Genome-Wide Analysis of the Mads-Box Transcription Factor Family in Solanum melongena
Abstract
1. Introduction
2. Results
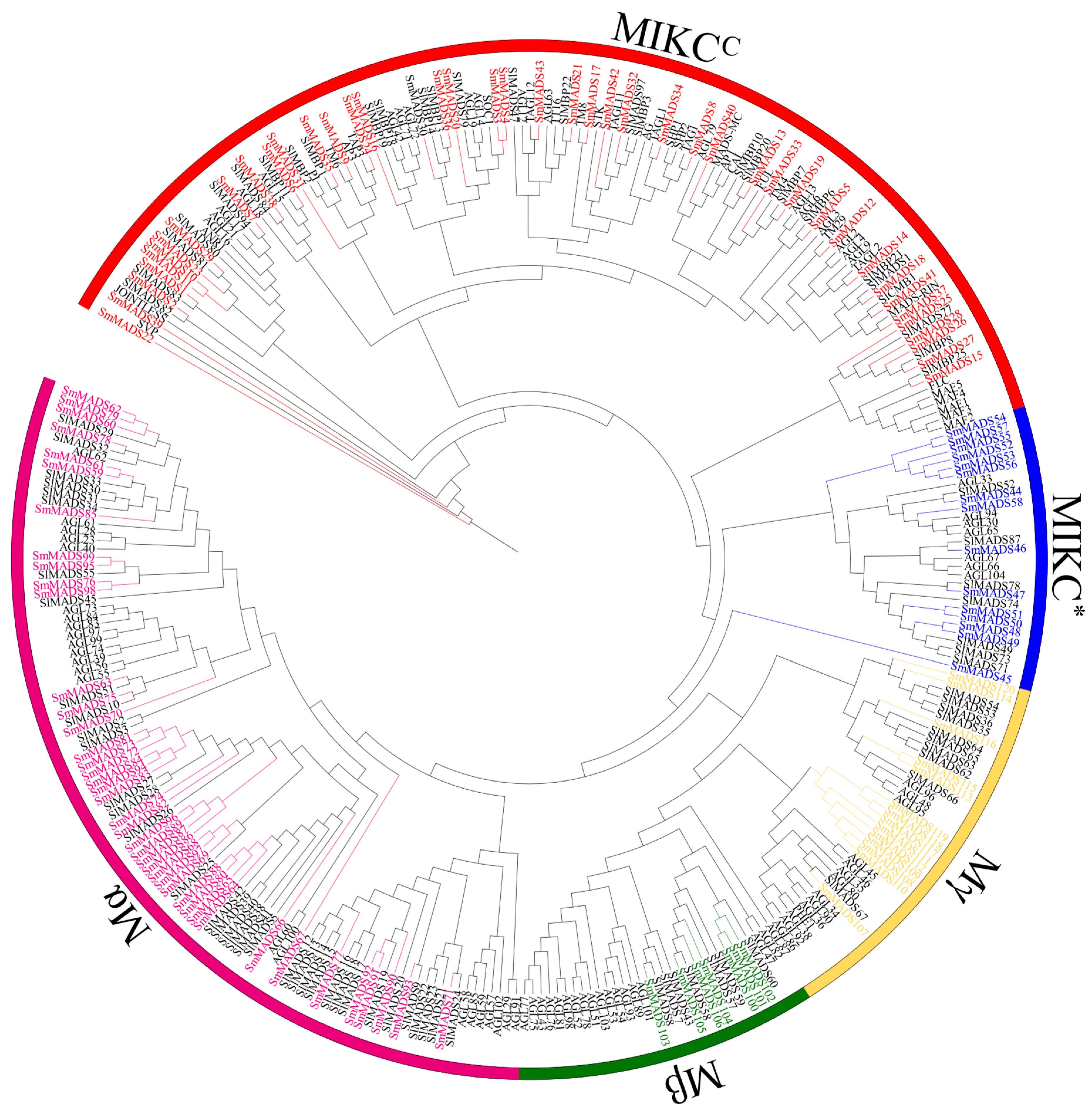
2.1. Identification and Phylogenetic Analysis of SmMADS-Box Genes in Eggplant
2.2. Gene Structure and Conserved Motif Analysis of the 120 SmMADS-Box Tfs
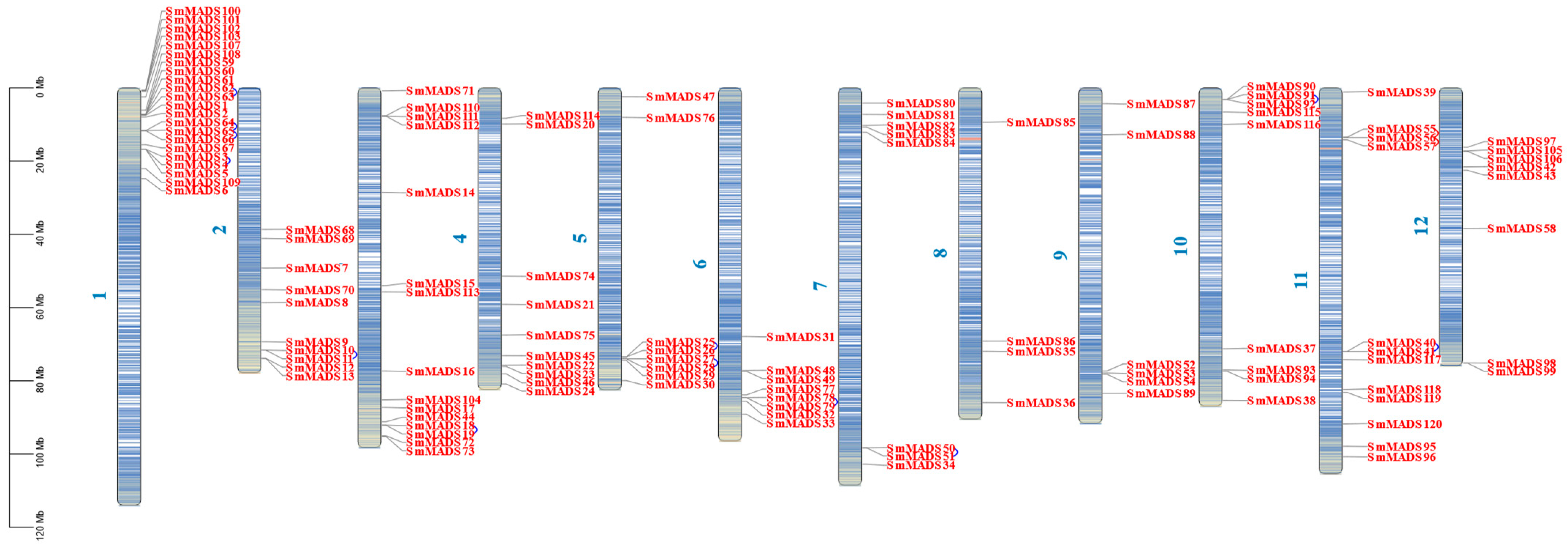
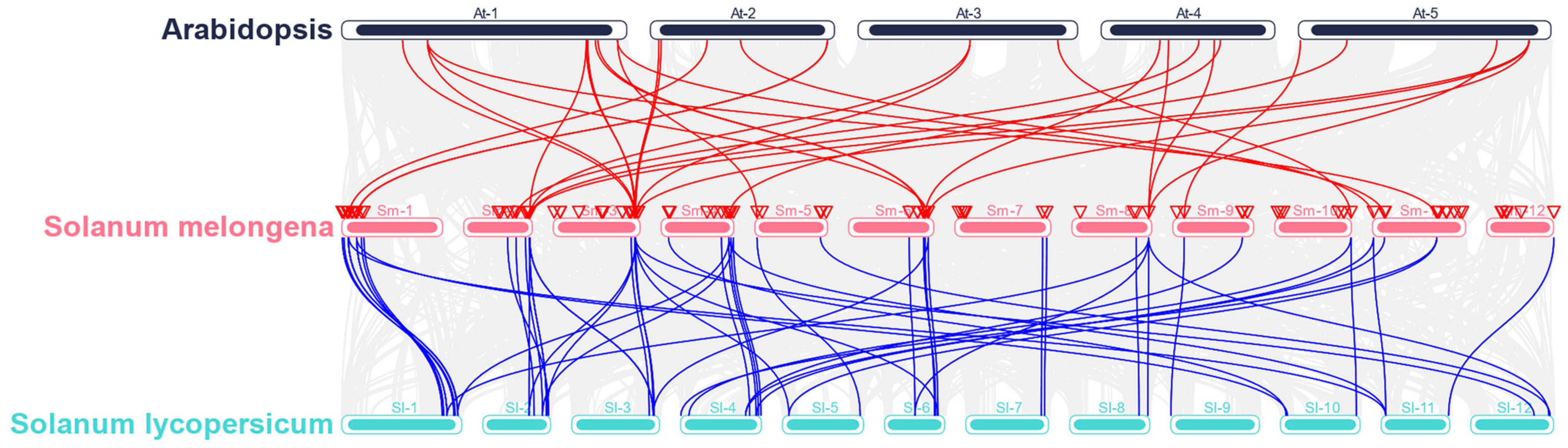
2.3. Chromosomal Location, Gene Duplication, and Homology Analysis of the 120 SmMADS-Box Genes
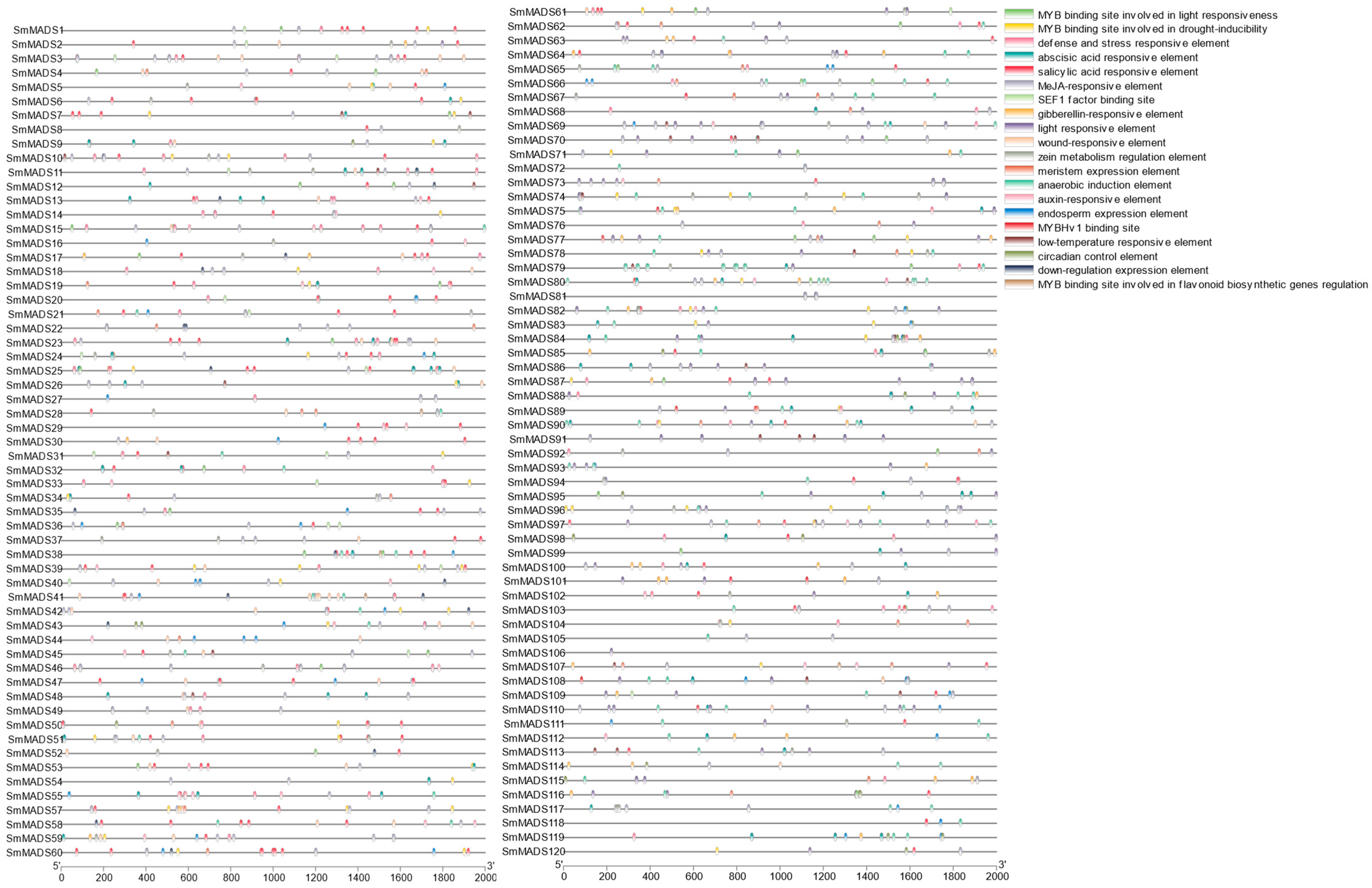
2.4. Cis-Regulatory Elements in the Promoters of the 120 SmMADS-Box Genes
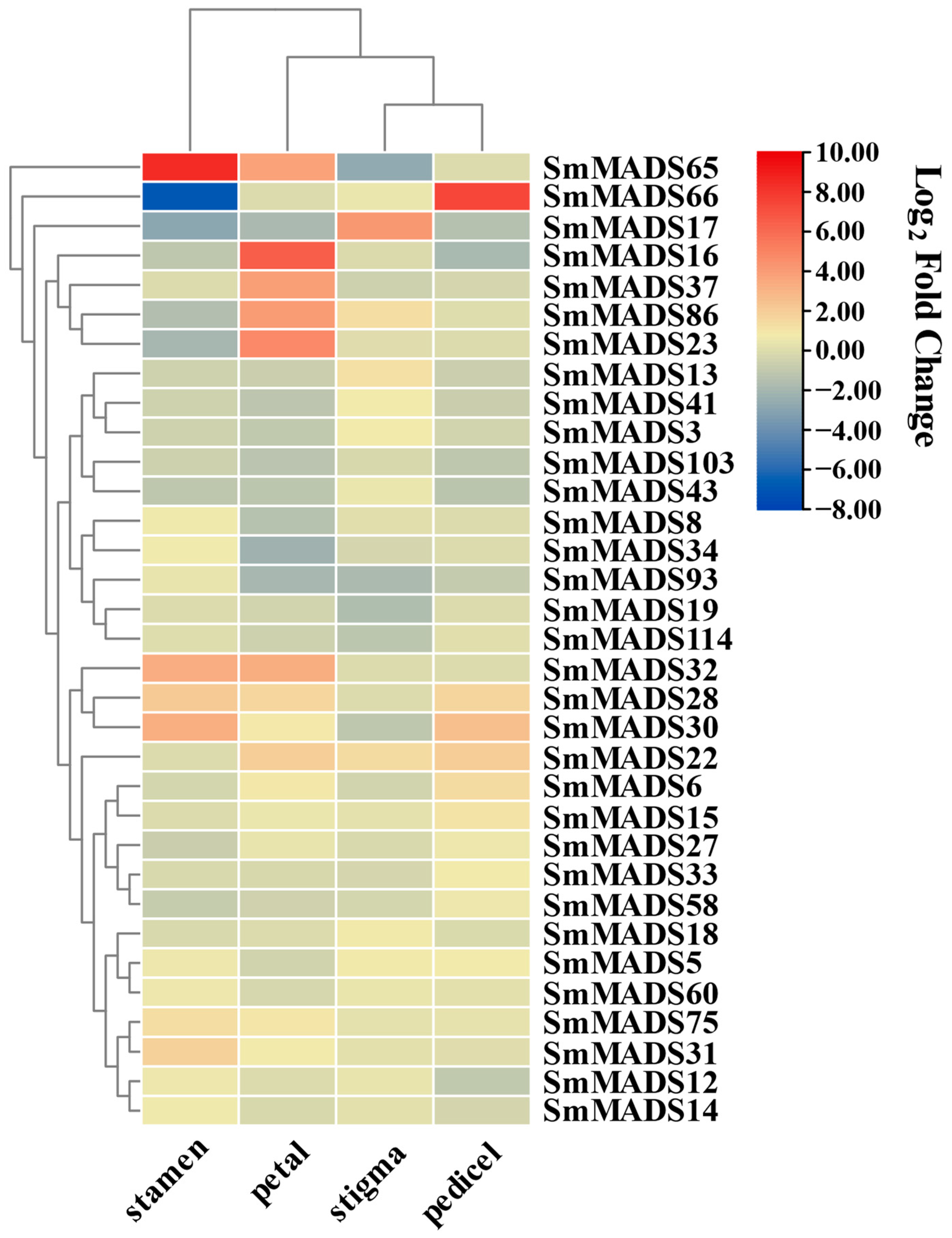
2.5. Flower Tissue-Specific Expression Patterns of SmMADS-Box Genes
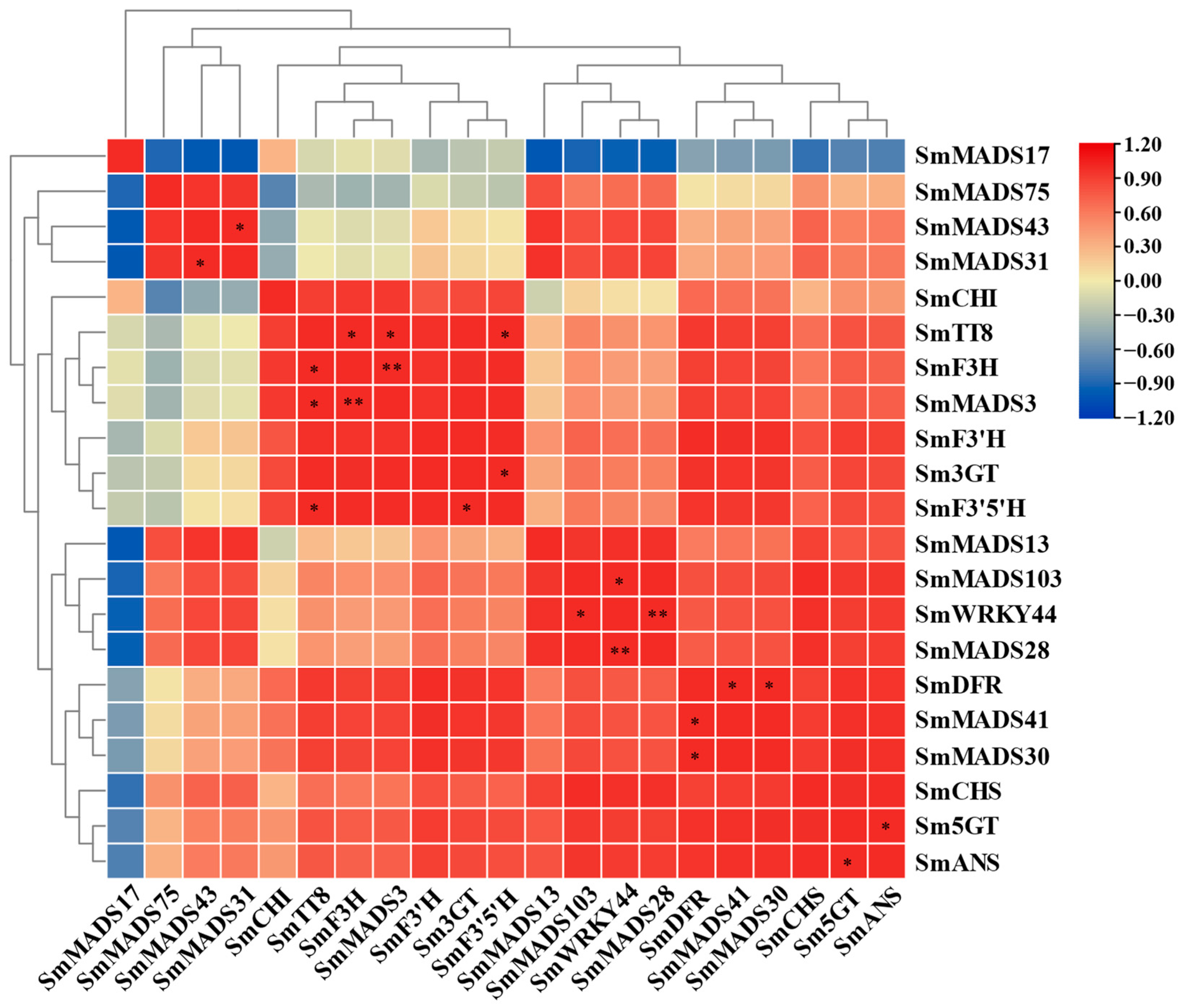
2.6. Correlation Analysis of Eggplant SmMADS-Box Genes and SMMYB113
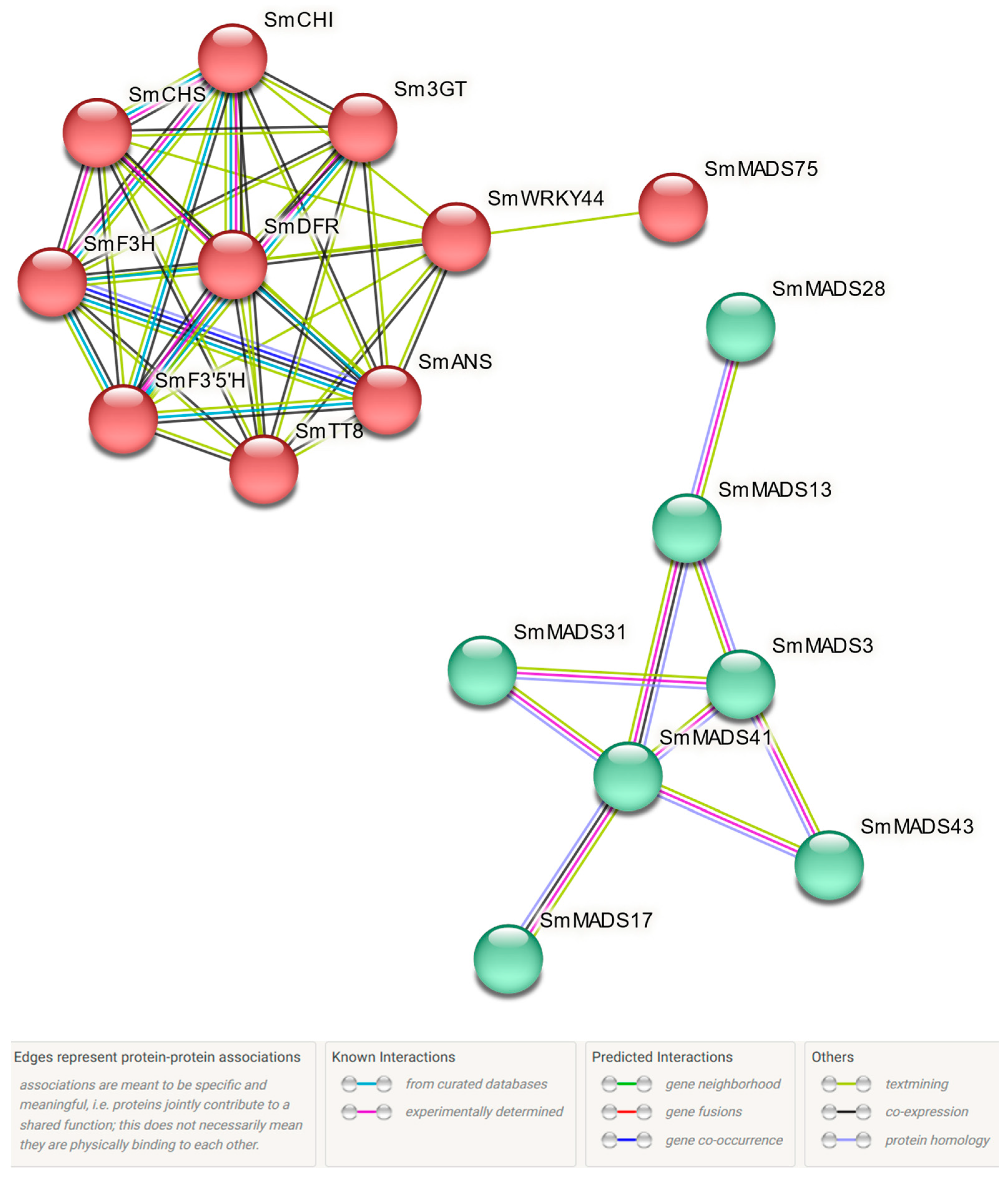
2.7. Protein Interaction Predictive Analysis of SmMADS-Box Tfs and Anthocyanin Biosynthesis-Related Genes
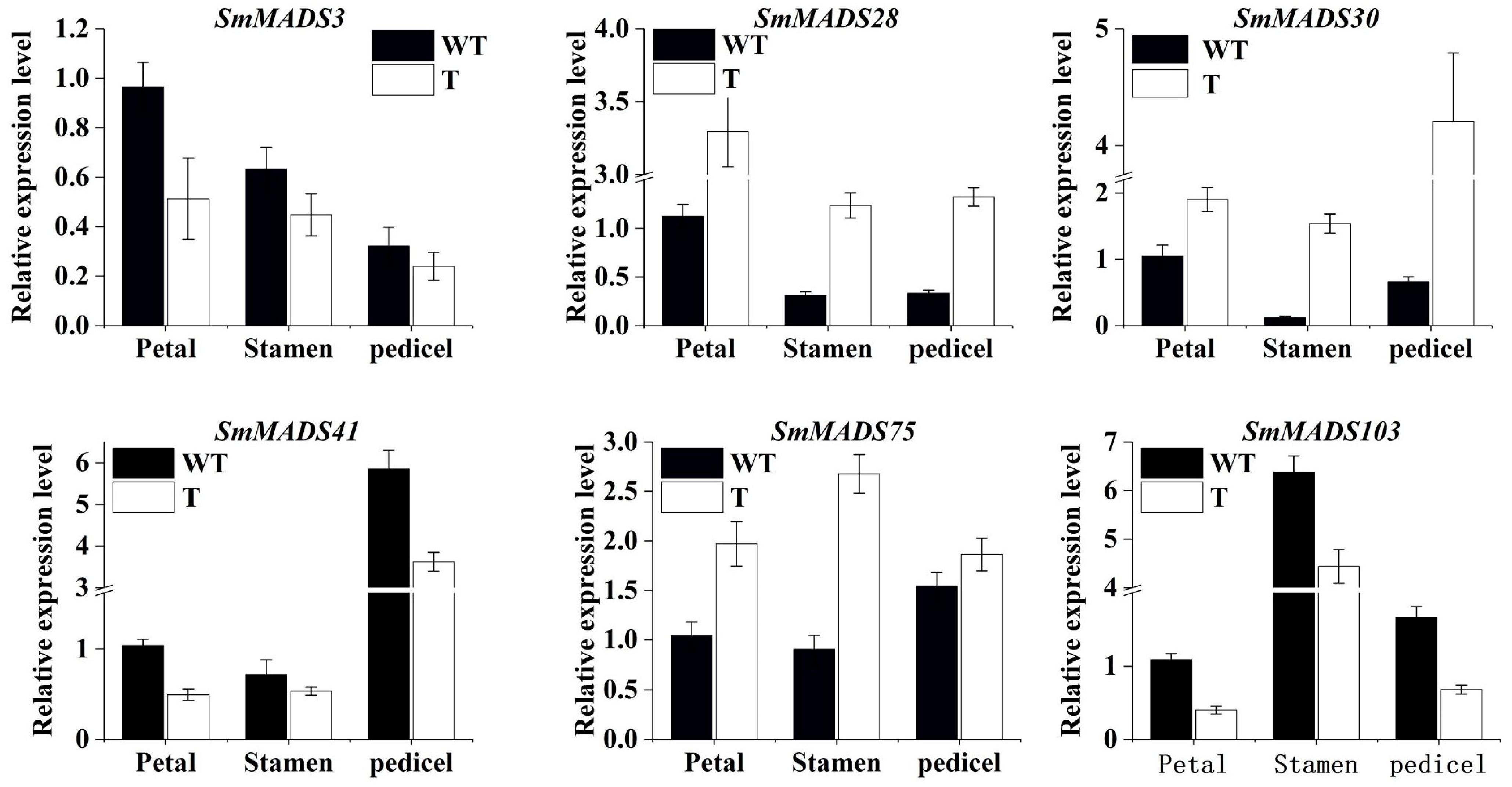
2.8. Effect of SMMYB113 Overexpression on the Expression Level of Six Selected Smmadx-Box Genes
3. Discussion
4. Materials and Methods
4.1. Identification of MADS-Box Genes in Eggplant
4.2. Phylogenetic, Conserved Motifs, and Gene Structure Analyses of MADS-Box Genes in Eggplant
4.3. Chromosomal Location, Gene Duplication, and Homology Analysiss
4.4. Cis-Element Analysis
4.5. Expression Pattern and Correlation Analysis
4.6. Interaction Network
4.7. Total RNA Extraction and qPCR Analysis
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Appendix A
| Gene Name | Gene Locus | Protein | Exon | Type | Subfamily | ||
|---|---|---|---|---|---|---|---|
| Length (aa) | MW (Da) | pI | |||||
| SmMADS1 | SMEL4.1_01g007770.1 | 89 | 10,077.78 | 10.14 | 3 | Type II | MIKCC |
| SmMADS2 | SMEL4.1_01g008380.1 | 161 | 18,252.78 | 7.84 | 5 | Type II | MIKCC |
| SmMADS3 | SMEL4.1_01g016420.1 | 191 | 21,929.28 | 9.13 | 6 | Type II | MIKCC |
| SmMADS4 | SMEL4.1_01g016430.1 | 61 | 6963.16 | 11.29 | 1 | Type II | MIKCC |
| SmMADS5 | SMEL4.1_01g016450.1 | 254 | 28,691.55 | 8.73 | 8 | Type II | MIKCC |
| SmMADS6 | SMEL4.1_01g022190.1 | 273 | 30,804.84 | 5.88 | 7 | Type II | MIKCC |
| SmMADS7 | SMEL4.1_02g006940.1 | 223 | 25,119.47 | 9.00 | 9 | Type II | MIKCC |
| SmMADS8 | SMEL4.1_02g011710.1 | 235 | 27,308.08 | 9.43 | 6 | Type II | MIKCC |
| SmMADS9 | SMEL4.1_02g021760.1 | 223 | 25,814.66 | 9.70 | 6 | Type II | MIKCC |
| SmMADS10 | SMEL4.1_02g024180.1 | 121 | 13,446.38 | 10.30 | 3 | Type II | MIKCC |
| SmMADS11 | SMEL4.1_02g024190.1 | 61 | 6888.07 | 10.65 | 1 | Type II | MIKCC |
| SmMADS12 | SMEL4.1_02g026490.1 | 125 | 28,475.31 | 8.25 | 8 | Type II | MIKCC |
| SmMADS13 | SMEL4.1_02g026500.1 | 158 | 18,427.18 | 8.83 | 4 | Type II | MIKCC |
| SmMADS14 | SMEL4.1_03g008110.1 | 224 | 25,730.65 | 9.80 | 7 | Type II | MIKCC |
| SmMADS15 | SMEL4.1_03g010360.1 | 77 | 8983.52 | 10.69 | 1 | Type II | MIKCC |
| SmMADS16 | SMEL4.1_03g015990.1 | 82 | 9441.02 | 10.02 | 2 | Type II | MIKCC |
| SmMADS17 | SMEL4.1_03g022770.1 | 141 | 16,105.54 | 9.98 | 3 | Type II | MIKCC |
| SmMADS18 | SMEL4.1_03g027790.1 | 246 | 28,209.90 | 8.94 | 8 | Type II | MIKCC |
| SmMADS19 | SMEL4.1_03g027800.1 | 248 | 28,728.36 | 9.10 | 8 | Type II | MIKCC |
| SmMADS20 | SMEL4.1_04g005970.1 | 61 | 7065.29 | 10.65 | 1 | Type II | MIKCC |
| SmMADS21 | SMEL4.1_04g011260.1 | 186 | 21,740.04 | 8.81 | 6 | Type II | MIKCC |
| SmMADS22 | SMEL4.1_04g017530.1 | 205 | 23,042.58 | 8.43 | 6 | Type II | MIKCC |
| SmMADS23 | SMEL4.1_04g017760.1 | 61 | 6892.02 | 10.43 | 1 | Type II | MIKCC |
| SmMADS24 | SMEL4.1_04g021980.1 | 207 | 23,958.04 | 8.91 | 5 | Type II | MIKCC |
| SmMADS25 | SMEL4.1_05g017100.1 | 66 | 7668.98 | 10.62 | 2 | Type II | MIKCC |
| SmMADS26 | SMEL4.1_05g017110.1 | 248 | 28,275.08 | 8.45 | 6 | Type II | MIKCC |
| SmMADS27 | SMEL4.1_05g017480.1 | 228 | 26,042.78 | 7.13 | 8 | Type II | MIKCC |
| SmMADS28 | SMEL4.1_05g017490.1 | 148 | 16,831.34 | 6.91 | 5 | Type II | MIKCC |
| SmMADS29 | SMEL4.1_05g017760.1 | 86 | 9562.26 | 11.39 | 2 | Type II | MIKCC |
| SmMADS30 | SMEL4.1_05g022920.1 | 68 | 7793.10 | 10.28 | 2 | Type II | MIKCC |
| SmMADS31 | SMEL4.1_06g010960.1 | 64 | 7347.62 | 10.01 | 2 | Type II | MIKCC |
| SmMADS32 | SMEL4.1_06g018550.1 | 233 | 27,010.92 | 9.31 | 7 | Type II | MIKCC |
| SmMADS33 | SMEL4.1_06g022050.1 | 218 | 25,088.34 | 8.88 | 7 | Type II | MIKCC |
| SmMADS34 | SMEL4.1_07g020500.1 | 219 | 24,525.82 | 8.70 | 5 | Type II | MIKCC |
| SmMADS35 | SMEL4.1_08g015360.1 | 185 | 21,477.57 | 9.21 | 5 | Type II | MIKCC |
| SmMADS36 | SMEL4.1_08g022290.1 | 272 | 31,277.04 | 9.36 | 9 | Type II | MIKCC |
| SmMADS37 | SMEL4.1_10g015470.1 | 234 | 26,809.40 | 7.76 | 7 | Type II | MIKCC |
| SmMADS38 | SMEL4.1_10g025220.1 | 201 | 22,745.80 | 6.55 | 6 | Type II | MIKCC |
| SmMADS39 | SMEL4.1_11g000980.1 | 292 | 33,368.72 | 8.72 | 8 | Type II | MIKCC |
| SmMADS40 | SMEL4.1_11g015290.1 | 185 | 21,873.14 | 9.37 | 7 | Type II | MIKCC |
| SmMADS41 | SMEL4.1_11g015300.1 | 214 | 24,679.20 | 8.84 | 8 | Type II | MIKCC |
| SmMADS42 | SMEL4.1_12g007250.1 | 229 | 26,734.45 | 9.25 | 7 | Type II | MIKCC |
| SmMADS43 | SMEL4.1_12g007390.1 | 111 | 12,556.75 | 9.25 | 3 | Type II | MIKCC |
| SmMADS44 | SMEL4.1_03g026850.1 | 255 | 29,039.15 | 8.96 | 7 | Type II | MIKC* |
| SmMADS45 | SMEL4.1_04g015770.1 | 53 | 6018.15 | 10.10 | 2 | Type II | MIKC* |
| SmMADS46 | SMEL4.1_04g019230.1 | 312 | 35,774.12 | 5.30 | 8 | Type II | MIKC* |
| SmMADS47 | SMEL4.1_05g001970.1 | 359 | 41,450.34 | 6.77 | 10 | Type II | MIKC* |
| SmMADS48 | SMEL4.1_06g013880.1 | 138 | 15,508.61 | 5.41 | 5 | Type II | MIKC* |
| SmMADS49 | SMEL4.1_06g013910.1 | 220 | 24,584.16 | 8.57 | 7 | Type II | MIKC* |
| SmMADS50 | SMEL4.1_07g017280.1 | 197 | 21,873.74 | 5.82 | 8 | Type II | MIKC* |
| SmMADS51 | SMEL4.1_07g017290.1 | 197 | 21,755.52 | 5.09 | 8 | Type II | MIKC* |
| SmMADS52 | SMEL4.1_09g016090.1 | 77 | 8866.39 | 9.00 | 2 | Type II | MIKC* |
| SmMADS53 | SMEL4.1_09g016160.1 | 121 | 13,988.34 | 9.42 | 3 | Type II | MIKC* |
| SmMADS54 | SMEL4.1_09g016200.1 | 82 | 9597.44 | 9.47 | 2 | Type II | MIKC* |
| SmMADS55 | SMEL4.1_11g007660.1 | 58 | 6872.10 | 9.78 | 1 | Type II | MIKC* |
| SmMADS56 | SMEL4.1_11g007670.1 | 103 | 11,804.79 | 9.51 | 2 | Type II | MIKC* |
| SmMADS57 | SMEL4.1_11g007680.1 | 82 | 9657.53 | 9.89 | 2 | Type II | MIKC* |
| SmMADS58 | SMEL4.1_12g008860.1 | 591 | 65,950.08 | 4.54 | 11 | Type II | MIKC* |
| SmMADS59 | SMEL4.1_01g007470.1 | 208 | 23,518.86 | 9.94 | 1 | Type I | Mα |
| SmMADS60 | SMEL4.1_01g007480.1 | 333 | 37,709.97 | 9.10 | 5 | Type I | Mα |
| SmMADS61 | SMEL4.1_01g007490.1 | 208 | 23,394.70 | 10.00 | 1 | Type I | Mα |
| SmMADS62 | SMEL4.1_01g007510.1 | 221 | 25,061.62 | 9.37 | 1 | Type I | Mα |
| SmMADS63 | SMEL4.1_01g007520.1 | 234 | 26,183.44 | 6.76 | 1 | Type I | Mα |
| SmMADS64 | SMEL4.1_01g011870.1 | 199 | 21,962.69 | 7.69 | 1 | Type I | Mα |
| SmMADS65 | SMEL4.1_01g011880.1 | 157 | 17,748.33 | 9.87 | 1 | Type I | Mα |
| SmMADS66 | SMEL4.1_01g011890.1 | 107 | 11,605.35 | 9.94 | 2 | Type I | Mα |
| SmMADS67 | SMEL4.1_01g015320.1 | 197 | 22,125.41 | 8.88 | 2 | Type I | Mα |
| SmMADS68 | SMEL4.1_02g004220.1 | 82 | 9327.69 | 9.81 | 1 | Type I | Mα |
| SmMADS69 | SMEL4.1_02g004750.1 | 88 | 9891.25 | 8.74 | 1 | Type I | Mα |
| SmMADS70 | SMEL4.1_02g009330.1 | 163 | 18,034.52 | 6.74 | 1 | Type I | Mα |
| SmMADS71 | SMEL4.1_03g000810.1 | 177 | 20,440.99 | 6.76 | 1 | Type I | Mα |
| SmMADS72 | SMEL4.1_03g030700.1 | 176 | 20,252.33 | 8.78 | 1 | Type I | Mα |
| SmMADS73 | SMEL4.1_03g030950.1 | 177 | 20,399.58 | 8.45 | 1 | Type I | Mα |
| SmMADS74 | SMEL4.1_04g009890.1 | 131 | 15,092.25 | 8.57 | 2 | Type I | Mα |
| SmMADS75 | SMEL4.1_04g013110.1 | 213 | 24,212.58 | 7.53 | 1 | Type I | Mα |
| SmMADS76 | SMEL4.1_05g006350.1 | 196 | 21,900.60 | 5.10 | 1 | Type I | Mα |
| SmMADS77 | SMEL4.1_06g017190.1 | 160 | 18,834.31 | 9.25 | 1 | Type I | Mα |
| SmMADS78 | SMEL4.1_06g017740.1 | 195 | 21,797.04 | 9.34 | 1 | Type I | Mα |
| SmMADS79 | SMEL4.1_06g017750.1 | 221 | 24,990.43 | 9.06 | 1 | Type I | Mα |
| SmMADS80 | SMEL4.1_07g002870.1 | 154 | 17,280.80 | 9.60 | 1 | Type I | Mα |
| SmMADS81 | SMEL4.1_07g004050.1 | 177 | 20,399.58 | 8.45 | 1 | Type I | Mα |
| SmMADS82 | SMEL4.1_07g005030.1 | 82 | 9232.37 | 6.13 | 1 | Type I | Mα |
| SmMADS83 | SMEL4.1_07g005100.1 | 98 | 11,440.09 | 9.19 | 1 | Type I | Mα |
| SmMADS84 | SMEL4.1_07g005320.1 | 136 | 15,978.29 | 7.75 | 1 | Type I | Mα |
| SmMADS85 | SMEL4.1_08g005230.1 | 194 | 22,151.55 | 9.26 | 1 | Type I | Mα |
| SmMADS86 | SMEL4.1_08g014240.1 | 183 | 21,181.86 | 5.90 | 1 | Type I | Mα |
| SmMADS87 | SMEL4.1_09g003500.1 | 158 | 18,165.39 | 5.77 | 1 | Type I | Mα |
| SmMADS88 | SMEL4.1_09g006790.1 | 485 | 53,474.31 | 9.86 | 11 | Type I | Mα |
| SmMADS89 | SMEL4.1_09g019190.1 | 135 | 15,697.91 | 9.42 | 2 | Type I | Mα |
| SmMADS90 | SMEL4.1_10g002530.1 | 166 | 18,817.01 | 4.69 | 1 | Type I | Mα |
| SmMADS91 | SMEL4.1_10g002560.1 | 157 | 18,298.85 | 5.81 | 1 | Type I | Mα |
| SmMADS92 | SMEL4.1_10g002570.1 | 172 | 19,826.53 | 8.92 | 1 | Type I | Mα |
| SmMADS93 | SMEL4.1_10g018710.1 | 154 | 17,739.07 | 5.37 | 2 | Type I | Mα |
| SmMADS94 | SMEL4.1_10g018900.1 | 144 | 16,898.56 | 9.62 | 1 | Type I | Mα |
| SmMADS95 | SMEL4.1_11g020700.1 | 162 | 18,672.25 | 6.32 | 1 | Type I | Mα |
| SmMADS96 | SMEL4.1_11g022720.1 | 159 | 18,373.66 | 5.71 | 1 | Type I | Mα |
| SmMADS97 | SMEL4.1_12g006270.1 | 88 | 10,060.66 | 9.95 | 1 | Type I | Mα |
| SmMADS98 | SMEL4.1_12g017590.1 | 216 | 23,832.56 | 5.33 | 1 | Type I | Mα |
| SmMADS99 | SMEL4.1_12g017640.1 | 176 | 19,816.48 | 5.57 | 1 | Type I | Mα |
| SmMADS100 | SMEL4.1_01g000700.1 | 320 | 37,011.97 | 6.85 | 1 | Type I | Mβ |
| SmMADS101 | SMEL4.1_01g000740.1 | 312 | 35,650.71 | 6.16 | 1 | Type I | Mβ |
| SmMADS102 | SMEL4.1_01g000760.1 | 308 | 35,469.51 | 7.60 | 1 | Type I | Mβ |
| SmMADS103 | SMEL4.1_01g001040.1 | 129 | 14,916.10 | 8.96 | 1 | Type I | Mβ |
| SmMADS104 | SMEL4.1_03g020940.1 | 329 | 37,220.89 | 5.01 | 1 | Type I | Mβ |
| SmMADS105 | SMEL4.1_12g006420.1 | 296 | 33,732.90 | 4.72 | 1 | Type I | Mβ |
| SmMADS106 | SMEL4.1_12g006460.1 | 315 | 35,613.13 | 5.00 | 1 | Type I | Mβ |
| SmMADS107 | SMEL4.1_01g002320.1 | 239 | 27,173.99 | 9.18 | 1 | Type I | Mγ |
| SmMADS108 | SMEL4.1_01g006240.1 | 397 | 44,217.68 | 5.66 | 1 | Type I | Mγ |
| SmMADS109 | SMEL4.1_01g020650.1 | 318 | 35,923.03 | 7.92 | 1 | Type I | Mγ |
| SmMADS110 | SMEL4.1_03g004740.1 | 331 | 36,957.31 | 5.93 | 1 | Type I | Mγ |
| SmMADS111 | SMEL4.1_03g004770.1 | 243 | 28,181.97 | 6.24 | 1 | Type I | Mγ |
| SmMADS112 | SMEL4.1_03g004780.1 | 231 | 26,834.53 | 6.35 | 1 | Type I | Mγ |
| SmMADS113 | SMEL4.1_03g010570.1 | 180 | 20,547.28 | 6.75 | 1 | Type I | Mγ |
| SmMADS114 | SMEL4.1_04g005570.1 | 174 | 19,849.64 | 8.56 | 1 | Type I | Mγ |
| SmMADS115 | SMEL4.1_10g005130.1 | 164 | 19,207.47 | 9.28 | 1 | Type I | Mγ |
| SmMADS116 | SMEL4.1_10g006300.1 | 232 | 26,200.63 | 6.37 | 1 | Type I | Mγ |
| SmMADS117 | SMEL4.1_11g015800.1 | 397 | 44,978.00 | 5.89 | 1 | Type I | Mγ |
| SmMADS118 | SMEL4.1_11g016930.1 | 399 | 45,259.92 | 5.60 | 1 | Type I | Mγ |
| SmMADS119 | SMEL4.1_11g017020.1 | 245 | 28,156.78 | 9.01 | 1 | Type I | Mγ |
| SmMADS120 | SMEL4.1_11g017890.1 | 137 | 15,902.54 | 9.23 | 1 | Type I | Mγ |
| Primers | Sequence(5′—3′) |
|---|---|
| qPCR_SmMADS3_F | GACACCGTCTGACCAGGC |
| qPCR_SmMADS3_R | TAGCGGCGCTATCTCCGT |
| qPCR_SmMADS28_F | TCGTCCTCACGCTCAACG |
| qPCR_SmMADS28_R | AGGAAATCCGGTGTGGCG |
| qPCR_SmMADS30_F | CCCACCCAACTTTTTACCCG |
| qPCR_SmMADS30_R | AGACGGACTGAGAGAGCACT |
| qPCR_SmMADS41_F | AGACCTCGAACCCGTCCA |
| qPCR_SmMADS41_R | CCGATCGATGTTCGCCGA |
| qPCR_SmMADS75_F | ACGGGTGCTGAAATCGCT |
| qPCR_SmMADS75_R | ACTGCGTCAGGGCTAGGA |
| qPCR_SmMADS103_F | GCCATGGCGGAACAAGATC |
| qPCR_SmMADS103_R | TTCCCCATGAGGACCGGT |


References
- Ng, M.; Yanofsky, M.F. Function and evolution of the plant MADS-box gene family. Nat. Rev. Genet. 2001, 2, 186–195. [Google Scholar] [CrossRef] [PubMed]
- Schilling, S.; Pan, S.; Kennedy, A.; Melzer, R. MADS-box genes and crop domestication: The jack of all traits. J. Exp. Bot. 2018, 69, 1447–1469. [Google Scholar] [CrossRef] [PubMed]
- Alvarez-Buylla, E.R.; García-Ponce, B.; Sánchez, M.P.; Espinosa-Soto, C.; García-Gómez, M.L.; Piñeyro-Nelson, A.; Garay-Arroyo, A. MADS-box genes underground becoming mainstream: Plant root developmental mechanisms. New Phytol. 2019, 223, 1143–1158. [Google Scholar] [CrossRef]
- Schwarz-Sommer, Z.; Huijser, P.; Nacken, W.; Saedler, H.; Sommer, H. Genetic Control of Flower Development by Homeotic Genes in Antirrhinum majus. Science 1990, 250, 931–936. [Google Scholar] [CrossRef] [PubMed]
- Shore, P.; Sharrocks, A.D. The MADS-box family of transcription factors. Eur. J. Biochem. 1995, 229, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Gramzow, L.; Theissen, G. A hitchhiker’s guide to the MADS world of plants. Genome Biol. 2010, 11, 214. [Google Scholar] [CrossRef]
- Henschel, K.; Kofuji, R.; Hasebe, M.; Saedler, H.; Münster, T.; Theissen, G. Two ancient classes of MIKC-type MADS-box genes are present in the moss Physcomitrella patens. Mol. Biol. Evol. 2002, 19, 801–814. [Google Scholar] [CrossRef]
- Nam, J.; Kim, J.; Lee, S.; An, G.; Ma, H.; Nei, M. Type I MADS-box genes have experienced faster birth-and-death evolution than type II MADS-box genes in angiosperms. Proc. Natl. Acad. Sci. USA 2004, 101, 1910–1915. [Google Scholar] [CrossRef]
- Wang, Y.; Jiang, H.; Wang, G. PHERES1 Controls Endosperm Gene Imprinting and Seed Development. Trends Plant Sci. 2020, 25, 517–519. [Google Scholar] [CrossRef]
- Coito, J.L.; Silva, H.; Ramos, M.; Montez, M.; Cunha, J.; Amâncio, S.; Costa, M.; Rocheta, M. Vitis Flower Sex Specification Acts Downstream and Independently of the ABCDE Model Genes. Front. Plant Sci. 2018, 9, 1029. [Google Scholar] [CrossRef]
- Amasino, R.M.; Michaels, S.D. The timing of flowering. Plant Physiol. 2010, 154, 516–520. [Google Scholar] [CrossRef] [PubMed]
- Bergamini, C.; Cardone, M.F.; Anaclerio, A.; Perniola, R.; Pichierri, A.; Genghi, R.; Alba, V.; Forleo, L.R.; Caputo, A.R.; Montemurro, C.; et al. Validation assay of p3_VvAGL11 marker in a wide range of genetic background for early selection of stenospermocarpy in Vitis vinifera L. Mol. Biotechnol. 2013, 54, 1021–1030. [Google Scholar] [CrossRef]
- Ocarez, N.; Mejia, N. Suppression of the D-class MADS-box AGL11 gene triggers seedlessness in fleshy fruits. Plant Cell Rep. 2016, 35, 239–254. [Google Scholar] [CrossRef]
- Singh, R.; Low, E.T.; Ooi, L.C.; Ong-Abdullah, M.; Ting, N.C.; Nagappan, J.; Nookiah, R.; Amiruddin, M.D.; Rosli, R.; Manaf, M.A.; et al. The oil palm SHELL gene controls oil yield and encodes a homologue of SEEDSTICK. Nature 2013, 500, 340–344. [Google Scholar] [CrossRef] [PubMed]
- Montiel, G.; Gaudet, M.; Laurans, F.; Rozenberg, P.; Simon, M.; Gantet, P.; Jay-Allemand, C.; Breton, C. Overexpression of MADS-box Gene AGAMOUS-LIKE 12 Activates Root Development in Juglans sp. and Arabidopsis thaliana. Plants 2020, 9, 444. [Google Scholar] [CrossRef] [PubMed]
- Parenicová, L.; de Folter, S.; Kieffer, M.; Horner, D.S.; Favalli, C.; Busscher, J.; Cook, H.E.; Ingram, R.M.; Kater, M.M.; Davies, B.; et al. Molecular and phylogenetic analyses of the complete MADS-box transcription factor family in Arabidopsis: New openings to the MADS world. Plant Cell 2003, 15, 1538–1551. [Google Scholar] [CrossRef]
- Arora, R.; Garwal, P.; Ray, S.; Singh, A.K.; Singh, V.P.; Tyagi, A.K.; Kapoor, S. MADS-box gene family in rice: Genome-wide identification, organization and expression profiling during reproductive development and stress. BMC Genom. 2007, 8, 242. [Google Scholar] [CrossRef]
- Wang, Y.; Zhang, J.; Hu, Z.; Guo, X.; Tian, S.; Chen, G. Genome-Wide Analysis of the MADS-Box Transcription Factor Family in Solanum lycopersicum. Int. J. Mol. Sci. 2019, 20, 2961. [Google Scholar] [CrossRef]
- Gao, H.; Wang, Z.; Li, S.; Hou, M.; Zhou, Y.; Zhao, Y.; Li, G.; Zhao, H.; Ma, H. Genome-wide survey of potato MADS-box genes reveals that StMADS1 and StMADS13 are putative downstream targets of tuberigen StSP6A. BMC Genom. 2018, 19, 726. [Google Scholar] [CrossRef]
- Hu, L.; Liu, S. Genome-wide analysis of the MADS-box gene family in cucumber. Genome 2012, 55, 245–256. [Google Scholar] [CrossRef]
- Añibarro-Ortega, M.; Pinela, J.; Alexopoulos, A.; Petropoulos, S.A.; Ferreira, I.; Barros, L. The powerful Solanaceae: Food and nutraceutical applications in a sustainable world. Adv. Food Nutr. Res. 2022, 100, 131–172. [Google Scholar] [PubMed]
- Weese, T.L.; Bohs, L. Eggplant origins: Out of Africa, into the Orient. Taxon 2010, 59, 49–56. [Google Scholar] [CrossRef]
- Gajewski, M.; Katarzyna, K.; Bajer, M. The Influence of Postharvest Storage on Quality Characteristics of Fruit of Eggplant Cultivars. Not. Bot. Horti Agrobot. Cluj-Napoca 2009, 37, 200–205. [Google Scholar]
- Hanson, P.M.; Yang, R.Y.; Tsou, S.C.S.; Ledesma, D.; Engle, L.; Lee, T.C. Diversity in eggplant (Solanum melongena) for superoxide scavenging activity, total phenolics, and ascorbic acid. J. Food Compos. Anal. 2006, 19, 594–600. [Google Scholar] [CrossRef]
- Xu, W.; Bobet, S.; Le Gourrierec, J.; Grain, D.; De Vos, D.; Berger, A.; Salsac, F.; Kelemen, Z.; Boucherez, J.; Rolland, A.; et al. TRANSPARENT TESTA 16 and 15 act through different mechanisms to control proanthocyanidin accumulation in Arabidopsis testa. J. Exp. Bot. 2017, 68, 2859–2870. [Google Scholar] [CrossRef] [PubMed]
- Su, M.; Wang, S.; Liu, W.; Yang, M.; Zhang, Z.; Wang, N.; Chen, X. MdJa2 Participates in the Brassinosteroid Signaling Pathway to Regulate the Synthesis of Anthocyanin and Proanthocyanidin in Red-Fleshed Apple. Front. Plant Sci. 2022, 13, 830349. [Google Scholar] [CrossRef]
- Cheng, L.; Li, R.; Wang, X.; Ge, S.; Wang, S.; Liu, X.; He, J.; Jiang, C.Z.; Qi, M.; Xu, T.; et al. A SlCLV3-SlWUS module regulates auxin and ethylene homeostasis in low light-induced tomato flower abscission. Plant Cell 2022, 34, 4388–4408. [Google Scholar] [CrossRef]
- Gonzalez, A.; Zhao, M.; Leavitt, J.M.; Lloyd, A. Regulation of the anthocyanin biosynthetic pathway by the TTG1/bHLH/Myb transcriptional complex in Arabidopsis seedlings. Plant J. 2008, 53, 814–827. [Google Scholar] [CrossRef]
- Yanofsky, M.F.; Ma, H.; Bowman, J.L.; Drews, G.N.; Feldmann, K.A.; Meyerowitz, E.M. The protein encoded by the Arabidopsis homeotic gene agamous resembles transcription factors. Nature 1990, 346, 35–39. [Google Scholar] [CrossRef]
- Krizek, B.A.; Meyerowitz, E.M. The Arabidopsis homeotic genes APETALA3 and PISTILLATA are sufficient to provide the B class organ identity function. Development 1996, 122, 11–22. [Google Scholar] [CrossRef]
- Pelaz, S.; Ditta, G.S.; Baumann, E.; Wisman, E.; Yanofsky, M.F. B and C floral organ identity functions require SEPALLATA MADS-box genes. Nature 2000, 405, 200–203. [Google Scholar] [CrossRef] [PubMed]
- Mizzotti, C.; Ezquer, I.; Paolo, D.; Rueda-Romero, P.; Guerra, R.F.; Battaglia, R.; Rogachev, I.; Aharoni, A.; Kater, M.M.; Caporali, E.; et al. SEEDSTICK is a master regulator of development and metabolism in the Arabidopsis seed coat. PLoS Genet. 2014, 10, e1004856. [Google Scholar] [CrossRef] [PubMed]
- Raza, Q.; Riaz, A.; Atif, R.M.; Hussain, B.; Rana, I.A.; Ali, Z.; Budak, H.; Alaraidh, I.A. Genome-Wide Diversity of MADS-Box Genes in Bread Wheat is Associated with its Rapid Global Adaptability. Front. Genet. 2022, 12, 818880. [Google Scholar] [CrossRef] [PubMed]
- Saha, G.; Park, J.I.; Jung, H.J.; Ahmed, N.U.; Kayum, M.A.; Chung, M.Y.; Hur, Y.; Cho, Y.G.; Watanabe, M.; Nou, I.S. Genome-wide identification and characterization of MADS-box family genes related to organ development and stress resistance in Brassica rapa. BMC Genom. 2015, 16, 178. [Google Scholar] [CrossRef]
- Li, C.; Wang, Y.; Xu, L.; Nie, S.; Chen, Y.; Liang, D.; Sun, X.; Karanja, B.K.; Luo, X.; Liu, L. Genome-Wide Characterization of the MADS-Box Gene Family in Radish (Raphanus sativus L.) and Assessment of Its Roles in Flowering and Floral Organogenesis. Front. Plant Sci. 2016, 7, 1390. [Google Scholar] [CrossRef]
- Tian, Y.; Dong, Q.; Ji, Z.; Chi, F.; Cong, P.; Zhou, Z. Genome-wide identification and analysis of the MADS-box gene family in apple. Gene 2015, 555, 277–290. [Google Scholar] [CrossRef]
- Wei, X.; Wang, L.; Yu, J.; Zhang, Y.; Li, D.; Zhang, X. Genome-wide identification and analysis of the MADS-box gene family in sesame. Gene 2015, 569, 66–76. [Google Scholar] [CrossRef]
- Won, S.Y.; Jung, J.A.; Kim, J.S. Genome-wide analysis of the MADS-Box gene family in Chrysanthemum. Comput. Biol. Chem. 2021, 90, 107424. [Google Scholar] [CrossRef]
- Wang, P.; Wang, S.; Chen, Y.; Xu, X.; Guang, X.; Zhang, Y. Genome-wide Analysis of the MADS-Box Gene Family in Watermelon. Comput. Biol. Chem. 2019, 80, 341–350. [Google Scholar] [CrossRef]
- Dorca-Fornell, C.; Gregis, V.; Grandi, V.; Coupland, G.; Colombo, L.; Kater, M.M. The Arabidopsis SOC1-like genes AGL42, AGL71 and AGL72 promote flowering in the shoot apical and axillary meristems. Plant J. 2011, 67, 1006–1017. [Google Scholar] [CrossRef]
- Chen, M.K.; Hsu, W.H.; Lee, P.F.; Thiruvengadam, M.; Chen, H.-I.; Yang, C.-H. The MADS box gene, FOREVER YOUNG FLOWER, acts as a repressor controlling floral organ senescence and abscission in Arabidopsis. Plant J. 2011, 68, 168–185. [Google Scholar] [CrossRef] [PubMed]
- Kang, I.H.; Steffen, J.G.; Portereiko, M.F.; Lloyd, A.; Drews, G.N. The AGL62 MADS domain protein regulates cellularization during endosperm development in Arabidopsis. Plant Cell 2008, 20, 635–647. [Google Scholar] [CrossRef] [PubMed]
- Liu, D.; Wang, D.; Qin, Z.; Zhang, D.; Yin, L.; Wu, L.; Colasanti, J.; Li, A.; Mao, L. The SEPALLATA MADS-box protein SLMBP21 forms protein complexes with JOINTLESS and MACROCALYX as a transcription activator for development of the tomato flower abscission zone. Plant J. 2014, 77, 284–296. [Google Scholar] [CrossRef]
- Liu, J.; Xu, B.; Hu, L.; Li, M.; Su, W.; Wu, J.; Yang, J.; Jin, Z. Involvement of a banana MADS-box transcription factor gene in ethylene-induced fruit ripening. Plant Cell Rep. 2009, 28, 103–111. [Google Scholar] [CrossRef] [PubMed]
- Nakatsuka, T.; Saito, M.; Yamada, E.; Fujita, K.; Yamagishi, N.; Yoshikawa, N.; Nishihara, M. Isolation and characterization of the C-class MADS-box gene involved in the formation of double flowers in Japanese gentian. BMC Plant Biol. 2015, 15, 182. [Google Scholar] [CrossRef]
- Figueiredo, D.D.; Batista, R.A.; Roszak, P.J.; Hennig, L.; Köhler, C. Auxin production in the endosperm drives seed coat development in Arabidopsis. eLife 2016, 5, e20542. [Google Scholar] [CrossRef]
- Hsu, H.F.; Chen, W.H.; Shen, Y.H.; Hsu, W.H.; Mao, W.T.; Yang, C.H. Multifunctional evolution of B and AGL6 MADS box genes in orchids. Nat. Commun. 2021, 12, 902. [Google Scholar] [CrossRef]
- Mao, W.T.; Hsu, H.F.; Hsu, W.H.; Li, J.Y.; Lee, Y.I.; Yang, C.H. The C-Terminal Sequence and PI motif of the Orchid (Oncidium Gower Ramsey) PISTILLATA (PI) Ortholog Determine its Ability to Bind AP3 Orthologs and Enter the Nucleus to Regulate Downstream Genes Controlling Petal and Stamen Formation. Plant Cell Physiol. 2015, 56, 2079–2099. [Google Scholar]
- Wu, R.; Wang, T.; McGie, T.; Voogd, C.; Allan, A.C.; Hellens, R.P.; Varkonyi-Gasic, E. Overexpression of the kiwifruit SVP3 gene affects reproductive development and suppresses anthocyanin biosynthesis in petals, but has no effect on vegetative growth, dormancy, or flowering time. J. Exp. Bot. 2014, 65, 4985–4995. [Google Scholar] [CrossRef]
- Qi, F.; Liu, Y.; Luo, Y.; Cui, Y.; Lu, C.; Li, H.; Huang, H.; Dai, S. Functional analysis of the ScAG and ScAGL11 MADS-box transcription factors for anthocyanin biosynthesis and bicolour pattern formation in Senecio cruentus ray florets. Hortic. Res. 2022, 9, uhac071. [Google Scholar] [CrossRef]
- Altschul, S.F.; Wootton, J.C.; Gertz, E.M.; Agarwala, R.; Morgulis, A.; Schäffer, A.A.; Yu, Y.K. Protein database searches using compositionally adjusted substitution matrices. FEBS J. 2005, 272, 5101–5109. [Google Scholar] [CrossRef] [PubMed]
- Mistry, J.; Chuguransky, S.; Williams, L.; Qureshi, M.; Salazar, G.A.; Sonnhammer, E.; Tosatto, S.; Paladin, L.; Raj, S.; Richardson, L.J.; et al. Pfam: The protein families database in 2021. Nucleic Acids Res. 2021, 49, D412–D419. [Google Scholar] [CrossRef] [PubMed]
- Tamura, K.; Stecher, G.; Kumar, S. MEGA11: Molecular Evolutionary Genetics Analysis Version 11. Mol. Biol. Evol. 2021, 38, 3022–3027. [Google Scholar] [CrossRef] [PubMed]
- Letunic, I.; Bork, P. Interactive Tree Of Life (iTOL) v5: An online tool for phylogenetic tree display and annotation. Nucleic Acids Res. 2021, 49, W293–W296. [Google Scholar] [CrossRef] [PubMed]
- Ayaz, A.; Huang, H.; Zheng, M.; Zaman, W.; Li, D.; Saqib, S.; Zhao, H.; Lü, S. Molecular Cloning and Functional Analysis of GmLACS2-3 Reveals Its Involvement in Cutin and Suberin Biosynthesis along with Abiotic Stress Tolerance. Int. J. Mol. Sci. 2021, 22, 9175. [Google Scholar] [CrossRef]
- Ayaz, A.; Saqib, S.; Huang, H.; Zaman, W.; Lü, S.; Zhao, H. Genome-wide comparative analysis of long-chain acyl-CoA synthetases (LACSs) gene family: A focus on identification, evolution and expression profiling related to lipid synthesis. Plant Physiol. Biochem. 2021, 161, 1–11. [Google Scholar] [CrossRef]
- Chen, C.; Chen, H.; Zhang, Y.; Thomas, H.R.; Frank, M.H.; He, Y.; Xia, R. TBtools: An Integrative Toolkit Developed for Interactive Analyses of Big Biological Data. Mol Plant. 2020, 13, 1194–1202. [Google Scholar] [CrossRef]
- Krzywinski, M.; Schein, J.; Birol, I.; Connors, J.; Gascoyne, R.; Horsman, D.; Jones, S.J.; Marra, M.A. Circos: An information aesthetic for comparative genomics. Genome Res. 2009, 19, 1639–1645. [Google Scholar] [CrossRef]
- Szklarczyk, D.; Franceschini, A.; Wyder, S.; Forslund, K.; Heller, D.; Huerta-Cepas, J.; Simonovic, M.; Roth, A.; Santos, A.; Tsafou, K.P.; et al. STRING v10: Protein-protein interaction networks, integrated over the tree of life. Nucleic Acids Res. 2015, 43, D447–D452. [Google Scholar] [CrossRef]
- Arocho, A.; Chen, B.; Ladanyi, M.; Pan, Q. Validation of the 2-DeltaDeltaCt calculation as an alternate method of data analysis for quantitative PCR of BCR-ABL P210 transcripts. Diagn. Mol. Pathol. 2006, 15, 56–61. [Google Scholar] [CrossRef]



Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chen, Q.; Li, J.; Yang, F. Genome-Wide Analysis of the Mads-Box Transcription Factor Family in Solanum melongena. Int. J. Mol. Sci. 2023, 24, 826. https://doi.org/10.3390/ijms24010826
Chen Q, Li J, Yang F. Genome-Wide Analysis of the Mads-Box Transcription Factor Family in Solanum melongena. International Journal of Molecular Sciences. 2023; 24(1):826. https://doi.org/10.3390/ijms24010826
Chicago/Turabian StyleChen, Qi, Jing Li, and Fengjuan Yang. 2023. "Genome-Wide Analysis of the Mads-Box Transcription Factor Family in Solanum melongena" International Journal of Molecular Sciences 24, no. 1: 826. https://doi.org/10.3390/ijms24010826
APA StyleChen, Q., Li, J., & Yang, F. (2023). Genome-Wide Analysis of the Mads-Box Transcription Factor Family in Solanum melongena. International Journal of Molecular Sciences, 24(1), 826. https://doi.org/10.3390/ijms24010826






