AML1-ETO-Related Fusion Circular RNAs Contribute to the Proliferation of Leukemia Cells
Abstract
:1. Introduction
2. Results
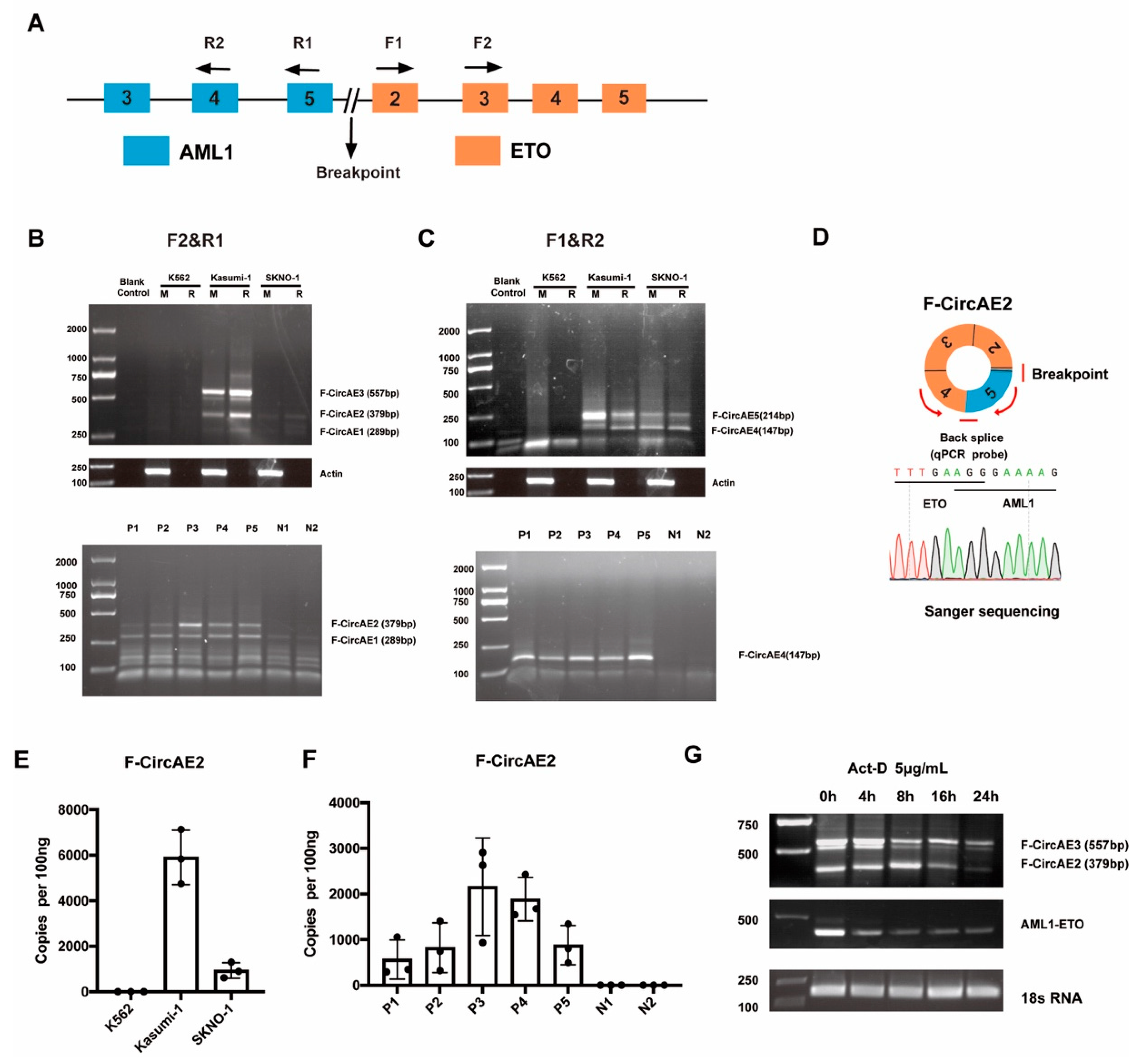
2.1. Identification of AML1-ETO Fusion Circular RNAs in AML1-ETO Positive Cell Lines and Primary AML Patients
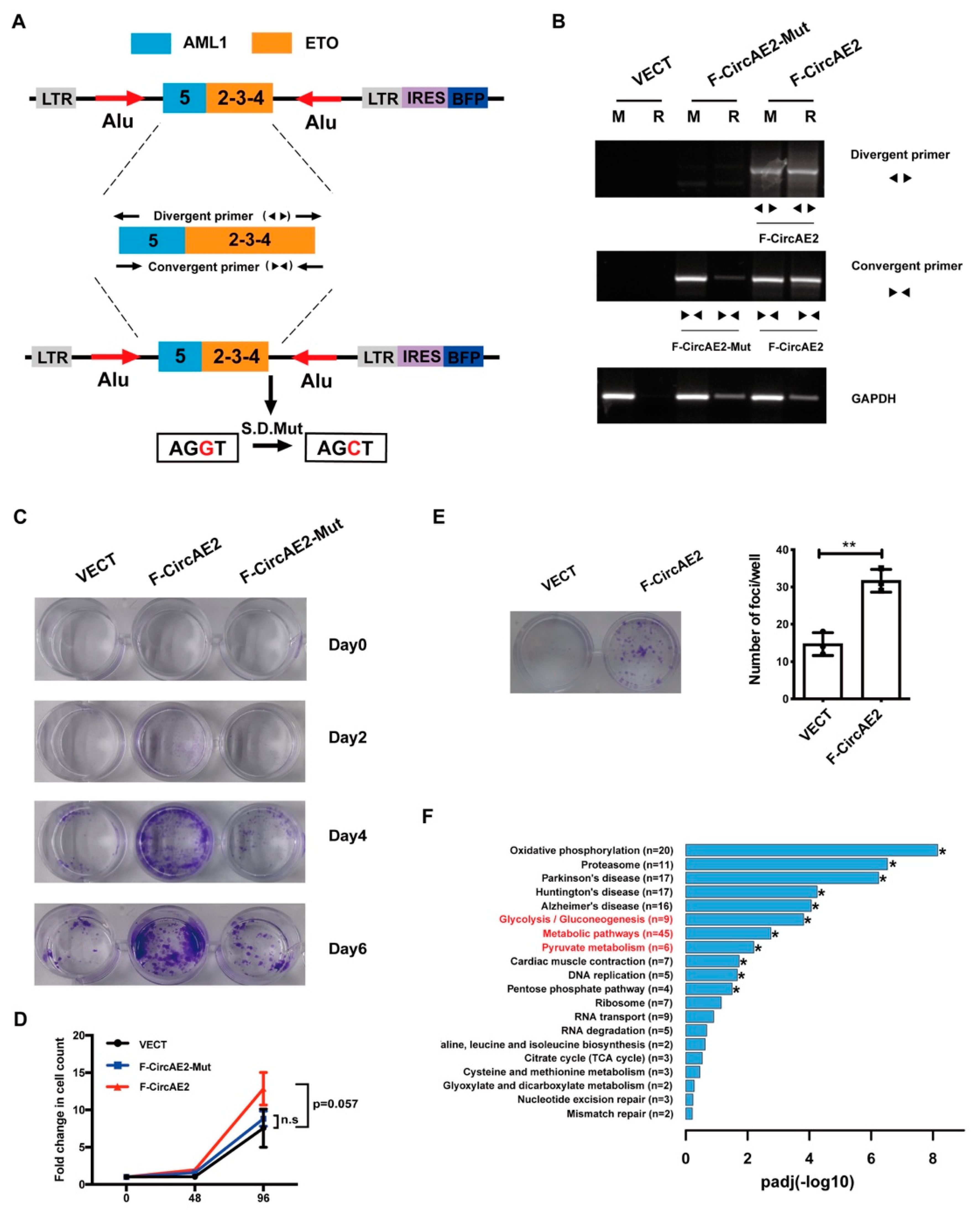
2.2. Over-Expression of F-CircAE2 Promoted the Proliferation of NIH3T3 Cells In Vitro
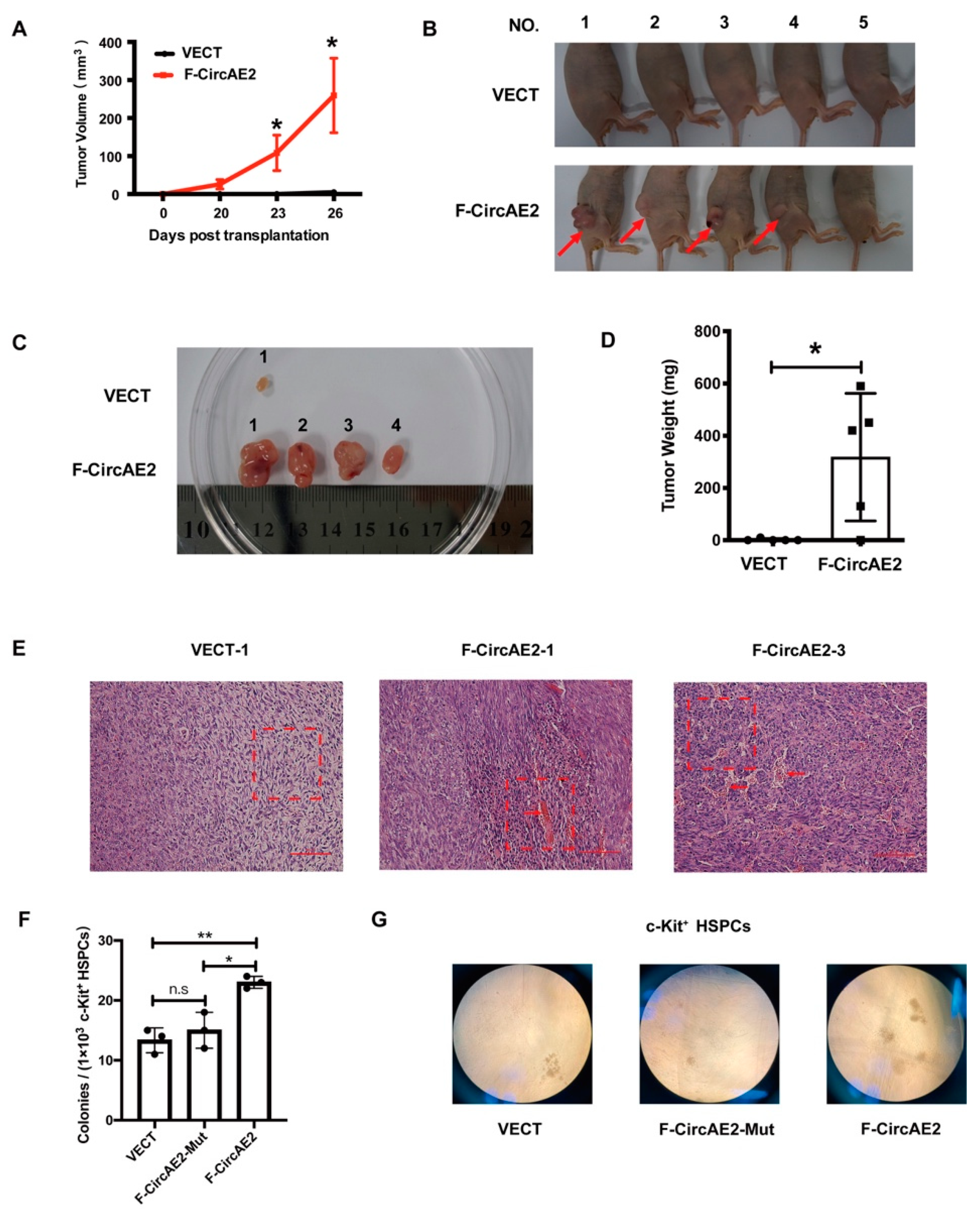
2.3. F-CircAE2 Promoted Tumorigenesis of NIH3T3 Cells In Vivo
2.4. F-CircAE2 Increased the Clonogenic Ability of c-Kit+ HSPCs
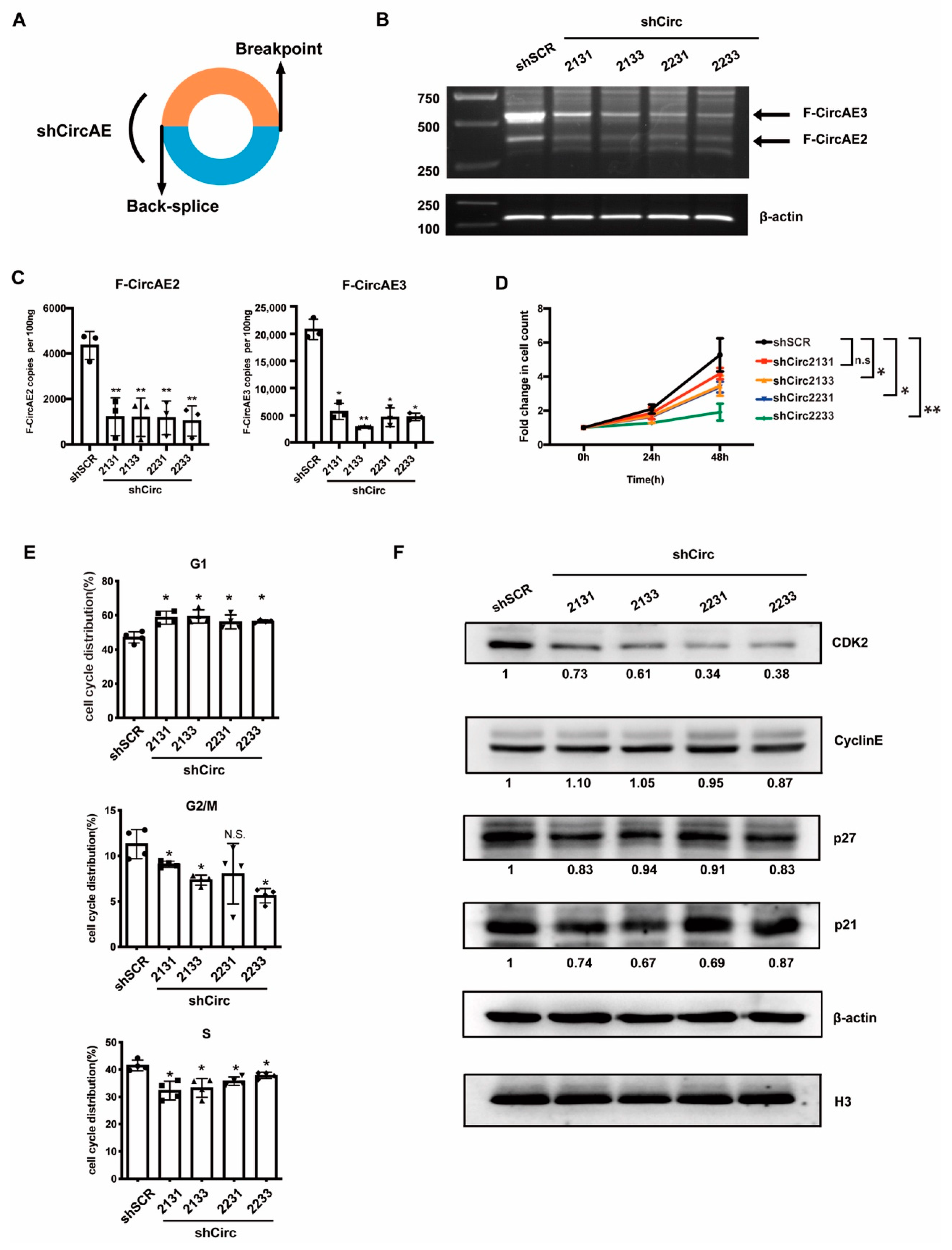
2.5. Endogenous F-CircAEs Expression Was Essential for the Maintenance of AML1-ETO-Positive Leukemia Cells
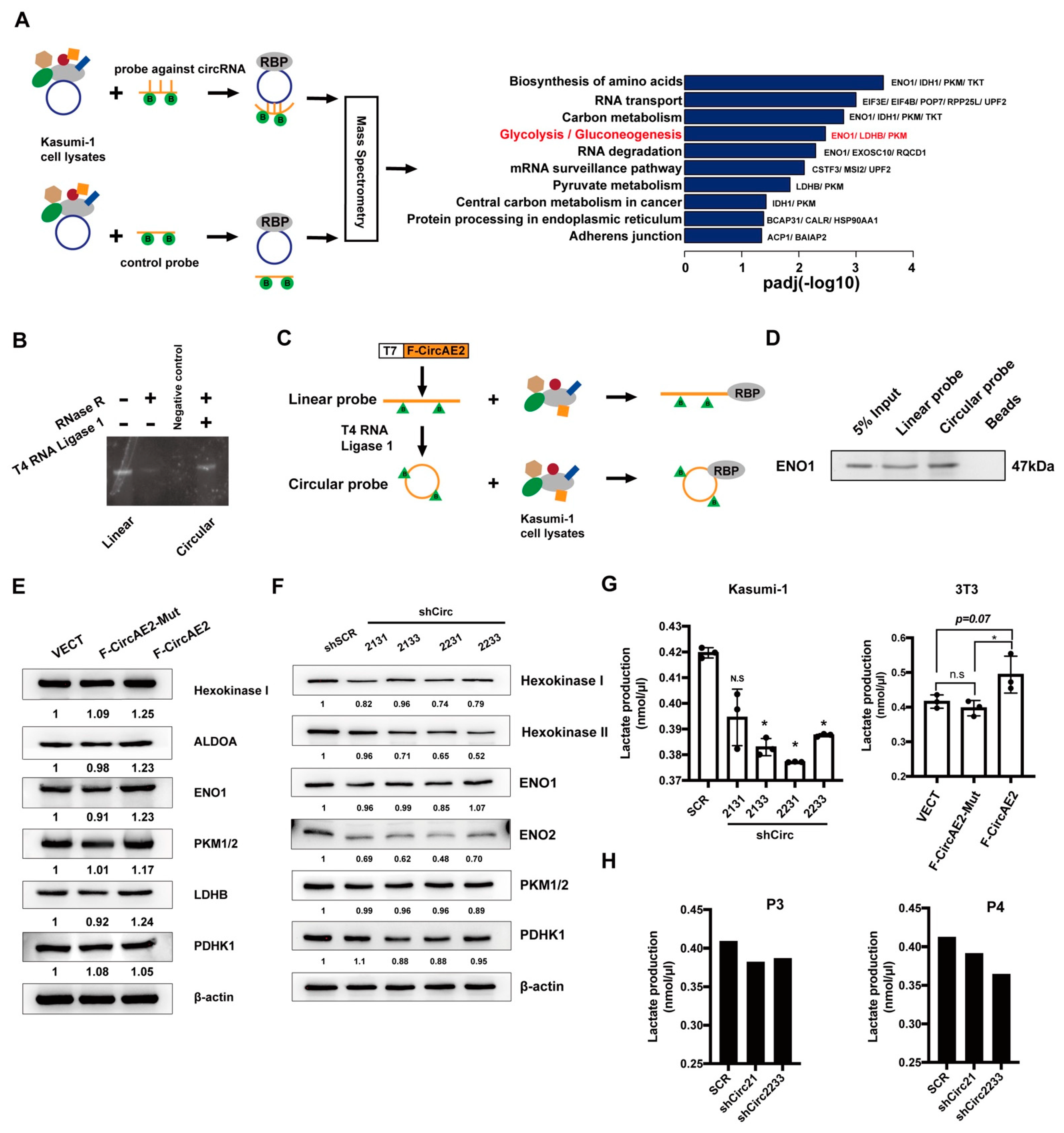
2.6. F-CircAE2 Bound to Proteins Involved in the Glycolysis Pathway and Promoted Aerobic Glycolysis of Leukemia Cells
3. Discussion
4. Materials and Methods
4.1. Cells and Culture Conditions, Actinomycin D Treatment
4.2. RNA Preparation, RNase-R Treatment, and PCR
4.3. F-CircAE2 and F-CircAE3 Copy Number Measurement
4.4. Proliferation Assay
4.5. NIH3T3 Growth and Foci Formation Assays, and Crystal Violet Staining
4.6. Tumorigenicity in Immunodeficient Mice
4.7. RNA Pulldown Assay
4.8. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Zhang, Y.; Strissel, P.; Strick, R.; Chen, J.; Nucifora, G.; Le Beau, M.M.; Larson, R.A.; Rowley, J.D. Genomic DNA breakpoints in AML1/RUNX1 and ETO cluster with topoisomerase II DNA cleavage and DNase I hypersensitive sites in t(8;21) leukemia. Proc. Natl. Acad. Sci. USA 2002, 99, 3070–3075. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, J.; Hoshino, T.; Redner, R.L.; Kajigaya, S.; Liu, J.M. ETO, fusion partner in t(8;21) acute myeloid leukemia, represses transcription by interaction with the human N-CoR/mSin3/HDAC1 complex. Proc. Natl. Acad. Sci. USA 1998, 95, 10860–10865. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liang, D.; Wilusz, J.E. Short intronic repeat sequences facilitate circular RNA production. Genes Dev. 2014, 28, 2233–2247. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, X.O.; Wang, H.B.; Zhang, Y.; Lu, X.; Chen, L.L.; Yang, L. Complementary sequence-mediated exon circularization. Cell 2014, 159, 134–147. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Barrett, S.P.; Wang, P.L.; Salzman, J. Circular RNA biogenesis can proceed through an exon-containing lariat precursor. Elife 2015, 4, e07540. [Google Scholar] [CrossRef]
- Hsu, M.T.; Coca-Prados, M. Electron microscopic evidence for the circular form of RNA in the cytoplasm of eukaryotic cells. Nature 1979, 280, 339–340. [Google Scholar] [CrossRef]
- Cocquerelle, C.; Mascrez, B.; Hetuin, D.; Bailleul, B. Mis-splicing yields circular RNA molecules. FASEB J. 1993, 7, 155–160. [Google Scholar] [CrossRef] [Green Version]
- Guo, J.U.; Agarwal, V.; Guo, H.; Bartel, D.P. Expanded identification and characterization of mammalian circular RNAs. Genome Biol. 2014, 15, 409. [Google Scholar] [CrossRef]
- Bachmayr-Heyda, A.; Reiner, A.T.; Auer, K.; Sukhbaatar, N.; Aust, S.; Bachleitner-Hofmann, T.; Mesteri, I.; Grunt, T.W.; Zeillinger, R.; Pils, D. Correlation of circular RNA abundance with proliferation--exemplified with colorectal and ovarian cancer, idiopathic lung fibrosis, and normal human tissues. Sci. Rep. 2015, 5, 8057. [Google Scholar] [CrossRef] [Green Version]
- Vo, J.N.; Cieslik, M.; Zhang, Y.; Shukla, S.; Xiao, L.; Zhang, Y.; Wu, Y.M.; Dhanasekaran, S.M.; Engelke, C.G.; Cao, X.; et al. The Landscape of Circular RNA in Cancer. Cell 2019, 176, 869–881.e13. [Google Scholar] [CrossRef]
- Hansen, T.B.; Jensen, T.I.; Clausen, B.H.; Bramsen, J.B.; Finsen, B.; Damgaard, C.K.; Kjems, J. Natural RNA circles function as efficient microRNA sponges. Nature 2013, 495, 384–388. [Google Scholar] [CrossRef] [PubMed]
- Du, W.W.; Yang, W.; Liu, E.; Yang, Z.; Dhaliwal, P.; Yang, B.B. Foxo3 circular RNA retards cell cycle progression via forming ternary complexes with p21 and CDK2. Nucleic Acids Res. 2016, 44, 2846–2858. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Salzman, J.; Gawad, C.; Wang, P.L.; Lacayo, N.; Brown, P.O. Circular RNAs are the predominant transcript isoform from hundreds of human genes in diverse cell types. PLoS ONE 2012, 7, e30733. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Memczak, S.; Jens, M.; Elefsinioti, A.; Torti, F.; Krueger, J.; Rybak, A.; Maier, L.; Mackowiak, S.D.; Gregersen, L.H.; Munschauer, M.; et al. Circular RNAs are a large class of animal RNAs with regulatory potency. Nature 2013, 495, 333–338. [Google Scholar] [CrossRef]
- Alhasan, A.A.; Izuogu, O.G.; Al-Balool, H.H.; Steyn, J.S.; Evans, A.; Colzani, M.; Ghevaert, C.; Mountford, J.C.; Marenah, L.; Elliott, D.J.; et al. Circular RNA enrichment in platelets is a signature of transcriptome degradation. Blood 2016, 127, e1–e11. [Google Scholar] [CrossRef]
- Li, W.; Zhong, C.; Jiao, J.; Li, P.; Cui, B.; Ji, C.; Ma, D. Characterization of hsa_circ_0004277 as a New Biomarker for Acute Myeloid Leukemia via Circular RNA Profile and Bioinformatics Analysis. Int. J. Mol. Sci. 2017, 18, 597. [Google Scholar] [CrossRef] [Green Version]
- Hirsch, S.; Blatte, T.J.; Grasedieck, S.; Cocciardi, S.; Rouhi, A.; Jongen-Lavrencic, M.; Paschka, P.; Kronke, J.; Gaidzik, V.I.; Dohner, H.; et al. Circular RNAs of the nucleophosmin (NPM1) gene in acute myeloid leukemia. Haematologica 2017, 102, 2039–2047. [Google Scholar] [CrossRef]
- Nicolet, B.P.; Engels, S.; Aglialoro, F.; van den Akker, E.; von Lindern, M.; Wolkers, M.C. Circular RNA expression in human hematopoietic cells is widespread and cell-type specific. Nucleic Acids Res. 2018, 46, 8168–8180. [Google Scholar] [CrossRef]
- Bonizzato, A.; Gaffo, E.; Te Kronnie, G.; Bortoluzzi, S. CircRNAs in hematopoiesis and hematological malignancies. Blood Cancer J. 2016, 6, e483. [Google Scholar] [CrossRef]
- Guarnerio, J.; Bezzi, M.; Jeong, J.C.; Paffenholz, S.V.; Berry, K.; Naldini, M.M.; Lo-Coco, F.; Tay, Y.; Beck, A.H.; Pandolfi, P.P. Oncogenic Role of Fusion-circRNAs Derived from Cancer-Associated Chromosomal Translocations. Cell 2016, 165, 289–302. [Google Scholar] [CrossRef]
- Smit, A.; Hubley, R.; Green, P. RepeatMasker Open-4.0. Available online: http://www.repeatmasker.or (accessed on 30 April 2017).
- Li, X.; Yang, L.; Chen, L.L. The Biogenesis, Functions, and Challenges of Circular RNAs. Mol. Cell 2018, 71, 428–442. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Subramanian, A.; Tamayo, P.; Mootha, V.K.; Mukherjee, S.; Ebert, B.L.; Gillette, M.A.; Paulovich, A.; Pomeroy, S.L.; Golub, T.R.; Lander, E.S.; et al. Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. USA 2005, 102, 15545–15550. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ganapathy-Kanniappan, S.; Geschwind, J.F. Tumor glycolysis as a target for cancer therapy: Progress and prospects. Mol. Cancer 2013, 12, 152. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zang, J.; Lu, D.; Xu, A. The interaction of circRNAs and RNA binding proteins: An important part of circRNA maintenance and function. J. Neurosci. Res. 2020, 98, 87–97. [Google Scholar] [CrossRef] [Green Version]
- Du, W.W.; Fang, L.; Yang, W.; Wu, N.; Awan, F.M.; Yang, Z.; Yang, B.B. Induction of tumor apoptosis through a circular RNA enhancing Foxo3 activity. Cell Death Differ. 2017, 24, 357–370. [Google Scholar] [CrossRef]
- Chen, E.Y.; Tan, C.M.; Kou, Y.; Duan, Q.; Wang, Z.; Meirelles, G.V.; Clark, N.R.; Ma’ayan, A. Enrichr: Interactive and collaborative HTML5 gene list enrichment analysis tool. BMC Bioinform. 2013, 14, 128. [Google Scholar] [CrossRef] [Green Version]
- Petkovic, S.; Muller, S. RNA circularization strategies in vivo and in vitro. Nucleic Acids Res. 2015, 43, 2454–2465. [Google Scholar] [CrossRef] [Green Version]
- Li, H.; Yang, F.; Hu, A.; Wang, X.; Fang, E.; Chen, Y.; Li, D.; Song, H.; Wang, J.; Guo, Y.; et al. Therapeutic targeting of circ-CUX1/EWSR1/MAZ axis inhibits glycolysis and neuroblastoma progression. EMBO Mol. Med. 2019, 11, e10835. [Google Scholar] [CrossRef]
- Singh, V.; Uddin, M.H.; Zonder, J.A.; Azmi, A.S.; Balasubramanian, S.K. Circular RNAs in acute myeloid leukemia. Mol. Cancer 2021, 20, 149. [Google Scholar] [CrossRef]
- Zhou, J.; Zhou, L.Y.; Tang, X.; Zhang, J.; Zhai, L.L.; Yi, Y.Y.; Yi, J.; Lin, J.; Qian, J.; Deng, Z.Q. Circ-Foxo3 is positively associated with the Foxo3 gene and leads to better prognosis of acute myeloid leukemia patients. BMC Cancer 2019, 19, 930. [Google Scholar] [CrossRef]
- Yi, Y.Y.; Yi, J.; Zhu, X.; Zhang, J.; Zhou, J.; Tang, X.; Lin, J.; Wang, P.; Deng, Z.Q. Circular RNA of vimentin expression as a valuable predictor for acute myeloid leukemia development and prognosis. J. Cell Physiol. 2019, 234, 3711–3719. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Liu, X.; Cai, M.; Luo, A.; He, Y.; Liu, S.; Zhang, X.; Yang, X.; Xu, L.; Jiang, H. CircRNF220, not its linear cognate gene RNF220, regulates cell growth and is associated with relapse in pediatric acute myeloid leukemia. Mol. Cancer 2021, 20, 139. [Google Scholar] [CrossRef] [PubMed]
- Sun, Y.M.; Wang, W.T.; Zeng, Z.C.; Chen, T.Q.; Han, C.; Pan, Q.; Huang, W.; Fang, K.; Sun, L.Y.; Zhou, Y.F.; et al. circMYBL2, a circRNA from MYBL2, regulates FLT3 translation by recruiting PTBP1 to promote FLT3-ITD AML progression. Blood 2019, 134, 1533–1546. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tan, S.; Gou, Q.; Pu, W.; Guo, C.; Yang, Y.; Wu, K.; Liu, Y.; Liu, L.; Wei, Y.Q.; Peng, Y. Circular RNA F-circEA produced from EML4-ALK fusion gene as a novel liquid biopsy biomarker for non-small cell lung cancer. Cell Res. 2018, 28, 693–695. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tan, S.; Sun, D.; Pu, W.; Gou, Q.; Guo, C.; Gong, Y.; Li, J.; Wei, Y.Q.; Liu, L.; Zhao, Y.; et al. Circular RNA F-circEA-2a derived from EML4-ALK fusion gene promotes cell migration and invasion in non-small cell lung cancer. Mol. Cancer 2018, 17, 138. [Google Scholar] [CrossRef] [Green Version]
- Yang, F.; Fang, E.; Mei, H.; Chen, Y.; Li, H.; Li, D.; Song, H.; Wang, J.; Hong, M.; Xiao, W.; et al. Cis-Acting circ-CTNNB1 Promotes beta-Catenin Signaling and Cancer Progression via DDX3-Mediated Transactivation of YY1. Cancer Res. 2019, 79, 557–571. [Google Scholar] [CrossRef] [Green Version]
- Koppenol, W.H.; Bounds, P.L.; Dang, C.V. Otto Warburg’s contributions to current concepts of cancer metabolism. Nat. Rev. Cancer 2011, 11, 325–337. [Google Scholar] [CrossRef]
- Peng, S.Y.; Lai, P.L.; Pan, H.W.; Hsiao, L.P.; Hsu, H.C. Aberrant expression of the glycolytic enzymes aldolase B and type II hexokinase in hepatocellular carcinoma are predictive markers for advanced stage, early recurrence and poor prognosis. Oncol. Rep. 2008, 19, 1045–1053. [Google Scholar] [CrossRef]
- Hwang, T.L.; Liang, Y.; Chien, K.Y.; Yu, J.S. Overexpression and elevated serum levels of phosphoglycerate kinase 1 in pancreatic ductal adenocarcinoma. Proteomics 2006, 6, 2259–2272. [Google Scholar] [CrossRef]
- Chen, E.I. Mitochondrial dysfunction and cancer metastasis. J. Bioenerg. Biomembr. 2012, 44, 619–622. [Google Scholar] [CrossRef]
- Boag, J.M.; Beesley, A.H.; Firth, M.J.; Freitas, J.R.; Ford, J.; Hoffmann, K.; Cummings, A.J.; de Klerk, N.H.; Kees, U.R. Altered glucose metabolism in childhood pre-B acute lymphoblastic leukaemia. Leukemia 2006, 20, 1731–1737. [Google Scholar] [CrossRef]
- Herst, P.M.; Howman, R.A.; Neeson, P.J.; Berridge, M.V.; Ritchie, D.S. The level of glycolytic metabolism in acute myeloid leukemia blasts at diagnosis is prognostic for clinical outcome. J. Leukoc. Biol. 2011, 89, 51–55. [Google Scholar] [CrossRef] [PubMed]
- Song, K.; Li, M.; Xu, X.J.; Xuan, L.; Huang, G.N.; Song, X.L.; Liu, Q.F. HIF-1alpha and GLUT1 gene expression is associated with chemoresistance of acute myeloid leukemia. Asian Pac. J. Cancer Prev. 2014, 15, 1823–1829. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhou, J.; Zhang, S.; Chen, Z.; He, Z.; Xu, Y.; Li, Z. CircRNA-ENO1 promoted glycolysis and tumor progression in lung adenocarcinoma through upregulating its host gene ENO1. Cell Death Dis. 2019, 10, 885. [Google Scholar] [CrossRef] [Green Version]
- Singh, S.; Narang, A.S.; Mahato, R.I. Subcellular fate and off-target effects of siRNA, shRNA, and miRNA. Pharm. Res. 2011, 28, 2996–3015. [Google Scholar] [CrossRef]
- Reischl, D.; Zimmer, A. Drug delivery of siRNA therapeutics: Potentials and limits of nanosystems. Nanomedicine 2009, 5, 8–20. [Google Scholar] [CrossRef] [PubMed]
- He, A.T.; Liu, J.; Li, F.; Yang, B.B. Targeting circular RNAs as a therapeutic approach: Current strategies and challenges. Signal Transduct. Target Ther. 2021, 6, 185. [Google Scholar] [CrossRef]
- Robbins, M.; Judge, A.; MacLachlan, I. siRNA and innate immunity. Oligonucleotides 2009, 19, 89–102. [Google Scholar] [CrossRef]
- Zhang, C.; Huo, S.T.; Wu, Z.; Chen, L.; Wen, C.; Chen, H.; Du, W.W.; Wu, N.; Guan, D.; Lian, S.; et al. Rapid Development of Targeting circRNAs in Cardiovascular Diseases. Mol. Ther. Nucleic Acids 2020, 21, 568–576. [Google Scholar] [CrossRef]
- Liu, C.; Ge, H.M.; Liu, B.H.; Dong, R.; Shan, K.; Chen, X.; Yao, M.D.; Li, X.M.; Yao, J.; Zhou, R.M.; et al. Targeting pericyte-endothelial cell crosstalk by circular RNA-cPWWP2A inhibition aggravates diabetes-induced microvascular dysfunction. Proc. Natl. Acad. Sci. USA 2019, 116, 7455–7464. [Google Scholar] [CrossRef]
- Zheng, Q.; Bao, C.; Guo, W.; Li, S.; Chen, J.; Chen, B.; Luo, Y.; Lyu, D.; Li, Y.; Shi, G.; et al. Circular RNA profiling reveals an abundant circHIPK3 that regulates cell growth by sponging multiple miRNAs. Nat. Commun. 2016, 7, 11215. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, Y.; Nguyen, T.M.; Zhang, X.O.; Wang, L.; Phan, T.; Clohessy, J.G.; Pandolfi, P.P. Optimized RNA-targeting CRISPR/Cas13d technology outperforms shRNA in identifying functional circRNAs. Genome Biol. 2021, 22, 41. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Zhang, Q.; Zhang, D.E.; Zhou, C.; Xing, H.; Tian, Z.; Rao, Q.; Wang, M.; Wang, J. Overexpression of an isoform of AML1 in acute leukemia and its potential role in leukemogenesis. Leukemia 2009, 23, 739–745. [Google Scholar] [CrossRef] [PubMed]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, Y.; Liu, Y.; Xu, Y.; Xing, H.; Tian, Z.; Tang, K.; Rao, Q.; Wang, M.; Wang, J. AML1-ETO-Related Fusion Circular RNAs Contribute to the Proliferation of Leukemia Cells. Int. J. Mol. Sci. 2023, 24, 71. https://doi.org/10.3390/ijms24010071
Wang Y, Liu Y, Xu Y, Xing H, Tian Z, Tang K, Rao Q, Wang M, Wang J. AML1-ETO-Related Fusion Circular RNAs Contribute to the Proliferation of Leukemia Cells. International Journal of Molecular Sciences. 2023; 24(1):71. https://doi.org/10.3390/ijms24010071
Chicago/Turabian StyleWang, Ying, Yu Liu, Yingxi Xu, Haiyan Xing, Zheng Tian, Kejing Tang, Qing Rao, Min Wang, and Jianxiang Wang. 2023. "AML1-ETO-Related Fusion Circular RNAs Contribute to the Proliferation of Leukemia Cells" International Journal of Molecular Sciences 24, no. 1: 71. https://doi.org/10.3390/ijms24010071
APA StyleWang, Y., Liu, Y., Xu, Y., Xing, H., Tian, Z., Tang, K., Rao, Q., Wang, M., & Wang, J. (2023). AML1-ETO-Related Fusion Circular RNAs Contribute to the Proliferation of Leukemia Cells. International Journal of Molecular Sciences, 24(1), 71. https://doi.org/10.3390/ijms24010071




