Essential Oils on Controlling Plant Pathogenic Bacteria Xanthomonas Species: A Review. Plant Pathol. J. 2011, 27, 207–224. [Google Scholar] [CrossRef] [Green Version]
- Liu, X.; Cai, J.; Chen, H.; Zhong, Q.; Hou, Y.; Chen, W.; Chen, W. Antibacterial activity and mechanism of linalool against Pseudomonas aeruginosa. Microb. Pathog. 2020, 141, 103980. [Google Scholar] [CrossRef]
- Khan, A.; Ahmad, A.; Akhtar, F.; Yousuf, S.; Xess, I.; Khan, L.A.; Manzoor, N. Ocimum sanctum essential oil and its active principles exert their antifungal activity by disrupting ergosterol biosynthesis and membrane integrity. Res. Microbiol. 2010, 161, 816–823. [Google Scholar] [CrossRef]
- Ren, X.; Xu, Z.; Deng, R.; Huang, L.; Zheng, R.; Kong, Q. Peppermint essential oil suppresses Geotrichum citri-aurantii growth by destructing the cell structure, internal homeostasis, and cell cycle. J. Agric. Food Chem. 2021, 69, 7786–7797. [Google Scholar] [CrossRef]
- Thangamani, S.; Eldesouky, H.E.; Mohammad, H.; Pascuzzi, P.E.; Avramova, L.; Hazbun, T.R.; Seleem, M.N. Ebselen exerts antifungal activity by regulating glutathione (GSH) and reactive oxygen species (ROS) production in fungal cells. Biochim. Biophys. Acta BBA Gen. Subj. 2017, 1861, 3002–3010. [Google Scholar] [CrossRef] [Green Version]
- Zhou, H.; Tao, N.; Jia, L. Antifungal activity of citral, octanal and α-terpineol against Geotrichum citri-aurantii. Food Control 2014, 37, 277–283. [Google Scholar] [CrossRef]
- Mate´s, J.M. Effects of antioxidant enzymes in the molecular control of reactive oxygen species toxicology. Toxicology 2000, 153, 83–104. [Google Scholar] [CrossRef]
- Yang, S.-K.; Yusoff, K.; Ajat, M.; Wee, C.-Y.; Yap, P.-S.; Lim, S.-H.; Lai, K.-S. Combinatorial Antimicrobial Efficacy and Mechanism of Linalool Against Clinically Relevant Klebsiella pneumoniae. Front. Microbiol. 2021, 12. [Google Scholar] [CrossRef]
- Belenky, P.; Camacho, D.; Collins, J.J. Fungicidal Drugs Induce a Common Oxidative-Damage Cellular Death Pathway. Cell Rep. 2013, 3, 350–358. [Google Scholar] [CrossRef] [Green Version]
- OuYang, Q.; Tao, N.; Zhang, M. A Damaged Oxidative Phosphorylation Mechanism Is Involved in the Antifungal Activity of Citral against Penicillium digitatum. Front. Microbiol. 2018, 9, 239. [Google Scholar] [CrossRef] [Green Version]
- Rodríguez-Saavedra, C.; Morgado-Martínez, L.E.; Burgos-Palacios, A.; King-Díaz, B.; López-Coria, M.; Sánchez-Nieto, S. Moonlighting Proteins: The Case of the Hexokinases. Front. Mol. Biosci. 2021, 8. [Google Scholar] [CrossRef]
- Ma, L.J.; Geiser, D.M.; Proctor, R.H.; Rooney, A.P.; O’Donnell, K.; Trail, F.; Gardiner, D.M.; Manners, J.M.; Kazan, K. Fusarium pathogenomics. Annu. Rev. Microbiol. 2013, 67, 399–416. [Google Scholar] [CrossRef] [PubMed]
- de Sain, M.; Rep, M. The role of pathogen-secreted proteins in fungal vascular wilt diseases. Int. J. Mol. Sci. 2015, 16, 23970–23993. [Google Scholar] [CrossRef] [PubMed]
- Kikot, G.E.; Hours, R.A.; Alconada, T.M. Contribution of cell wall degrading enzymes to pathogenesis of Fusarium graminearum : A review. J. Basic Microbiol. 2008, 49, 231–241. [Google Scholar] [CrossRef] [PubMed]
- Husaini, A.M.; Sakina, A.; Cambay, S.R. Host-pathogen interaction in Fusarium oxysporum infections: Where do we stand? Mol. Plant Microbe. Interact. 2018, 31, 889–898. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Di, X.; Gomila, J.; Takken, F.L.W. Involvement of salicylic acid, ethylene and jasmonic acid signalling pathways in the sus-ceptibility of tomato to Fusarium oxysporum. Mol. Plant Pathol. 2017, 18, 1024–1035. [Google Scholar] [CrossRef] [Green Version]
- Liu, B.; Hong, Y.-B.; Zhang, Y.-F.; Li, X.-H.; Huang, L.; Zhang, H.-J.; Li, D.-Y.; Song, F.-M. Tomato WRKY transcriptional factor SlDRW1 is required for disease resistance against Botrytis cinerea and tolerance to oxidative stress. Plant Sci. 2014, 227, 145–156. [Google Scholar] [CrossRef]
- Farmer, E.E.; Johnson, R.R.; Ryan, C.A. Regulation of expression of proteinase inhibitor genes by methyl jasmonate and jasmonic acid. Plant Physiol. 1992, 98, 995–1002. [Google Scholar] [CrossRef] [Green Version]
- Wu, K.; Tian, L.; Hollingworth, J.; Brown, D.C.W.; Miki, B. Functional analysis of tomato Pti4 in Arabidopsis. Plant Physiol. 2002, 128, 30–37. [Google Scholar] [CrossRef]
- Song, Y.; Chen, D.; Lu, K.; Sun, Z.; Zeng, R. Enhanced tomato disease resistance primed by arbuscular mycorrhizal fungus. Front. Plant Sci. 2015, 6, 786. [Google Scholar] [CrossRef] [Green Version]
- Elsharkawy, M.M.; Al-Askar, A.A.; Behiry, S.I.; Abdelkhalek, A.; Saleem, M.H.; Kamran, M.; Derbalah, A. Resistance induction and nematicidal activity of certain monoterpenes against tomato root-knot caused by Meloidogyne incognita. Front. Plant Sci. 2022, 13, 982414. [Google Scholar] [CrossRef]
- Singh, V.K.; Singh, H.B.; Upadhyay, R.S. Role of fusaric acid in the development of 'Fusarium wilt' symptoms in tomato: Physiological, biochemical and proteomic perspectives. Plant Physiol. Biochem. 2017, 118, 320–332. [Google Scholar] [CrossRef]
- Nogués, S.; Cotxarrera, L.; Alegre, L.; Trillas, M.I. Limitations to photosynthesis in tomato leaves induced by Fusarium wilt. New Phytol. 2002, 154, 461–470. [Google Scholar] [CrossRef]
- Zhang, X.; Wang, H.; Zhu, W.; Li, W.; Wang, F. Transcriptome Analysis Reveals the Effects of Chinese Chive (Allium tuberosum R.) Extract on Fusarium oxysporum f. sp. radicis-lycopersici Spore Germination. Curr. Microbiol. 2020, 77, 855–864. [Google Scholar] [CrossRef]
- Montenegro, I.; Said, B.; Godoy, P.; Besoain, X.; Parra, C.; Díaz, K.; Madrid, A. Antifungal Activity of Essential Oil and Main Components from Mentha pulegium Growing Wild on the Chilean Central Coast. Agronomy 2020, 10, 254. [Google Scholar] [CrossRef] [Green Version]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef] [Green Version]
- Gong, B.; Zhang, C.; Li, X.; Wen, D.; Wang, S.; Shi, Q.; Wang, X. Identification of NaCl and NaHCO3 stress responsive proteins in tomato roots using iTRAO-based analysis. Biochem. Biophys. Res. Commun. 2014, 446, 417–422. [Google Scholar] [CrossRef]
- Duan, M.; Feng, H.-L.; Wang, L.-Y.; Li, D.; Meng, Q.-W. Overexpression of thylakoidal ascorbate peroxidase shows enhanced resistance to chilling stress in tomato. J. Plant Physiol. 2012, 169, 867–877. [Google Scholar] [CrossRef]
- Bardford, M.M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein dye binding. Anal. Biochem. 1976, 72, 248–254. [Google Scholar] [CrossRef]
- Li, K.; Zhong, C.; Shi, Q.; Bi, H.; Gong, B. Cold plasma seed treatment improves chilling resistance of tomato plants through hydrogen peroxide and abscisic acid signaling pathway. Free. Radic. Biol. Med. 2021, 172, 286–297. [Google Scholar] [CrossRef]
- Gawehns, F.; Houterman, P.M.; Ichou, F.A.; Michielse, C.B.; Hijdra, M.; Cornelissen, B.J.C.; Rep, M.; Takken, F.L.W. The Fusarium oxysporum Effector Six6 Contributes to Virulence and Suppresses I-2-Mediated Cell Death. Mol. Plant-Microbe Interact. 2014, 27, 336–348. [Google Scholar] [CrossRef] [Green Version]
- Ding, S.; Zhang, Y.; Hu, Z.; Huang, X.; Zhang, B.; Lu, Q.; Wen, X.; Wang, Y.; Lu, C. mTERF5 Acts as a Transcriptional Pausing Factor to Positively Regulate Transcription of Chloroplast psbEFLJ. Mol. Plant 2019, 12, 1259–1277. [Google Scholar] [CrossRef] [PubMed]
- Lichtenthaler, H.K. Chlorophylls and carotenoids: Pigments of photosynthetic biomembranes. Methods Enzymol. 1987, 148, 350–382. [Google Scholar]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative pcr and the 2(-Delta Delta C(T)) method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]
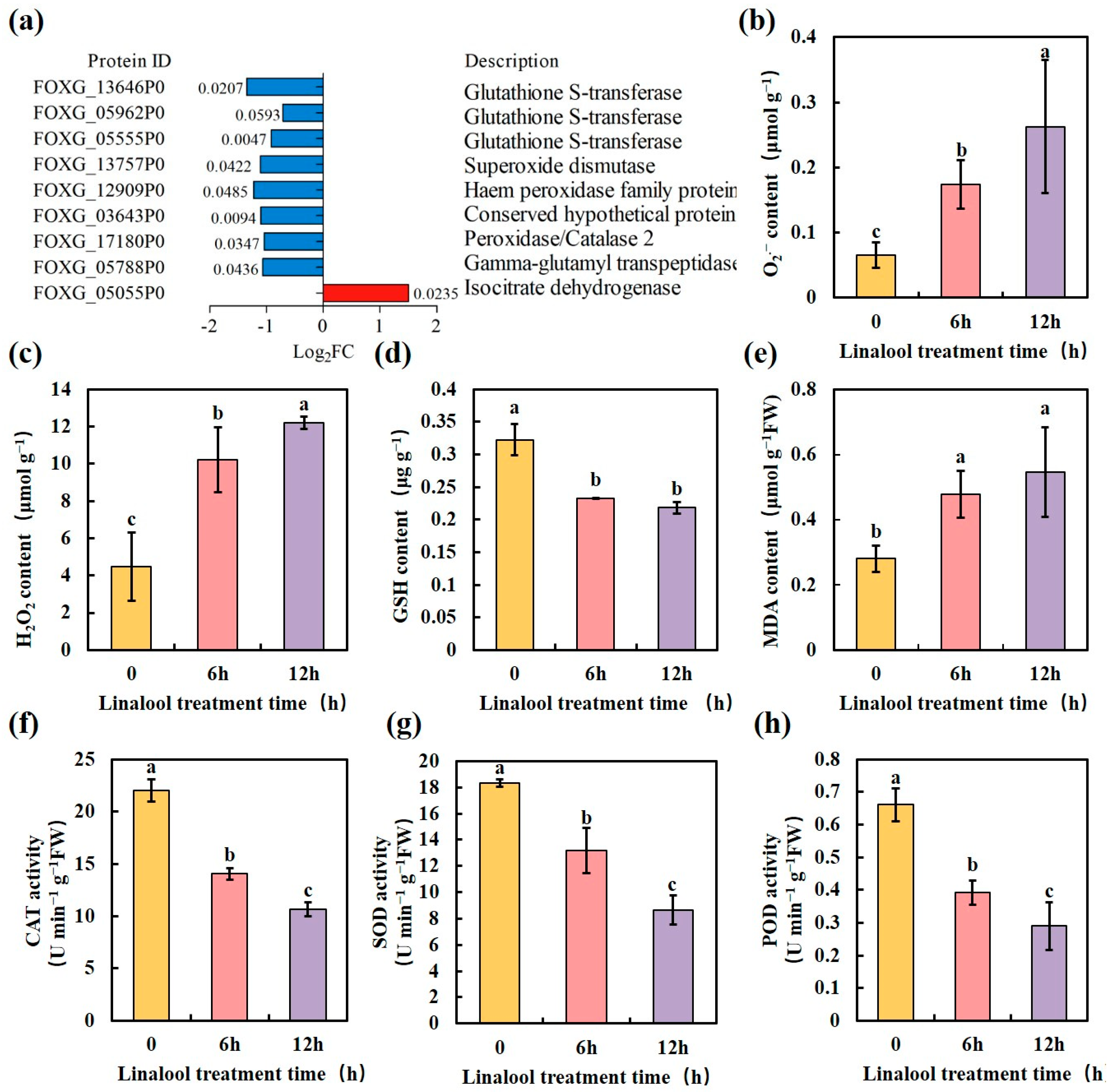
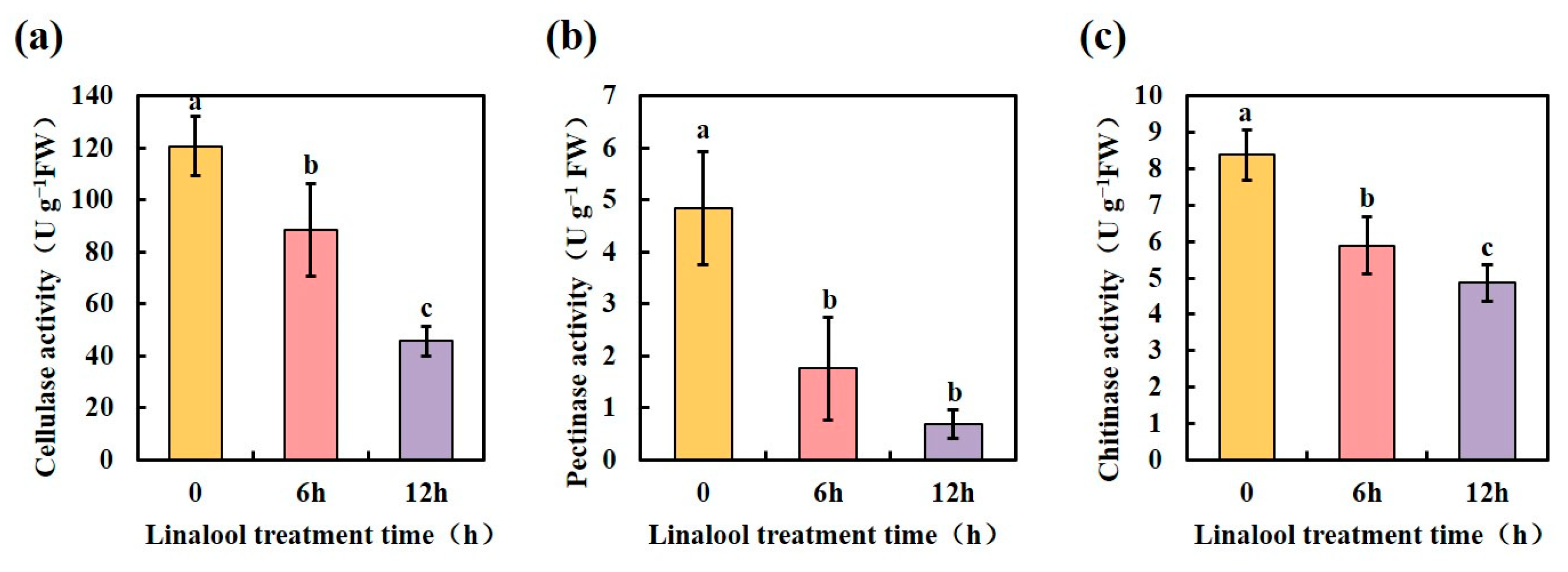
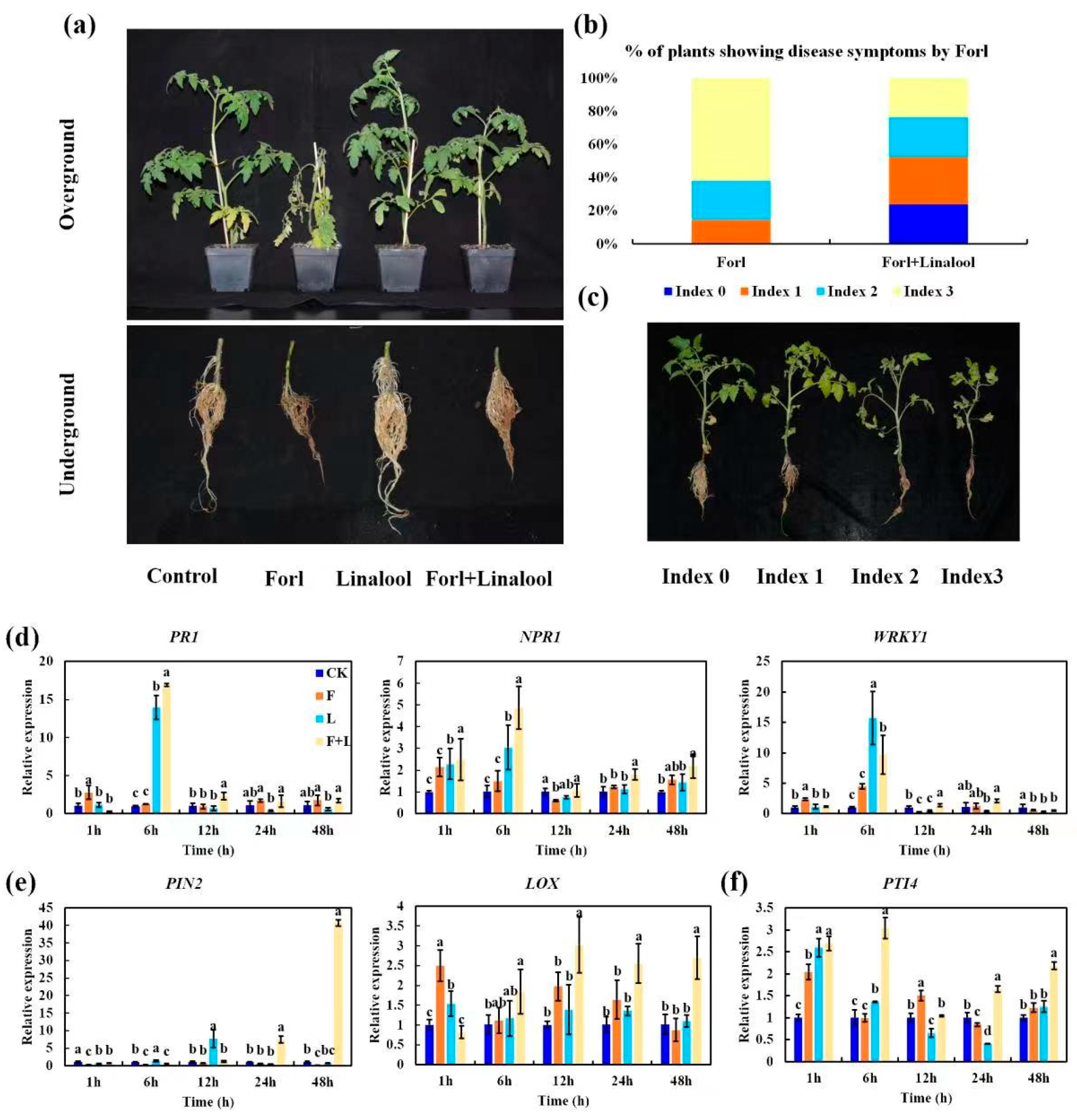

| Protein ID | Log2FC | p-Value | COG_Function_Description |
|---|---|---|---|
| Energy production and conversion | |||
| FOXG_05055P0 | 1.50 | 0.0235 | Isocitrate dehydrogenase |
| FOXG_14637P0 | −1.36 | 0.0205 | Malate/lactate dehydrogenase |
| FOXG_08948P0 | −1.14 | 0.0140 | FAD/FMN-containing dehydrogenase |
| FOXG_03411P0 | 1.56 | 0.0252 | Acyl-CoA reductase or other NAD-dependent aldehyde dehydrogenase |
| FOXG_08681P0 | −1.19 | 0.0128 | Glycerol-3-phosphate dehydrogenase |
| FOXG_02187P0 | −1.03 | 0.0129 | Pyruvate/2-oxoglutarate dehydrogenase complex, dihydrolipoamide acyltransferase (E2) component |
| FOXG_13538P0 | 1.44 | 0.0134 | Trans-aconitate methyltransferase |
| FOXG_02524P0 | 1.44 | 0.0041 | FAD/FMN-containing dehydrogenase |
| Carbohydrate transport and metabolism | |||
| FOXG_15373P0 ** | −1.37 | 0.0050 | Chitinase, GH18 family |
| FOXG_00102P0 | −1.56 | 0.0205 | N-acetyl-beta-hexosaminidase |
| FOXG_14504P0 ** | −1.85 | 0.0018 | Endo-1,4-beta-xylanase, GH35 family |
| FOXG_03723P0 ** | −1.03 | 0.0224 | Exo-beta-1,3-glucanase, GH17 family |
| FOXG_10748P0 ** | −1.48 | 0.0019 | Chitinase, GH18 family |
| FOXG_13407P0 | −1.57 | 0.0268 | 6-phosphogluconolactonase, cycloisomerase 2 family |
| FOXG_02349P0 ** | −1.18 | 0.0175 | Periplasmic beta-glucosidase and related glycosidases |
| FOXG_05841P0 | −1.47 | 0.0394 | D-arabinose 1-dehydrogenase, Zn-dependent alcohol dehydrogenase family |
| FOXG_15329P0 ** | −1.37 | 0.0028 | Chitinase, GH18 family |
| FOXG_10052P0 ** | −1.84 | 0.0010 | Polygalacturonase |
| FOXG_10867P0 | −1.39 | 0.0072 | N-acetyl-beta-hexosaminidase |
| FOXG_05948P0 ** | −1.76 | 0.0018 | Pectate lyase |
| FOXG_03195P0 | −1.47 | 0.0101 | 6-phosphogluconolactonase/Glucosamine-6-phosphate isomerase/deaminase |
| FOXG_06401P0 | −1.03 | 0.0023 | Mannose-6-phosphate isomerase, class I |
| FOXG_03963P0 | −1.11 | 0.0266 | 6-phosphogluconolactonase, cycloisomerase 2 family |
| FOXG_07873P0 * | −1.20 | 0.0102 | Na+/melibiose symporter or related transporter |
| FOXG_11081P0 ** | −2.07 | 0.0462 | Aryl-phospho-beta-D-glucosidase BglC, GH1 family |
| FOXG_03194P0 * | −1.04 | 0.0076 | Hexokinase |
| FOXG_03843P0 * | −1.04 | 0.0278 | Predicted arabinose efflux permease, MFS family |
| FOXG_08305P0 | −1.79 | 0.0230 | Predicted alpha-1,6-mannanase, GH76 family |
| FOXG_15351P0 | −2.38 | 0.0087 | Alpha-glucosidase, glycosyl hydrolase family GH31 |
| FOXG_08602P0 ** | −1.33 | 0.0233 | Oxalate decarboxylase/archaeal phosphoglucose isomerase, cupin superfamily |
| FOXG_06388P0 * | −1.40 | 0.0065 | Predicted arabinose efflux permease, MFS family |
| FOXG_10189P0 | −1.45 | 0.0183 | TPP-dependent 2-oxoacid decarboxylase, includes indolepyruvate decarboxylase |
| Lipid transport and metabolism | |||
| FOXG_15474P0 | −1.65 | 0.0188 | Glycerophosphoryl diester phosphodiesterase |
| FOXG_12721P0 | −1.11 | 0.0027 | Acyl-CoA dehydrogenase related to the alkylation response protein AidB |
| FOXG_09686P0 | −1.37 | 0.0057 | Carboxylesterase type B |
| FOXG_13474P0 | −1.29 | 0.0035 | Carboxylesterase type B |
| FOXG_10416P0 | −1.75 | 0.0017 | Lysophospholipase, alpha-beta hydrolase superfamily |
| FOXG_11716P0 | 1.05 | 0.0022 | Cyclopropane fatty-acyl-phospholipid synthase and related methyltransferases |
| FOXG_08523P0 | 1.09 | 0.0482 | Fatty-acid desaturase |
| FOXG_11948P0 | −1.36 | 0.0200 | Lysophospholipase, alpha-beta hydrolase superfamily |
| FOXG_05822P0 | −1.25 | 0.0011 | Lysophospholipase, alpha-beta hydrolase superfamily |
| FOXG_12687P0 | 1.08 | 0.0109 | Phosphatidate phosphatase APP1 |
| FOXG_13507P0 | 2.35 | 0.0109 | Acyl CoA:acetate/3-ketoacid CoA transferase, beta subunit |
| Pathogenicity-associated proteins | |||
| FOXG_06095P0 * | −0.79 | 0.0098 | Arylamine N-acetyltransferase 2 |
| FOXG_01310P0 * | −0.53 | 0.0338 | GTPase RHO3 |
| FOXG_07946P0 * | −0.77 | 0.0424 | GTPase-activating protein GYP7 |
| FOXG_12808P0 * | −0.59 | 0.0145 | GTP-binding protein ypt1 |
| FOXG_13835P0 * | −0.70 | 0.0464 | Small GTPase-binding protein |
| FOXG_01420P0 * | −0.56 | 0.0137 | GTPase SAR1 |
| FOXG_09867P0 * | −1.48 | 0.0024 | Glutathione-dependent formaldehyde-activating enzyme |
| FOXG_06321P0 * | −0.56 | 0.0450 | G-protein alpha subunit |
| FOXG_10547P0 * | −0.89 | 0.0111 | Lipase ATG15 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, X.; Wang, Q.; Li, H.; Wang, X.; Zhang, R.; Yang, X.; Jiang, Q.; Shi, Q. Revealing the Mechanisms for Linalool Antifungal Activity against Fusarium oxysporum and Its Efficient Control of Fusarium Wilt in Tomato Plants. Int. J. Mol. Sci. 2023, 24, 458. https://doi.org/10.3390/ijms24010458
Li X, Wang Q, Li H, Wang X, Zhang R, Yang X, Jiang Q, Shi Q. Revealing the Mechanisms for Linalool Antifungal Activity against Fusarium oxysporum and Its Efficient Control of Fusarium Wilt in Tomato Plants. International Journal of Molecular Sciences. 2023; 24(1):458. https://doi.org/10.3390/ijms24010458
Chicago/Turabian StyleLi, Xiuming, Qifang Wang, Haosen Li, Xiaoyun Wang, Ruimin Zhang, Xiaoyu Yang, Qiwei Jiang, and Qinghua Shi. 2023. "Revealing the Mechanisms for Linalool Antifungal Activity against Fusarium oxysporum and Its Efficient Control of Fusarium Wilt in Tomato Plants" International Journal of Molecular Sciences 24, no. 1: 458. https://doi.org/10.3390/ijms24010458
APA StyleLi, X., Wang, Q., Li, H., Wang, X., Zhang, R., Yang, X., Jiang, Q., & Shi, Q. (2023). Revealing the Mechanisms for Linalool Antifungal Activity against Fusarium oxysporum and Its Efficient Control of Fusarium Wilt in Tomato Plants. International Journal of Molecular Sciences, 24(1), 458. https://doi.org/10.3390/ijms24010458







