Expression and Function of StAR in Cancerous and Non-Cancerous Human and Mouse Breast Tissues: New Insights into Diagnosis and Treatment of Hormone-Sensitive Breast Cancer
Abstract
:1. Introduction
2. Results
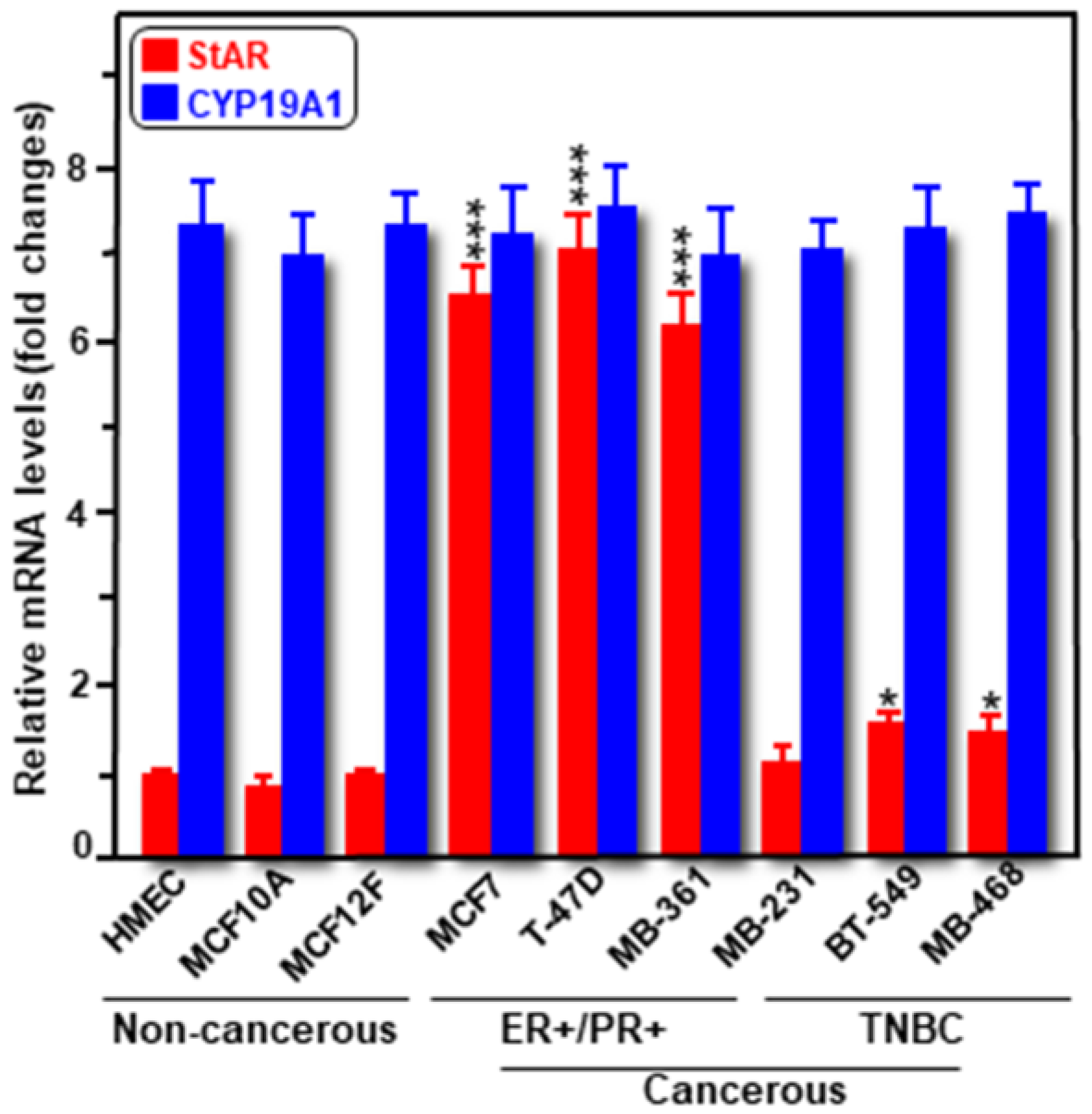
2.1. Expression of StAR and CYP19A1 mRNA and Protein Levels in Cancerous and Non-Cancerous Human Breast Cell Lines
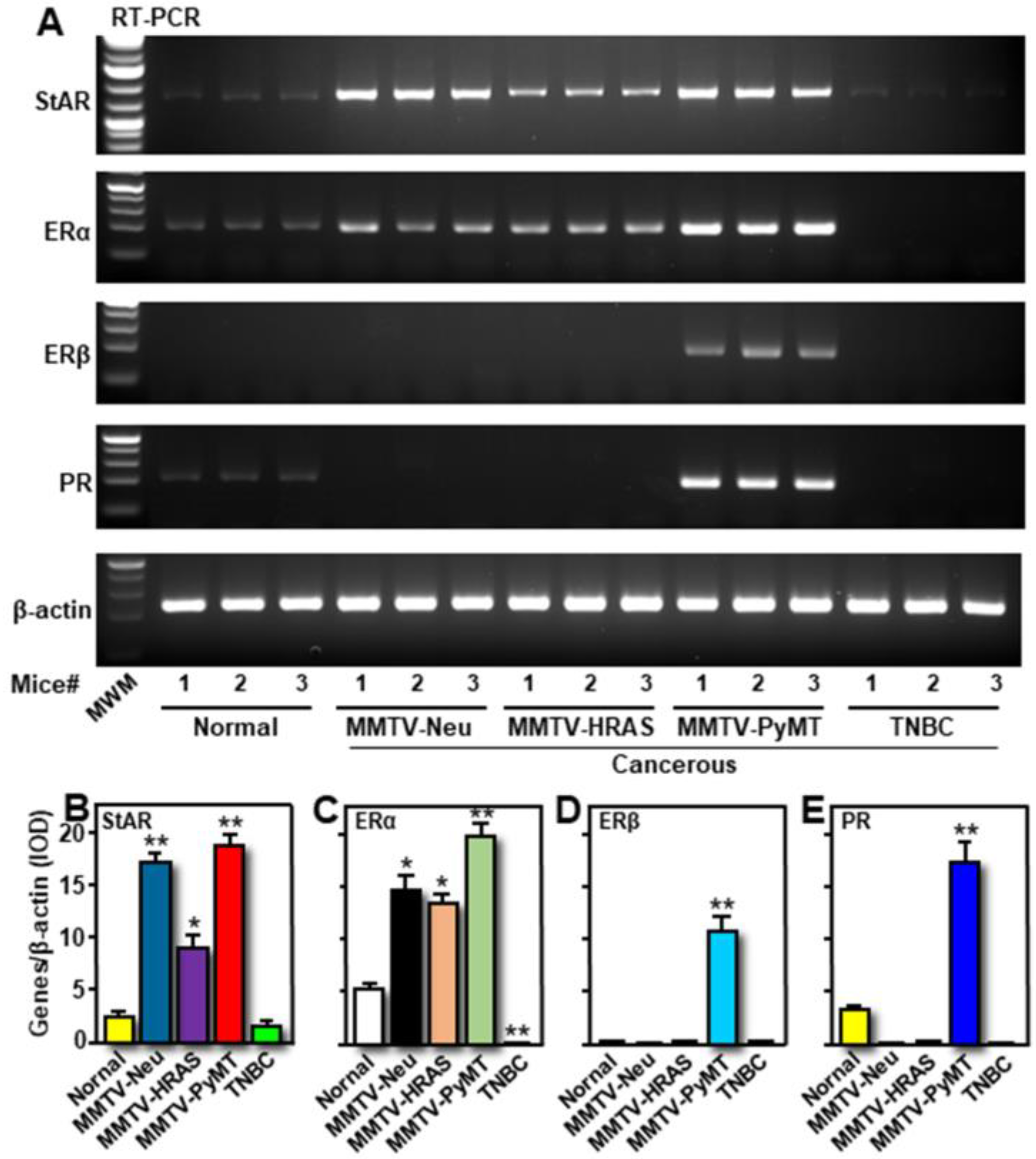
2.2. Differential Expression of StAR, ERα, ERβ, and PR mRNAs, and E2 Levels in Transgenic Mouse Models of Breast Tumors
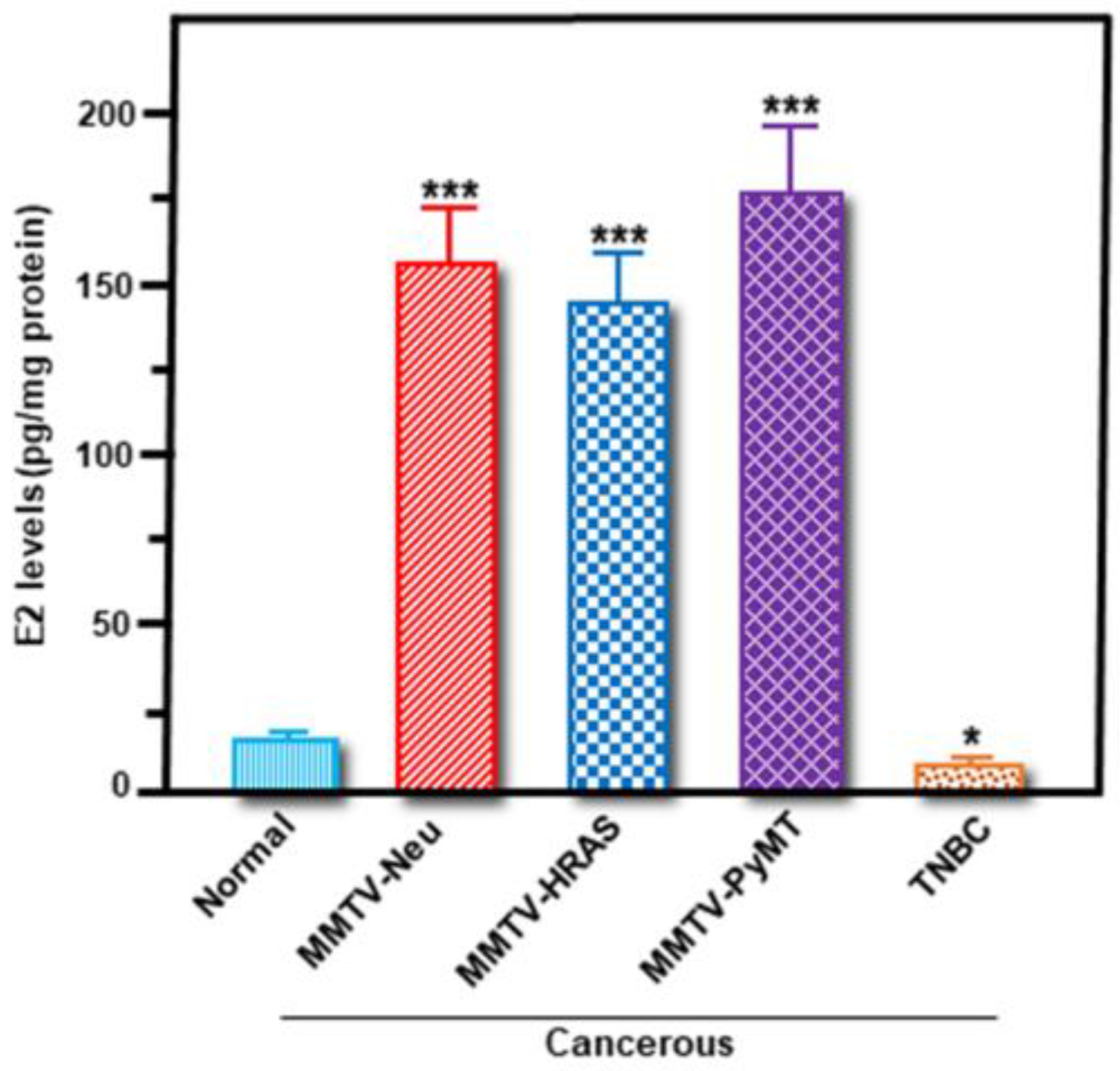
2.3. Functional Relevance of StAR and Aromatase Expression, and Their Correlation to E2 Accumulation, in Breast Tumor Tissue and Plasma of MMTV-PyMT Mice
2.4. Inhibition of a Variety of HDACs on StAR and E2 Levels in Primary Cultures of Enriched Breast Tumor Epithelial Cells from MMTV-PyMT Mice
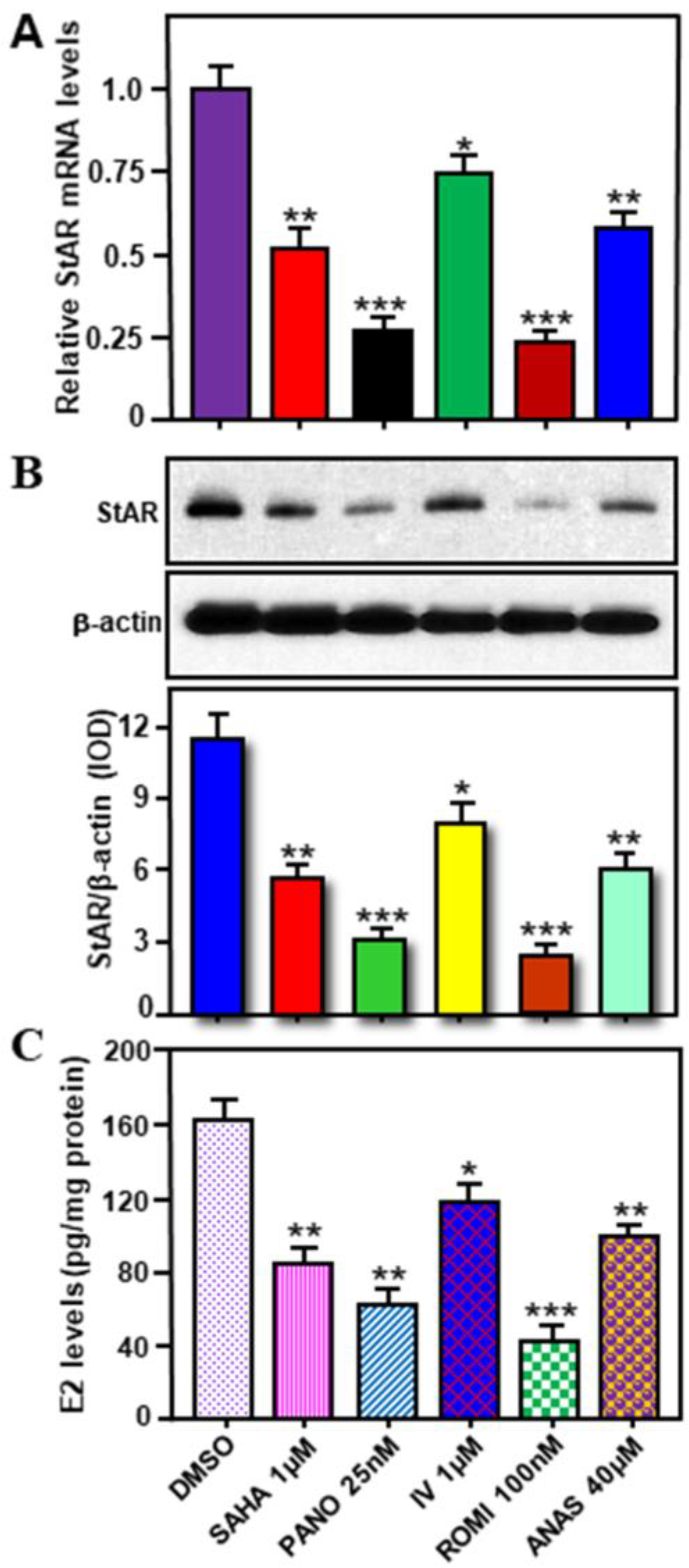
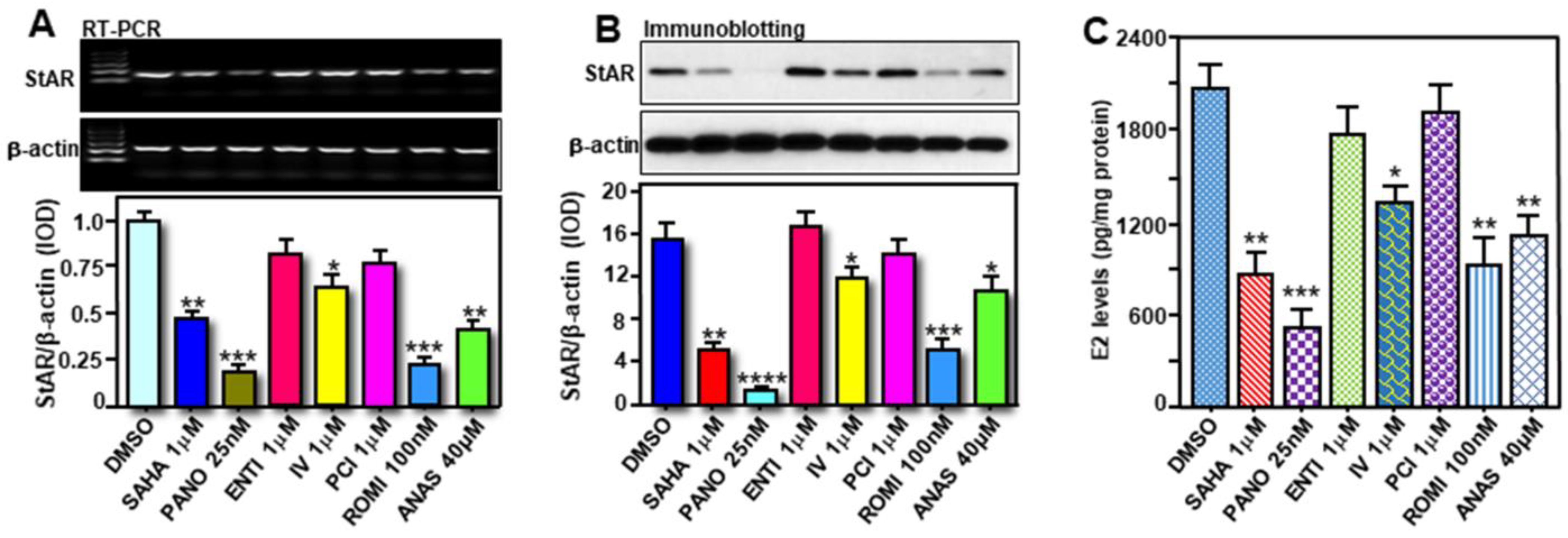
2.5. Effects of Various HDACIs on StAR Expression and E2 Biosynthesis in Hormone-Sensitive MCF7 Cells
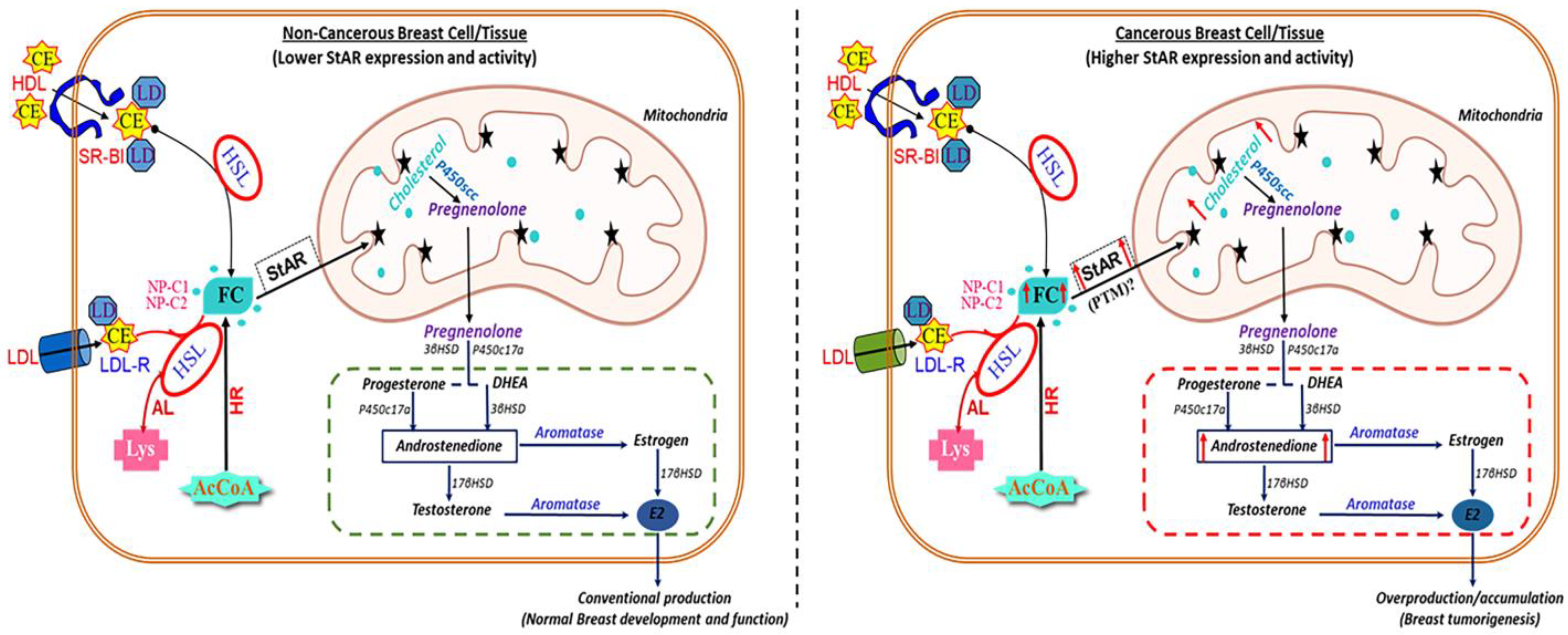
3. Discussion
4. Materials and Methods
4.1. Cell Lines, Animals, and Reagents
4.2. Immunoblotting
4.3. Immunofluorescence
4.4. Extraction of RNA and, Semi-Quantitative RT-PCR and Real-Time PCR
4.5. Determination of E2 by Enzyme-Linked Immunosorbent Assay (ELISA)
4.6. Isolation and Primary Culture of Breast Tumor Epithelial Cells from MMTV-PyMT Mice and Treatment of These Cells with HDACIs
4.7. Statistical Analysis
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| ATCC | American type culture collection |
| BC | breast cancer |
| StAR | steroidogenic acute regulatory protein |
| E2 | 17β-estradiol |
| ER+/PR+ | estrogen progesterone receptor positive |
| TNBC | triple negative BC |
| HDACIs | histone deacetylase inhibitors |
| PANO | Panobinostat |
| ROMI | Romidepsin |
| ENTI | Entinostat |
| PCI | PCI-34051 |
| IV | inhibitor IV |
| ELISA | enzyme-linked immunosorbent assay |
| MMTV | mouse mammary tumor virous |
| PyMT | polyoma middle T antigen |
References
- Miller, W.L.; Bose, H.S. Early steps in steroidogenesis: Intracellular cholesterol trafficking. J. Lipid. Res. 2011, 52, 2111–2135. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Manna, P.R.; Stetson, C.L.; Slominski, A.T.; Pruitt, K. Role of the steroidogenic acute regulatory protein in health and disease. Endocrine 2016, 51, 7–21. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Stocco, D.M.; Wang, X.; Jo, Y.; Manna, P.R. Multiple signaling pathways regulating steroidogenesis and steroidogenic acute regulatory protein expression: More complicated than we thought. Mol. Endocrinol. 2005, 19, 2647–2659. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Castillo, A.F.; Orlando, U.; Helfenberger, K.E.; Poderoso, C.; Podesta, E.J. The role of mitochondrial fusion and StAR phosphorylation in the regulation of StAR activity and steroidogenesis. Mol. Cell. Endocrinol. 2015, 408, 73–79. [Google Scholar] [CrossRef] [PubMed]
- Manna, P.R.; Cohen-Tannoudji, J.; Counis, R.; Garner, C.W.; Huhtaniemi, I.; Kraemer, F.B.; Stocco, D.M. Mechanisms of action of hormone-sensitive lipase in mouse Leydig cells: Its role in the regulation of the steroidogenic acute regulatory protein. J. Biol. Chem. 2013, 288, 8505–8518. [Google Scholar] [CrossRef] [Green Version]
- Manna, P.R.; Dyson, M.T.; Stocco, D.M. Role of basic leucine zipper proteins in transcriptional regulation of the steroidogenic acute regulatory protein gene. Mol. Cell. Endocrinol. 2009, 302, 1–11. [Google Scholar] [CrossRef] [Green Version]
- Manna, P.R.; Ahmed, A.U.; Yang, S.; Narasimhan, M.; Cohen-Tannoudji, J.; Slominski, A.T.; Pruitt, K. Genomic profiling of the steroidogenic acute regulatory protein in breast cancer: In silico assessments and a mechanistic perspective. Cancers 2019, 11, 623. [Google Scholar] [CrossRef] [Green Version]
- Castro-Piedras, I.; Sharma, M.; Brelsfoard, J.; Vartak, D.; Martinez, E.G.; Rivera, C.; Molehin, D.; Bright, R.K.; Fokar, M.; Guindon, J.; et al. Nuclear Dishevelled targets gene regulatory regions and promotes tumor growth. EMBO Rep. 2021, 22, e50600. [Google Scholar] [CrossRef]
- Manna, P.R.; Ahmed, A.U.; Molehin, D.; Narasimhan, M.; Pruitt, K.; Reddy, P.H. Hormonal and genetic regulatory events in breast cancer and its therapeutics: Importance of the steroidogenic acute regulatory protein. Biomedicines 2022, 10, 1313. [Google Scholar] [CrossRef]
- Manna, P.R.; Molehin, D.; Ahmed, A.U. Dysregulation of aromatase in breast, endometrial, and ovarian cancers: An overview of therapeutic strategies. Prog. Mol. Biol. Transl. Sci. 2016, 144, 487–537. [Google Scholar]
- Molehin, D.; Castro-Piedras, I.; Sharma, M.; Sennoune, S.R.; Arena, D.; Manna, P.R.; Pruitt, K. Aromatase acetylation patterns and altered activity in response to Sirtuin inhibition. Mol. Cancer Res. 2018, 16, 1530–1542. [Google Scholar] [CrossRef]
- Simpson, E.; Santen, R.J. Celebrating 75 years of oestradiol. J. Mol. Endocrinol. 2015, 55, T1–T20. [Google Scholar] [CrossRef] [Green Version]
- Khan, M.Z.I.; Uzair, M.; Nazli, A.; Chen, J.Z. An overview on estrogen receptors signaling and its ligands in breast cancer. Eur. J. Med. Chem. 2022, 241, 114658. [Google Scholar] [CrossRef] [PubMed]
- Sakach, E.; O’Regan, R.; Meisel, J.; Li, X. Molecular classification of triple negative breast cancer and the emergence of targeted therapies. Clin. Breast Cancer 2021, 21, 509–520. [Google Scholar] [CrossRef] [PubMed]
- Bou Zerdan, M.; Ghorayeb, T.; Saliba, F.; Allam, S.; Bou Zerdan, M.; Yaghi, M.; Bilani, N.; Jaafar, R.; Nahleh, Z. Triple negative breast cancer: Updates on classification and treatment in 2021. Cancers 2022, 14, 1253. [Google Scholar] [CrossRef] [PubMed]
- Renoir, J.M.; Marsaud, V.; Lazennec, G. Estrogen receptor signaling as a target for novel breast cancer therapeutics. Biochem. Pharmacol. 2013, 85, 449–465. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bulun, S.E.; Lin, Z.; Zhao, H.; Lu, M.; Amin, S.; Reierstad, S.; Chen, D. Regulation of aromatase expression in breast cancer tissue. Ann. N. Y. Acad. Sci. 2009, 1155, 121–131. [Google Scholar] [CrossRef] [PubMed]
- Zhao, H.; Zhou, L.; Shangguan, A.J.; Bulun, S.E. Aromatase expression and regulation in breast and endometrial cancer. J. Mol. Endocrinol. 2016, 57, R19–R33. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sjoquist, K.M.; Martyn, J.; Edmondson, R.J.; Friedlander, M.L. The role of hormonal therapy in gynecological cancers-current status and future directions. Int. J. Gynecol. Cancer 2011, 21, 1328–1333. [Google Scholar] [CrossRef] [Green Version]
- Jahan, N.; Jones, C.; Rahman, R.L. Endocrine prevention of breast cancer. Mol. Cell. Endocrinol. 2021, 530, 111284. [Google Scholar] [CrossRef]
- Manna, P.R.; Ahmed, A.U.; Vartak, D.; Molehin, D.; Pruitt, K. Overexpression of the steroidogenic acute regulatory protein in breast cancer: Regulation by histone deacetylase inhibition. Biochem. Biophys. Res. Commun. 2019, 509, 476–482. [Google Scholar] [CrossRef]
- Castro-Piedras, I.; Sharma, M.; den Bakker, M.; Molehin, D.; Martinez, E.G.; Vartak, D.; Pruitt, W.M.; Deitrick, J.; Almodovar, S.; Pruitt, K. DVL1 and DVL3 differentially localize to CYP19A1 promoters and regulate aromatase mRNA in breast cancer cells. Oncotarget 2018, 9, 35639–35654. [Google Scholar] [CrossRef] [Green Version]
- Sharma, M.; Molehin, D.; Castro-Piedras, I.; Martinez, E.G.; Pruitt, K. Acetylation of conserved DVL-1 lysines regulates its nuclear translocation and binding to gene promoters in triple-negative breast cancer. Sci. Rep. 2019, 9, 16257. [Google Scholar] [CrossRef] [Green Version]
- West, A.C.; Johnstone, R.W. New and emerging HDAC inhibitors for cancer treatment. J. Clin. Invest. 2014, 124, 30–39. [Google Scholar] [CrossRef] [Green Version]
- Yoon, S.; Eom, G.H. HDAC and HDAC inhibitor: From cancer to cardiovascular diseases. Chonnam. Med. J. 2016, 52, 1–11. [Google Scholar] [CrossRef] [Green Version]
- Abderrahman, B.; Maximov, P.Y.; Curpan, R.F.; Hanspal, J.S.; Fan, P.; Xiong, R.; Tonetti, D.A.; Thatcher, G.R.J.; Jordan, V.C. Pharmacology and molecular mechanisms of clinically relevant estrogenestetrol and estrogen mimic BMI-135 for the treatment of endocrine-resistant breast bancer. Mol. Pharmacol. 2020, 98, 364–381. [Google Scholar] [CrossRef]
- Verza, F.A.; Das, U.; Fachin, A.L.; Dimmock, J.R.; Marins, M. Roles of histone deacetylases and inhibitors in anticancer therapy. Cancers 2020, 12, 1664. [Google Scholar] [CrossRef]
- Brown, T.P.; Bhattacharjee, P.; Ramachandran, S.; Sivaprakasam, S.; Ristic, B.; Sikder, M.O.F.; Ganapathy, V. The lactate receptor GPR81 promotes breast cancer growth via a paracrine mechanism involving antigen-presenting cells in the tumor microenvironment. Oncogene 2020, 39, 3292–3304. [Google Scholar] [CrossRef]
- Ramachandran, S.; RSennoune, S.; Sharma, M.; Thangaraju, M.; VSuresh, V.; Sneigowski, T.; DBhutia, Y.; Pruitt, K.; Ganapathy, V. Expression and function of SLC38A5, an amino acid-coupled Na+/H+ exchanger, in triple-negative breast cancer and its relevance to macropinocytosis. Biochem. J. 2021, 478, 3957–3976. [Google Scholar] [CrossRef]
- Lin, E.Y.; Jones, J.G.; Li, P.; Zhu, L.; Whitney, K.D.; Muller, W.J.; Pollard, J.W. Progression to malignancy in the polyoma middle T oncoprotein mouse breast cancer model provides a reliable model for human diseases. Am. J. Pathol. 2003, 163, 2113–2126. [Google Scholar] [CrossRef] [Green Version]
- Holloway, K.R.; Barbieri, A.; Malyarchuk, S.; Saxena, M.; Nedeljkovic-Kurepa, A.; Cameron Mehl, M.; Wang, A.; Gu, X.; Pruitt, K. SIRT1 positively regulates breast cancer associated human aromatase (CYP19A1) expression. Mol. Endocrinol. 2013, 27, 480–490. [Google Scholar] [CrossRef]
- Nelson, E.R.; Wardell, S.E.; Jasper, J.S.; Park, S.; Suchindran, S.; Howe, M.K.; Carver, N.J.; Pillai, R.V.; Sullivan, P.M.; Sondhi, V.; et al. 27-Hydroxycholesterol links hypercholesterolemia and breast cancer pathophysiology. Science 2013, 342, 1094–1098. [Google Scholar] [CrossRef] [Green Version]
- Silvente-Poirot, S.; Poirot, M. Cancer. Cholesterol and cancer, in the balance. Science 2014, 343, 1445–1446. [Google Scholar] [CrossRef]
- Poirot, M.; Soules, R.; Mallinger, A.; Dalenc, F.; Silvente-Poirot, S. Chemistry, biochemistry, metabolic fate and mechanism of action of 6-oxo-cholestan-3beta,5alpha-diol (OCDO), a tumor promoter and cholesterol metabolite. Biochimie 2018, 153, 139–149. [Google Scholar] [CrossRef]
- de Medina, P.; Diallo, K.; Huc-Claustre, E.; Attia, M.; Soules, R.; Silvente-Poirot, S.; Poirot, M. The 5,6-epoxycholesterol metabolic pathway in breast cancer: Emergence of new pharmacological targets. Br. J. Pharmacol. 2021, 178, 3248–3260. [Google Scholar] [CrossRef]
- Capper, C.P.; Rae, J.M.; Auchus, R.J. The metabolism, analysis, and targeting of steroid hormones in breast and prostate cancer. Horm. Cancer 2016, 7, 149–164. [Google Scholar] [CrossRef] [Green Version]
- Molehin, D.; Rasha, F.; Rahman, R.L.; Pruitt, K. Regulation of aromatase in cancer. Mol. Cell. Biochem. 2021, 476, 2449–2464. [Google Scholar] [CrossRef]
- Simpson, E.R.; Misso, M.; Hewitt, K.N.; Hill, R.A.; Boon, W.C.; Jones, M.E.; Kovacic, A.; Zhou, J.; Clyne, C.D. Estrogen—The good, the bad, and the unexpected. Endocr. Rev. 2005, 26, 322–330. [Google Scholar] [CrossRef] [Green Version]
- Fuseler, J.W.; Robichaux, J.P.; Atiyah, H.I.; Ramsdell, A.F. Morphometric and fractal dimension analysis identifies early neoplastic changes in mammary epithelium of MMTV-cNeu mice. Anticancer Res. 2014, 34, 1171–1177. [Google Scholar]
- Ionkina, A.A.; Balderrama-Gutierrez, G.; Ibanez, K.J.; Phan, S.H.D.; Cortez, A.N.; Mortazavi, A.; Prescher, J.A. Transcriptome analysis of heterogeneity in mouse model of metastatic breast cancer. Breast Cancer Res. 2021, 23, 93. [Google Scholar] [CrossRef]
- Attalla, S.; Taifour, T.; Bui, T.; Muller, W. Insights from transgenic mouse models of PyMT-induced breast cancer: Recapitulating human breast cancer progression in vivo. Oncogene 2021, 40, 475–491. [Google Scholar] [CrossRef]
- Strauss, J.F., III; Kishida, T.; Christenson, L.K.; Fujimoto, T.; Hiroi, H. START domain proteins and the intracellular trafficking of cholesterol in steroidogenic cells. Mol. Cell. Endocrinol. 2003, 202, 59–65. [Google Scholar] [CrossRef]
- Alpy, F.; Legueux, F.; Bianchetti, L.; Tomasetto, C. START domain-containing proteins: A review of their role in lipid transport and exchange. Med. Sci. 2009, 25, 181–191. [Google Scholar]
- Tomasetto, C.; Regnier, C.; Moog-Lutz, C.; Mattei, M.G.; Chenard, M.P.; Lidereau, R.; Basset, P.; Rio, M.C. Identification of four novel human genes amplified and overexpressed in breast carcinoma and localized to the q11-q21.3 region of chromosome 17. Genomics 1995, 28, 367–376. [Google Scholar] [CrossRef]
- Akiyama, N.; Sasaki, H.; Ishizuka, T.; Kishi, T.; Sakamoto, H.; Onda, M.; Hirai, H.; Yazaki, Y.; Sugimura, T.; Terada, M. Isolation of a candidate gene, CAB1, for cholesterol transport to mitochondria from the c-ERBB-2 amplicon by a modified cDNA selection method. Cancer Res. 1997, 57, 3548–3553. [Google Scholar]
- Manna, P.R.; Huhtaniemi, I.T.; Stocco, D.M. Mechanisms of protein kinase C signaling in the modulation of 3′,5′-cyclic adenosine monophosphate-mediated steroidogenesis in mouse gonadal cells. Endocrinology 2009, 150, 3308–3317. [Google Scholar] [CrossRef] [Green Version]
- Manna, P.R.; Soh, J.W.; Stocco, D.M. The involvement of specific PKC isoenzymes in phorbol ester-mediated regulation of steroidogenic acute regulatory protein expression and steroid synthesis in mouse Leydig cells. Endocrinology 2011, 152, 313–325. [Google Scholar] [CrossRef] [Green Version]
- Manna, P.R.; Slominski, A.T.; King, S.R.; Stetson, C.L.; Stocco, D.M. Synergistic activation of steroidogenic acute regulatory protein expression and steroid biosynthesis by retinoids: Involvement of cAMP/PKA signaling. Endocrinology 2014, 155, 576–591. [Google Scholar] [CrossRef] [Green Version]
- Arakane, F.; King, S.R.; Du, Y.; Kallen, C.B.; Walsh, L.P.; Watari, H.; Stocco, D.M.; Strauss, J.F., III. Phosphorylation of steroidogenic acute regulatory protein (StAR) modulates its steroidogenic activity. J. Biol. Chem. 1997, 272, 32656–32662. [Google Scholar] [CrossRef] [Green Version]
- Clark, B.J.; Ranganathan, V.; Combs, R. Steroidogenic acute regulatory protein expression is dependent upon post-translational effects of cAMP-dependent protein kinase A. Mol. Cell. Endocrinol. 2001, 173, 183–192. [Google Scholar] [CrossRef]
- Manna, P.R.; Stetson, C.L.; Daugherty, C.; Shimizu, I.; Syapin, P.J.; Garrel, G.; Cohen-Tannoudji, J.; Huhtaniemi, I.; Slominski, A.T.; Pruitt, K.; et al. Up-regulation of steroid biosynthesis by retinoid signaling: Implications for aging. Mech. Ageing Dev. 2015, 150, 74–82. [Google Scholar] [CrossRef] [Green Version]
- Dunbier, A.K.; Hong, Y.; Masri, S.; Brown, K.A.; Sabnis, G.J.; Palomares, M.R. Progress in aromatase research and identification of key future directions. J. Steroid Biochem. Mol. Biol. 2010, 118, 311–315. [Google Scholar] [CrossRef]
- Lonning, P.E.; Haynes, B.P.; Straume, A.H.; Dunbier, A.; Helle, H.; Knappskog, S.; Dowsett, M. Exploring breast cancer estrogen disposition: The basis for endocrine manipulation. Clin. Cancer Res. 2011, 17, 4948–4958. [Google Scholar] [CrossRef] [Green Version]
- Manna, P.R.; Sennoune, S.R.; Martinez-Zaguilan, R.; Slominski, A.T.; Pruitt, K. Regulation of retinoid mediated cholesterol efflux involves liver X receptor activation in mouse macrophages. Biochem. Biophys. Res. Commun. 2015, 464, 312–317. [Google Scholar] [CrossRef]
- Jechlinger, M. Organotypic culture of untransformed and tumorigenic primary mammary epithelial cells. Cold Spring Harb. Protoc. 2015, 2015, 457–461. [Google Scholar] [CrossRef]
- Cui, J.; Guo, W. Establishment and long-term culture of mouse mammary stem cell organoids and breast tumor organoids. STAR Protoc. 2021, 2, 100577. [Google Scholar] [CrossRef]


| Genes | Forward Primer | Reverse Primer |
|---|---|---|
| Human StAR | CTG GGA GCT CCT ACA GAC AC | AGC CGA GAA CCG AGT AGA GAG |
| Mouse StAR | #1: GGA GCT CTC TGC TTG GTT CTC, or #2: GAC CTT GAA AGG CTC AGG AAG AAC | #1: TTA GCA CTT CGT CCC CGT TC, or #2: TAG CTG AAG ATG GAC AGA CTT GC |
| Mouse ERα (ESR1) | ATT ATG GGG TCT GGT CCT GC | CTT TCC GTA TGC CGC CTT TC |
| Mouse ERβ (ESR1) | AGA CGA AGA GTG CTG TCC CA | GGG GTA CAT ACT GGA GTT GAG G |
| Mouse PR (PR) | ATC TGG CTG TCA CTA TGG CG | ACT TAC GAC CTC CAA GGA CCA |
| Human CYP19A1 | GGC AGT GCC TGC AAC TAC TA | GTC ACC TCC TCC AAC CTG TC |
| Mouse CYP19A1 | AAC ATG CTC TTC CTG GGG AT | GTC CTT GAC GGA TCG TTC ATA C |
| Human β-actin | GGA CTT CGA GCA AGA GAT GG | AGC ACT GTG TTG GCG TAC AG |
| Mouse β-actin | CTG GCA CCA CAC CTT CTA | GGG CAC AGT GTG GGT GAC |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Manna, P.R.; Ramachandran, S.; Pradeepkiran, J.A.; Molehin, D.; Castro-Piedras, I.; Pruitt, K.; Ganapathy, V.; Reddy, P.H. Expression and Function of StAR in Cancerous and Non-Cancerous Human and Mouse Breast Tissues: New Insights into Diagnosis and Treatment of Hormone-Sensitive Breast Cancer. Int. J. Mol. Sci. 2023, 24, 758. https://doi.org/10.3390/ijms24010758
Manna PR, Ramachandran S, Pradeepkiran JA, Molehin D, Castro-Piedras I, Pruitt K, Ganapathy V, Reddy PH. Expression and Function of StAR in Cancerous and Non-Cancerous Human and Mouse Breast Tissues: New Insights into Diagnosis and Treatment of Hormone-Sensitive Breast Cancer. International Journal of Molecular Sciences. 2023; 24(1):758. https://doi.org/10.3390/ijms24010758
Chicago/Turabian StyleManna, Pulak R., Sabarish Ramachandran, Jangampalli Adi Pradeepkiran, Deborah Molehin, Isabel Castro-Piedras, Kevin Pruitt, Vadivel Ganapathy, and P. Hemachandra Reddy. 2023. "Expression and Function of StAR in Cancerous and Non-Cancerous Human and Mouse Breast Tissues: New Insights into Diagnosis and Treatment of Hormone-Sensitive Breast Cancer" International Journal of Molecular Sciences 24, no. 1: 758. https://doi.org/10.3390/ijms24010758





