GPR19 Coordinates Multiple Molecular Aspects of Stress Responses Associated with the Aging Process
Abstract
1. Introduction
2. Results
2.1. Coordinated Protein Expression Profiles of GPR19 in Advanced Aging Murine Models
2.2. Ectopic Expression Induced Human GPR19 Perturbagen Responses in Human Cells
2.3. GPR19 Perturbagen Responses Are Reminiscent of a Cancer-Associated Functional Network
2.4. GPR19 Perturbagen Responses Demonstrate a Complex “Dose-Dependent” Functional Diversity
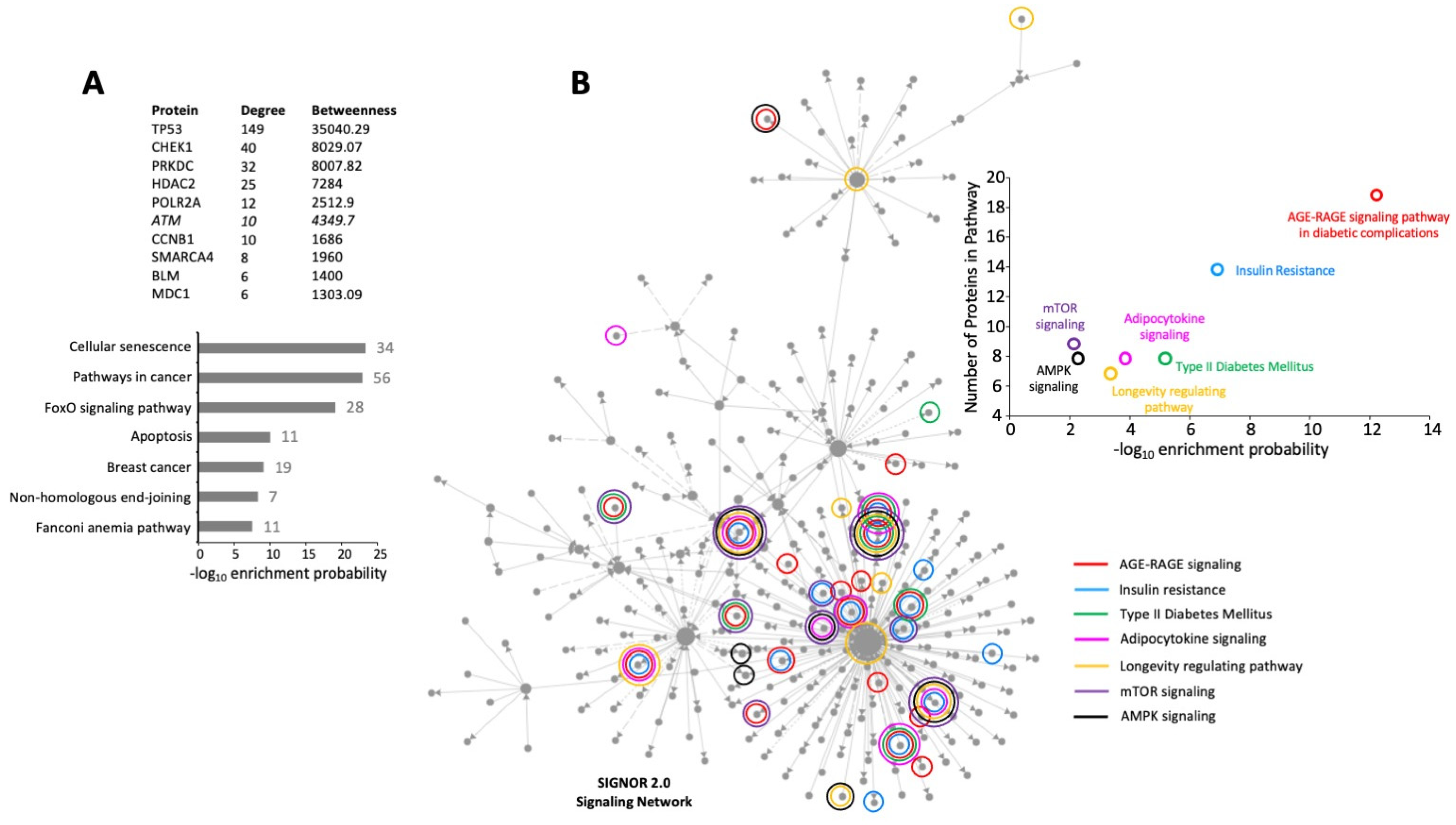
2.5. Distinctive Compartment-Based Interpretation of Gpr19 Perturbagen Response Indicates a Persistent Molecular Signature of DNA Damage Management, Energy Regulation, and Cancer Physiology
2.6. Extracting MDC1-Associated Signaling Components from the Global GPR19 Perturbagen Signature
2.7. Protein Subcomplex Analysis of the GPR19 Perturbagen Response Phenotype
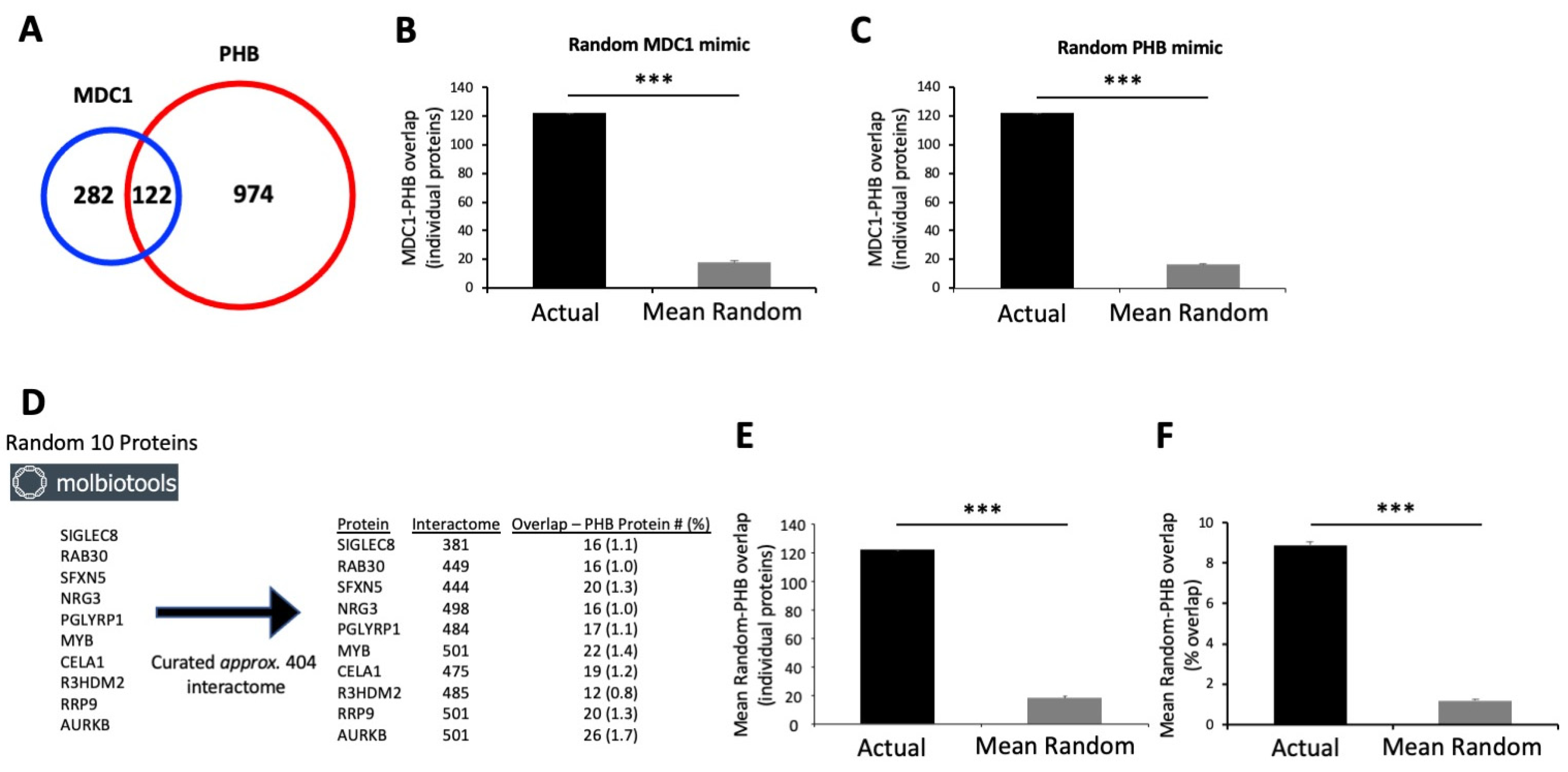
2.8. Verification of the Potential Biological Significance of an MDC1-PHB Functional Interaction
2.9. Interaction Analysis of Potential MDC1 PHB Functional Intersection
2.10. Expression of GPR19 Can Attenuate Loss of Cell Viability Induced by Exogenous Cell Stressors
3. Discussion
4. Materials and Methods
4.1. Cell Culture, Transfection, and Treatment
4.2. Cellular Protein Extraction
4.3. Quantitative Proteomic Analyses
4.4. Bioinformatic Analyses
4.5. Immunoblots and, Immunoprecipitation
4.6. Murine Tissue RT-PCR
4.7. Statistical Analyses
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Maudsley, S.; Patel, S.A.; Park, S.S.; Luttrell, L.M.; Martin, B. Functional signaling biases in G protein-coupled receptors: Game Theory and receptor dynamics. Mini Rev. Med. Chem. 2012, 12, 831–840. [Google Scholar] [CrossRef] [PubMed]
- Maudsley, S.; Siddiqui, S.; Martin, B. Systems analysis of arrestin pathway functions. Prog. Mol. Biol. Transl. Sci. 2013, 118, 431–467. [Google Scholar] [CrossRef] [PubMed]
- Maudsley, S.; Martin, B.; Janssens, J.; Etienne, H.; Jushaj, A.; van Gastel, J.; Willemsen, A.; Chen, H.; Gesty-Palmer, D.; Luttrell, L.M. Informatic deconvolution of biased GPCR signaling mechanisms from in vivo pharmacological experimentation. Methods 2016, 92, 51–63. [Google Scholar] [CrossRef]
- Maudsley, S.; Leysen, H.; van Gastel, J.; Martin, B. Systems Pharmacology: Enabling Multidimensional Therapeutics; Elsevier: Amsterdam, The Netherlands, 2021. [Google Scholar] [CrossRef]
- Leysen, H.; van Gastel, J.; Hendrickx, J.O.; Santos-Otte, P.; Martin, B.; Maudsley, S. G Protein-Coupled Receptor Systems as Crucial Regulators of DNA Damage Response Processes. Int. J. Mol. Sci. 2018, 19, 2919. [Google Scholar] [CrossRef] [PubMed]
- Van Gastel, J.; Leysen, H.; Santos-Otte, P.; Hendrickx, J.O.; Azmi, A.; Martin, B.; Maudsley, S. The RXFP3 receptor is functionally associated with cellular responses to oxidative stress and DNA damage. Aging 2019, 11, 11268–11313. [Google Scholar] [CrossRef]
- Santos-Otte, P.; Leysen, H.; van Gastel, J.; Hendrickx, J.O.; Martin, B.; Maudsley, S. G Protein-Coupled Receptor Systems and Their Role in Cellular Senescence. Comput. Struct. Biotechnol. J. 2019, 17, 1265–1277. [Google Scholar] [CrossRef]
- Van Gastel, J.; Leysen, H.; Boddaert, J.; Vangenechten, L.; Luttrell, L.M.; Martin, B.; Maudsley, S. Aging-related modifications to G protein-coupled receptor signaling diversity. Pharm. Ther. 2021, 223, 107793. [Google Scholar] [CrossRef]
- Leysen, H.; Walter, D.; Christiaenssen, B.; Vandoren, R.; Harputluoğlu, İ.; Van Loon, N.; Maudsley, S. GPCRs Are Optimal Regulators of Complex Biological Systems and Orchestrate the Interface between Health and Disease. Int. J. Mol. Sci. 2021, 22, 13387. [Google Scholar] [CrossRef]
- Guo, S.; Zhao, T.; Yun, Y.; Xie, X. Recent progress in assays for GPCR drug discovery. Am. J. Physiol. Cell Physiol. 2022, 323, C583–C594. [Google Scholar] [CrossRef]
- Hendrickx, J.O.; van Gastel, J.; Leysen, H.; Martin, B.; Maudsley, S. High-dimensionality Data Analysis of Pharmacological Systems Associated with Complex Diseases. Pharm. Rev. 2020, 72, 191–217. [Google Scholar] [CrossRef]
- Moran, B.M.; McKillop, A.M.; O’Harte, F.P. Development of novel ligands for peptide GPCRs. Curr. Opin. Pharm. 2016, 31, 57–62. [Google Scholar] [CrossRef] [PubMed]
- Luttrell, L.M.; Maudsley, S.; Bohn, L.M. Fulfilling the Promise of “Biased” G Protein-Coupled Receptor Agonism. Mol. Pharmacol. 2015, 88, 579–588. [Google Scholar] [CrossRef] [PubMed]
- Janssens, J.; Etienne, H.; Idriss, S.; Azmi, A.; Martin, B.; Maudsley, S. Systems-Level G Protein-Coupled Receptor Therapy Across a Neurodegenerative Continuum by the GLP-1 Receptor System. Front. Endocrinol. 2014, 5, 142. [Google Scholar] [CrossRef] [PubMed]
- McDonald, P.H.; Lefkowitz, R.J. Beta-Arrestins: New roles in regulating heptahelical receptors’ functions. Cell. Signal. 2001, 13, 683–689. [Google Scholar] [CrossRef] [PubMed]
- Maudsley, S.; Martin, B.; Luttrell, L.M. The origins of diversity and specificity in g protein-coupled receptor signaling. J. Pharm. Exp. 2005, 314, 485–494. [Google Scholar] [CrossRef] [PubMed]
- Bohn, L.M.; McDonald, P.H. Seeking Ligand Bias: Assessing GPCR Coupling to Beta-Arrestins for Drug Discovery. Drug Discov. Today Technol. 2010, 7, e37–e42. [Google Scholar] [CrossRef]
- Van Gastel, J.; Boddaert, J.; Jushaj, A.; Premont, R.T.; Luttrell, L.M.; Janssens, J.; Martin, B.; Maudsley, S. GIT2-A keystone in ageing and age-related disease. Ageing Res. Rev. 2018, 43, 46–63. [Google Scholar] [CrossRef]
- Van Gastel, J.; Cai, H.; Cong, W.N.; Chadwick, W.; Daimon, C.; Leysen, H.; Hendrickx, J.O.; De Schepper, R.; Vangenechten, L.; Van Turnhout, J.; et al. Multidimensional informatic deconvolution defines gender-specific roles of hypothalamic GIT2 in aging trajectories. Mech. Ageing Dev. 2019, 184, 111150. [Google Scholar] [CrossRef]
- Chadwick, W.; Zhou, Y.; Park, S.S.; Wang, L.; Mitchell, N.; Stone, M.D.; Becker, K.G.; Martin, B.; Maudsley, S. Minimal peroxide exposure of neuronal cells induces multifaceted adaptive responses. PLoS ONE 2010, 5, e14352. [Google Scholar] [CrossRef]
- Chadwick, W.; Martin, B.; Chapter, M.C.; Park, S.S.; Wang, L.; Daimon, C.M.; Brenneman, R.; Maudsley, S. GIT2 acts as a potential keystone protein in functional hypothalamic networks associated with age-related phenotypic changes in rats. PLoS ONE 2012, 7, e36975. [Google Scholar] [CrossRef]
- Lu, D.; Cai, H.; Park, S.S.; Siddiqui, S.; Premont, R.T.; Schmalzigaug, R.; Paramasivam, M.; Seidman, M.; Bodogai, I.; Biragyn, A.; et al. Nuclear GIT2 is an ATM substrate and promotes DNA repair. Mol. Cell Biol. 2015, 35, 1081–1096, Erratum in Mol. Cell Biol. 2017, 37. [Google Scholar] [CrossRef] [PubMed]
- Martin, B.; Chadwick, W.; Janssens, J.; Premont, R.T.; Schmalzigaug, R.; Becker, K.G.; Lehrmann, E.; Wood, W.H.; Zhang, Y.; Siddiqui, S.; et al. GIT2 Acts as a Systems-Level Coordinator of Neurometabolic Activity and Pathophysiological Aging. Front. Endocrinol. 2016, 6, 191. [Google Scholar] [CrossRef] [PubMed]
- Siddiqui, S.; Lustig, A.; Carter, A.; Sankar, M.; Daimon, C.M.; Premont, R.T.; Etienne, H.; van Gastel, J.; Azmi, A.; Janssens, J.; et al. Genomic deletion of GIT2 induces a premature age-related thymic dysfunction and systemic immune system disruption. Aging 2017, 9, 706–740. [Google Scholar] [CrossRef] [PubMed]
- Anckaerts, C.; van Gastel, J.; Leysen, V.; Hinz, R.; Azmi, A.; Simoens, P.; Shah, D.; Kara, F.; Langbeen, A.; Bols, P.; et al. Image-guided phenotyping of ovariectomized mice: Altered functional connectivity, cognition, myelination, and dopaminergic functionality. Neurobiol. Aging 2019, 74, 77–89. [Google Scholar] [CrossRef] [PubMed]
- Zabaneh, D.; Balding, D.J. A genome-wide association study of the metabolic syndrome in Indian Asian men. PLoS ONE 2010, 5, e11961. [Google Scholar] [CrossRef]
- Van Gastel, J.; Janssens, J.; Etienne, H.; Azmi, A.; Maudsley, S. The synergistic GIT2-RXFP3 system in the brain and its importance in age-related disorders. Front. Aging NeuroSci. 2016, 3, 8. [Google Scholar]
- Van Gastel, J.; Hendrickx, J.O.; Leysen, H.; Luttrell, L.M.; Lee, M.-H.; Azmi, A.; Janssens, J.; Maudsley, S. The RXFP3-GIT2 signaling system represents a potential multidimensional therapeutic target in age-related disorders. FASEB J. 2018, 32, 533. [Google Scholar] [CrossRef]
- O’Dowd, B.F.; Nguyen, T.; Lynch, K.R.; Kolakowski, L.F., Jr.; Thompson, M.; Cheng, R.; Marchese, A.; Ng, G.; Heng, H.H.; George, S.R. A novel gene codes for a putative G protein-coupled receptor with an abundant expression in brain. FEBS Lett. 1996, 394, 325–329. [Google Scholar] [CrossRef]
- Montpetit, A.; Sinnett, D. Physical mapping of the G-protein coupled receptor 19 (GPR19) in the chromosome 12p12.3 region frequently rearranged in cancer cells. Hum. Genet. 1999, 105, 162–164. [Google Scholar] [CrossRef]
- Bresnick, J.N.; Skynner, H.A.; Chapman, K.L.; Jack, A.D.; Zamiara, E.; Negulescu, P.; Beaumont, K.; Patel, S.; McAllister, G. Identification of signal transduction pathways used by orphan g protein-coupled receptors. Assay Drug Dev. Technol. 2003, 1, 239–249. [Google Scholar] [CrossRef]
- Stein, L.M.; Yosten, G.L.; Samson, W.K. Adropin acts in brain to inhibit water drinking: Potential interaction with the orphan G protein-coupled receptor, GPR19. Am. J. Physiol. Regul. Integr. Comp. Physiol. 2016, 310, R476–R480. [Google Scholar] [CrossRef] [PubMed]
- Maudsley, S.; Walter, D.; Schrauwen, C.; Van Loon, N.; Harputluoğlu, İ.; Lenaerts, J.; McDonald, P. Intersection of the Orphan G Protein-Coupled Receptor, GPR19, with the Aging Process. Int. J. Mol. Sci. 2022, 23, 13598. [Google Scholar] [CrossRef] [PubMed]
- Rao, A.; Herr, D.R. G protein-coupled receptor GPR19 regulates E-cadherin expression and invasion of breast cancer cells. Biochim. Biophys Acta Mol. Cell Res. 2017, 1864, 1318–1327. [Google Scholar] [CrossRef] [PubMed]
- Southern, C.; Cook, J.M.; Neetoo-Isseljee, Z.; Taylor, D.L.; Kettleborough, C.A.; Merritt, A.; Bassoni, D.L.; Raab, W.J.; Quinn, E.; Wehrman, T.S.; et al. Screening β-arrestin recruitment for the identification of natural ligands for orphan G-protein-coupled receptors. J. Biomol. Screen. 2013, 18, 599–609. [Google Scholar] [CrossRef]
- Park, S.S.; Wu, W.W.; Zhou, Y.; Shen, R.F.; Martin, B.; Maudsley, S. Effective correction of experimental errors in quantitative proteomics using stable isotope labeling by amino acids in cell culture (SILAC). J. Proteom. 2012, 75, 3720–3732. [Google Scholar] [CrossRef]
- Martin, B.; Brenneman, R.; Golden, E.; Walent, T.; Becker, K.G.; Prabhu, V.V.; Wood, W., 3rd; Ladenheim, B.; Cadet, J.L.; Maudsley, S. Growth factor signals in neural cells: Coherent patterns of interaction control multiple levels of molecular and phenotypic responses. J. Biol. Chem. 2009, 284, 2493–2511. [Google Scholar] [CrossRef]
- Chadwick, W.; Boyle, J.P.; Zhou, Y.; Wang, L.; Park, S.S.; Martin, B.; Wang, R.; Becker, K.G.; Wood, W.H., 3rd; Zhang, Y.; et al. Multiple oxygen tension environments reveal diverse patterns of transcriptional regulation in primary astrocytes. PLoS ONE 2011, 6, e21638. [Google Scholar] [CrossRef]
- Pujana, M.A.; Han, J.D.; Starita, L.M.; Stevens, K.N.; Tewari, M.; Ahn, J.S.; Rennert, G.; Moreno, V.; Kirchhoff, T.; Gold, B. Network modeling links breast cancer susceptibility and centrosome dysfunction. Nat. Genet. 2007, 39, 1338–1349. [Google Scholar] [CrossRef]
- Guo, Q.Q.; Wang, S.S.; Zhang, S.S.; Xu, H.D.; Li, X.M.; Guan, Y.; Yi, F.; Zhou, T.T.; Jiang, B.; Bai, N.; et al. ATM-CHK2-Beclin 1 axis promotes autophagy to maintain ROS homeostasis under oxidative stress. EMBO J. 2020, 39, e103111. [Google Scholar] [CrossRef]
- Kwon, J.; Lee, S.; Kim, Y.N.; Lee, I.H. Deacetylation of CHK2 by SIRT1 protects cells from oxidative stress-dependent DNA damage response. Exp. Mol. Med. 2019, 51, 1–9. [Google Scholar] [CrossRef]
- Beliveau, A.; Yaswen, P. Soothing the watchman: Telomerase reduces the p53-dependent cellular stress response. Cell Cycle 2007, 6, 1284–1287. [Google Scholar] [CrossRef] [PubMed]
- Turan, V.; Oktay, K. BRCA-related ATM-mediated DNA double-strand break repair and ovarian aging. Hum. Reprod. Update 2020, 26, 43–57. [Google Scholar] [CrossRef] [PubMed]
- White, R.R.; Vijg, J. Do DNA Double-Strand Breaks Drive Aging? Mol. Cell 2016, 63, 729–738. [Google Scholar] [CrossRef] [PubMed]
- Vurusaner, B.; Poli, G.; Basaga, H. Tumor suppressor genes and ROS: Complex networks of interactions. Free Radic. Biol. Med. 2012, 52, 7–18. [Google Scholar] [CrossRef] [PubMed]
- Jourquin, J.; Duncan, D.; Shi, Z.; Zhang, B. GLAD4U: Deriving and prioritizing gene lists from PubMed literature. BMC Genom. 2012, 13, S20. [Google Scholar] [CrossRef]
- Lachmann, A.; Schilder, B.M.; Wojciechowicz, M.L.; Torre, D.; Kuleshov, M.V.; Keenan, A.B.; Ma’ayan, A. Geneshot: Search engine for ranking genes from arbitrary text queries. Nucleic. Acids Res. 2019, 47, W571–W577. [Google Scholar] [CrossRef]
- Lam, M.P.; Venkatraman, V.; Xing, Y.; Lau, E.; Cao, Q.; Ng, D.C.; Su, A.I.; Ge, J.; Van Eyk, J.E.; Ping, P. Data-Driven Approach To Determine Popular Proteins for Targeted Proteomics Translation of Six Organ Systems. J. Proteome Res. 2016, 15, 4126–4134. [Google Scholar] [CrossRef]
- Min, H.Y.; Lee, H.Y. Oncogene-Driven Metabolic Alterations in Cancer. Biomol. Ther. 2018, 26, 45–56. [Google Scholar] [CrossRef]
- Chen, Z.W.; Hu, J.F.; Wang, Z.W.; Liao, C.Y.; Kang, F.P.; Lin, C.F.; Huang, Y.; Huang, L.; Tian, Y.F.; Chen, S. Circular RNA circ-MTHFD1L induces HR repair to promote gemcitabine resistance via the miR-615-3p/RPN6 axis in pancreatic ductal adenocarcinoma. J. Exp. Clin. Cancer Res. 2022, 41, 153. [Google Scholar] [CrossRef]
- Zhuge, C.; Sun, X.; Chen, Y.; Lei, J. PDCD5 functions as a regulator of p53 dynamics in the DNA damage response. J. Theor. Biol. 2016, 388, 1–10, Erratum in J. Theor. Biol. 2016, 396, 210. [Google Scholar] [CrossRef]
- Banfi, F.; Rubio, A.; Zaghi, M.; Massimino, L.; Fagnocchi, G.; Bellini, E.; Luoni, M.; Cancellieri, C.; Bagliani, A.; Di Resta, C.; et al. SETBP1 accumulation induces P53 inhibition and genotoxic stress in neural progenitors underlying neurodegeneration in Schinzel-Giedion syndrome. Nat. Commun. 2021, 12, 4050. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Liao, K.; Zuo, W.; Liu, X.; Qiu, Z.; Gong, Z.; Liu, C.; Zeng, Q.; Qian, Y.; Jiang, L.; et al. Depletion of NFBD1/MDC1 Induces Apoptosis in Nasopharyngeal Carcinoma Cells Through the p53-ROS-Mitochondrial Pathway. Oncol. Res. 2017, 25, 123–136. [Google Scholar] [CrossRef] [PubMed]
- Zhao, L.; Cheng, Z.; Lu, Z.; Jin, J. NAD-dependent methylenetetrahydrofolate dehydrogenase inhibits oral squamous cell carcinoma cell proliferation and promotes apoptosis. Transl. Cancer Res. 2021, 10, 1457–1469. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Zhang, X.; Song, X.; Zhu, F.; Wang, Q.; Guo, C.; Liu, C.; Shi, Y.; Ma, C.; Wang, X.; et al. PDCD5 promotes cisplatin-induced apoptosis of glioma cells via activating mitochondrial apoptotic pathway. Cancer Biol. Ther. 2012, 13, 822–830. [Google Scholar] [CrossRef]
- Reilly, P.T.; Yu, Y.; Hamiche, A.; Wang, L. Cracking the ANP32 whips: Important functions, unequal requirement, and hints at disease implications. Bioessays 2014, 36, 1062–1071. [Google Scholar] [CrossRef]
- Pagesy, P.; Bouaboud, A.; Feng, Z.; Hulin, P.; Issad, T. Short O-GlcNAcase Is Targeted to the Mitochondria and Regulates Mitochondrial Reactive Oxygen Species Level. Cells 2022, 11, 1827. [Google Scholar] [CrossRef]
- Kvaløy, K.; Page, C.M.; Holmen, T.L. Epigenome-wide methylation differences in a group of lean and obese women—A HUNT Study. Sci. Rep. 2018, 8, 16330. [Google Scholar] [CrossRef]
- Yang, Y.R.; Jang, H.J.; Choi, S.S.; Lee, Y.H.; Lee, G.H.; Seo, Y.K.; Choi, J.H.; Park, D.; Koh, A.; Kim, I.S.; et al. Obesity resistance and increased energy expenditure by white adipose tissue browning in Oga(+/−) mice. Diabetologia 2015, 58, 2867–2876. [Google Scholar] [CrossRef]
- Ruff, S.E.; Logan, S.K.; Garabedian, M.J.; Huang, T.T. Roles for MDC1 in cancer development and treatment. DNA Repair 2020, 95, 102948. [Google Scholar] [CrossRef]
- Quevedo-Ocampo, J.; Escobedo-Calvario, A.; Souza-Arroyo, V.; Miranda-Labra, R.U.; Bucio-Ortiz, L.; Gutiérrez-Ruiz, M.C.; Chávez-Rodríguez, L.; Gomez-Quiroz, L.E. Folate Metabolism in Hepatocellular Carcinoma. What Do We Know So Far? Technol. Cancer Res. Treat. 2022, 21. [Google Scholar] [CrossRef]
- Wang, W.; Song, X.W.; Zhao, C.H. Roles of programmed cell death protein 5 in inflammation and cancer (Review). Int. J. Oncol. 2016, 49, 1801–1806. [Google Scholar] [CrossRef] [PubMed]
- Buddaseth, S.; Göttmann, W.; Blasczyk, R.; Huyton, T. Overexpression of the pp32r1 (ANP32C) oncogene or its functional mutant pp32r1Y140H confers enhanced resistance to FTY720 (Finguimod). Cancer Biol. Ther. 2014, 15, 289–296. [Google Scholar] [CrossRef] [PubMed]
- Antonescu, C.R.; Zhang, L.; Nielsen, G.P.; Rosenberg, A.E.; Dal Cin, P.; Fletcher, C.D. Consistent t(1;10) with rearrangements of TGFBR3 and MGEA5 in both myxoinflammatory fibroblastic sarcoma and hemosiderotic fibrolipomatous tumor. Genes Chromosomes Cancer 2011, 50, 757–764. [Google Scholar] [CrossRef] [PubMed]
- Makishima, H. Somatic SETBP1 mutations in myeloid neoplasms. Int. J. Hematol. 2017, 105, 732–742, Erratum in Int. J. Hematol. 2021, 114, 742. [Google Scholar] [CrossRef]
- Ma, W.; Wang, X.; Sun, H.; Xu, B.; Song, R.; Tian, Y.; Zhao, L.; Xu, Y.; Zhao, Y.; Yang, F.; et al. Oxidant stress-sensitive circRNA Mdc1 controls cardiomyocyte chromosome stability and cell cycle re-entry during heart regeneration. Pharm. Res. 2022, 184, 106422. [Google Scholar] [CrossRef]
- Motoyama, N.; Naka, K. DNA damage tumor suppressor genes and genomic instability. Curr. Opin. Genet. Dev. 2004, 14, 11–16. [Google Scholar] [CrossRef]
- Nishi, K.; Matsumoto, H.; Tashima, N.; Terada, S.; Nomura, N.; Kogo, M.; Morimoto, C.; Sunadome, H.; Nagasaki, T.; Oguma, T.; et al. Impacts of lipid-related metabolites, adiposity, and genetic background on blood eosinophil counts: The Nagahama study. Sci. Rep. 2021, 11, 15373. [Google Scholar] [CrossRef]
- Wilson, K.A.; Colavito, S.A.; Schulz, V.; Wakefield, P.H.; Sessa, W.; Tuck, D.; Stern, D.F. NFBD1/MDC1 regulates Cav1 and Cav2 independently of DNA damage and p53. Mol. Cancer Res. 2011, 9, 766–781. [Google Scholar] [CrossRef]
- Cai, H.; Cong, W.N.; Ji, S.; Rothman, S.; Maudsley, S.; Martin, B. Metabolic dysfunction in Alzheimer’s disease and related neurodegenerative disorders. Curr. Alzheimer Res. 2012, 9, 5–17. [Google Scholar] [CrossRef]
- Amorim, J.A.; Coppotelli, G.; Rolo, A.P.; Palmeira, C.M.; Ross, J.M.; Sinclair, D.A. Mitochondrial and metabolic dysfunction in ageing and age-related diseases. Nat. Rev. Endocrinol. 2022, 18, 243–258. [Google Scholar] [CrossRef]
- Hamzé, R.; Delangre, E.; Tolu, S.; Moreau, M.; Janel, N.; Bailbé, D.; Movassat, J. Type 2 Diabetes Mellitus and Alzheimer’s Disease: Shared Molecular Mechanisms and Potential Common Therapeutic Targets. Int. J. Mol. Sci. 2022, 23, 15287. [Google Scholar] [CrossRef] [PubMed]
- Semchyshyn, H. Is carbonyl/AGE/RAGE stress a hallmark of the brain aging? Pflug. Arch. 2021, 473, 723–734. [Google Scholar] [CrossRef] [PubMed]
- Muthyalaiah, Y.S.; Jonnalagadda, B.; John, C.M.; Arockiasamy, S. Impact of Advanced Glycation End products (AGEs) and its receptor (RAGE) on cancer metabolic signaling pathways and its progression. Glycoconj. J. 2021, 38, 717–734. [Google Scholar] [CrossRef] [PubMed]
- Singh, A.; Kukreti, R.; Saso, L.; Kukreti, S. Mechanistic Insight into Oxidative Stress-Triggered Signaling Pathways and Type 2 Diabetes. Molecules 2022, 27, 950. [Google Scholar] [CrossRef] [PubMed]
- Zamboni, M.; Mazzali, G.; Brunelli, A.; Saatchi, T.; Urbani, S.; Giani, A.; Rossi, A.P.; Zoico, E.; Fantin, F. The Role of Crosstalk between Adipose Cells and Myocytes in the Pathogenesis of Sarcopenic Obesity in the Elderly. Cells 2022, 11, 3361. [Google Scholar] [CrossRef]
- Ou, M.Y.; Zhang, H.; Tan, P.C.; Zhou, S.B.; Li, Q.F. Adipose tissue aging: Mechanisms and therapeutic implications. Cell Death Dis. 2022, 13, 300. [Google Scholar] [CrossRef]
- Carosi, J.M.; Fourrier, C.; Bensalem, J.; Sargeant, T.J. The mTOR-lysosome axis at the centre of ageing. FEBS Open Bio 2022, 12, 739–757. [Google Scholar] [CrossRef]
- Yu, M.; Zhang, H.; Wang, B.; Zhang, Y.; Zheng, X.; Shao, B.; Zhuge, Q.; Jin, K. Key Signaling Pathways in Aging and Potential Interventions for Healthy Aging. Cells 2021, 10, 660. [Google Scholar] [CrossRef]
- Maudsley, S.; Martin, B.; Gesty-Palmer, D.; Cheung, H.; Johnson, C.; Patel, S.; Becker, K.G.; Wood, W.H., 3rd; Zhang, Y.; Lehrmann, E.; et al. Delineation of a conserved arrestin-biased signaling repertoire in vivo. Mol. Pharm. 2015, 87, 706–717. [Google Scholar] [CrossRef]
- Mishra, S.; Nyomba, B.G. Prohibitin—At the crossroads of obesity-linked diabetes and cancer. Exp. Biol. Med. 2017, 242, 1170–1177. [Google Scholar] [CrossRef]
- Signorile, A.; Sgaramella, G.; Bellomo, F.; De Rasmo, D. Prohibitins: A Critical Role in Mitochondrial Functions and Implication in Diseases. Cells 2019, 8, 71. [Google Scholar] [CrossRef] [PubMed]
- Thuaud, F.; Ribeiro, N.; Nebigil, C.G.; Desaubry, L. Prohibitin ligands in cell death and survival: Mode of action and therapeutic potential. Chem. Biol. 2013, 20, 316–331. [Google Scholar] [CrossRef] [PubMed]
- Lourenço, A.B.; Rodríguez-Palero, M.J.; Doherty, M.K.; Cabrerizo Granados, D.; Hernando-Rodríguez, B.; Salas, J.J.; Venegas-Calerón, M.; Whitfield, P.D.; Artal-Sanz, M. The Mitochondrial PHB Complex Determines Lipid Composition and Interacts With the Endoplasmic Reticulum to Regulate Ageing. Front. Physiol. 2021, 12, 696275. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; He, Y. Prohibitin inhibits high glucose-induced apoptosis via maintaining mitochondrial function in human retinal capillary endothelial cells. Exp. Ther. Med. 2022, 23, 427. [Google Scholar] [CrossRef]
- Nijtmans, L.G.; Artal, S.M.; Grivell, L.A.; Coates, P.J. The mitochondrial PHB complex: Roles in mitochondrial respiratory complex assembly, ageing and degenerative disease. Cell Mol. Life Sci. 2002, 59, 143–155. [Google Scholar] [CrossRef]
- Koushyar, S.; Jiang, W.G.; Dart, D.A. Unveiling the potential of prohibitin in cancer. Cancer Lett. 2015, 369, 316–322. [Google Scholar] [CrossRef]
- Wang, D.; Tabti, R.; Elderwish, S.; Abou-Hamdan, H.; Djehal, A.; Yu, P.; Yurugi, H.; Rajalingam, K.; Nebigil, C.G.; Désaubry, L. Prohibitin ligands: A growing armamentarium to tackle cancers, osteoporosis, inflammatory, cardiac, and neurological diseases. Cell Mol. Life Sci. 2020, 77, 3525–3546. [Google Scholar] [CrossRef]
- Jakubowska, A.; Rozkrut, D.; Antoniou, A.; Hamann, U.; Scott, R.J.; McGuffog, L.; Healy, S.; Sinilnikova, O.M.; Rennert, G.; Lejbkowicz, F.; et al. Association of PHB 1630 C>T and MTHFR 677 C>T polymorphisms with breast and ovarian cancer risk in BRCA1/2 mutation carriers: Results from a multicenter study. Br. J. Cancer 2012, 106, 2016–2024. [Google Scholar] [CrossRef]
- Zhang, W.; Luo, J.; Chen, F.; Yang, F.; Song, W.; Zhu, A.; Guan, X. BRCA1 regulates PIG3-mediated apoptosis in a p53-dependent manner. Oncotarget 2015, 6, 7608–7618. [Google Scholar] [CrossRef]
- Hubel, P.; Urban, C.; Bergant, V.; Schneider, W.M.; Knauer, B.; Stukalov, A.; Scaturro, P.; Mann, A.; Brunotte, L.; Hoffmann, H.H.; et al. A protein-interaction network of interferon-stimulated genes extends the innate immune system landscape. Nat. Immunol. 2019, 20, 493–502. [Google Scholar] [CrossRef]
- Roy, S.J.; Glazkova, I.; Fréchette, L.; Iorio-Morin, C.; Binda, C.; Pétrin, D.; Trieu, P.; Robitaille, M.; Angers, S.; Hébert, T.E.; et al. Novel, gel-free proteomics approach identifies RNF5 and JAMP as modulators of GPCR stability. Mol. Endocrinol. 2013, 27, 1245–1266. [Google Scholar] [CrossRef] [PubMed]
- Havugimana, P.C.; Hart, G.T.; Nepusz, T.; Yang, H.; Turinsky, A.L.; Li, Z.; Wang, P.I.; Boutz, D.R.; Fong, V.; Phanse, S.; et al. A census of human soluble protein complexes. Cell 2012, 150, 1068–1081. [Google Scholar] [CrossRef] [PubMed]
- Sokolina, K.; Kittanakom, S.; Snider, J.; Kotlyar, M.; Maurice, P.; Gandía, J.; Benleulmi-Chaachoua, A.; Tadagaki, K.; Oishi, A.; Wong, V. Systematic protein-protein interaction mapping for clinically relevant human GPCRs. Mol. Syst. Biol. 2017, 13, 918. [Google Scholar] [CrossRef] [PubMed]
- Huttlin, E.L.; Bruckner, R.J.; Navarrete-Perea, J.; Cannon, J.R.; Baltier, K.; Gebreab, F.; Gygi, M.P.; Thornock, A.; Zarraga, G.; Tam, S.; et al. Dual proteome-scale networks reveal cell-specific remodeling of the human interactome. Cell 2021, 184, 3022–3040.e28. [Google Scholar] [CrossRef]
- Huttlin, E.L.; Pontano-Vaites, L.; Navarrete-Perea, J.; Bruckner, R.J.; Gebreab, F.; Gygi, M.P.; Thornock, A.; Fu, S.; Maenpaa, E.; Golbazi, A.; et al. The BioPlex Network of Human Protein Interactions: Additional Unpublished AP-MS Results (Pre-Publication). Available online: https://thebiogrid.org/166968/publication/the-bioplex-network-of-human-protein-interactions-additional-unpublished-ap-ms-results-pre-publication.html (accessed on 20 February 2023).
- Cong, W.N.; Wang, R.; Cai, H.; Daimon, C.M.; Scheibye-Knudsen, M.; Bohr, V.A.; Turkin, R.; Wood, W.H., 3rd; Becker, K.G.; Moaddel, R.; et al. Long-term artificial sweetener acesulfame potassium treatment alters neurometabolic functions in C57BL/6J mice. PLoS ONE 2013, 8, e70257. [Google Scholar] [CrossRef]
- He, B.; Yu, H.; Liu, S.; Wan, H.; Fu, S.; Liu, S.; Yang, J.; Zhang, Z.; Huang, H.; Li, Q.; et al. Mitochondrial cristae architecture protects against mtDNA release and inflammation. Cell Rep. 2022, 41, 111774. [Google Scholar] [CrossRef]
- Coster, G.; Goldberg, M. The cellular response to DNA damage: A focus on MDC1 and its interacting proteins. Nucleus 2010, 1, 166–178. [Google Scholar] [CrossRef]
- Sunadome, H.; Matsumoto, H.; Izuhara, Y.; Nagasaki, T.; Kanemitsu, Y.; Ishiyama, Y.; Morimoto, C.; Oguma, T.; Ito, I.; Murase, K.; et al. Correlation between eosinophil count, its genetic background and body mass index: The Nagahama Study. Allergol. Int. 2020, 69, 46–52. [Google Scholar] [CrossRef]
- Raut, G.K.; Chakrabarti, M.; Pamarthy, D.; Bhadra, M.P. Glucose starvation-induced oxidative stress causes mitochondrial dysfunction and apoptosis via Prohibitin 1 upregulation in human breast cancer cells. Free Radic. Biol. Med. 2019, 145, 428–441. [Google Scholar] [CrossRef]
- Montalvo-Javé, E.E.; Olguín-Martínez, M.; Hernández-Espinosa, D.R.; Sánchez-Sevilla, L.; Mendieta-Condado, E.; Contreras-Zentella, M.L.; Oñate-Ocaña, L.F.; Escalante-Tatersfield, T.; Echegaray-Donde, A.; Ruiz-Molina, J.M.; et al. Role of NADPH oxidases in inducing a selective increase of oxidant stress and cyclin D1 and checkpoint 1 over-expression during progression to human gastric adenocarcinoma. Eur J. Cancer 2016, 57, 50–57. [Google Scholar] [CrossRef]
- Greene, C.S.; Krishnan, A.; Wong, A.K.; Ricciotti, E.; Zelaya, R.A.; Himmelstein, D.S.; Zhang, R.; Hartmann, B.M.; Zaslavsky, E.; Sealfon, S.C.; et al. Understanding multicellular function and disease with human tissue-specific networks. Nat. Genet. 2015, 47, 569–576. [Google Scholar] [CrossRef] [PubMed]
- Ogrodnik, M.; Salmonowicz, H.; Gladyshev, V.N. Integrating cellular senescence with the concept of damage accumulation in aging: Relevance for clearance of senescent cells. Aging Cell 2019, 18, e12841, Erratum in Aging Cell 2019, 18, e12942. [Google Scholar] [CrossRef] [PubMed]
- Yousefzadeh, M.; Henpita, C.; Vyas, R.; Soto-Palma, C.; Robbins, P.; Niedernhofer, L. DNA damage-how and why we age? eLife 2021, 10, e62852. [Google Scholar] [CrossRef]
- Barja, G. Towards a unified mechanistic theory of aging. Exp. Gerontol. 2019, 124, 110627. [Google Scholar] [CrossRef] [PubMed]
- Chaudhari, S.N.; Kipreos, E.T. The Energy Maintenance Theory of Aging: Maintaining Energy Metabolism to Allow Longevity. Bioessays 2018, 40, e1800005. [Google Scholar] [CrossRef] [PubMed]
- Fang, Y.; Wang, X.; Yang, D.; Lu, Y.; Wei, G.; Yu, W.; Liu, X.; Zheng, Q.; Ying, J.; Hua, F. Relieving Cellular Energy Stress in Aging, Neurodegenerative, and Metabolic Diseases, SIRT1 as a Therapeutic and Promising Node. Front. Aging Neurosci. 2021, 13, 738686. [Google Scholar] [CrossRef] [PubMed]
- Mezhnina, V.; Ebeigbe, O.P.; Poe, A.; Kondratov, R.V. Circadian Control of Mitochondria in Reactive Oxygen Species Homeostasis. Antioxid. Redox. Signal. 2022, 37, 647–663. [Google Scholar] [CrossRef]
- Son, J.M.; Lee, C. Aging: All roads lead to mitochondria. Semin. Cell Dev. Biol. 2021, 116, 160–168. [Google Scholar] [CrossRef]
- Ayala, J.C.; Grismaldo, A.; Sequeda-Castañeda, L.G.; Aristizábal-Pachón, A.F.; Morales, L. Oxidative Stress in ICU Patients: ROS as Mortality Long-Term Predictor. Antioxidants 2021, 10, 1912. [Google Scholar] [CrossRef]
- Davalli, P.; Mitic, T.; Caporali, A.; Lauriola, A.; D’Arca, D. ROS, Cell Senescence, and Novel Molecular Mechanisms in Aging and Age-Related Diseases. Oxid. Med. Cell Longev. 2016, 2016, 3565127. [Google Scholar] [CrossRef]
- Warraich, U.E.; Hussain, F.; Kayani, H.U.R. Aging—Oxidative stress, antioxidants, and computational modeling. Heliyon 2020, 6, e04107. [Google Scholar] [CrossRef] [PubMed]
- Kenyon, C.J. The genetics of ageing. Nature 2010, 464, 504–512, Erratum in Nature 2010, 467, 622. [Google Scholar] [CrossRef] [PubMed]
- Séité, S.; Harrison, M.C.; Sillam-Dussès, D.; Lupoli, R.; Van Dooren, T.J.M.; Robert, A.; Poissonnier, L.A.; Lemainque, A.; Renault, D.; Acket, S.; et al. Lifespan prolonging mechanisms and insulin upregulation without fat accumulation in long-lived reproductives of a higher termite. Commun Biol. 2022, 5, 44. [Google Scholar] [CrossRef] [PubMed]
- Li, W.J.; Wang, C.W.; Tao, L.; Yan, Y.H.; Zhang, M.J.; Liu, Z.X.; Li, Y.X.; Zhao, H.Q.; Li, X.M.; He, X.D.; et al. Insulin signaling regulates longevity through protein phosphorylation in Caenorhabditis elegans. Nat. Commun. 2021, 12, 4568. [Google Scholar] [CrossRef]
- Wijsman, C.A.; Rozing, M.P.; Streefland, T.C.; le Cessie, S.; Mooijaart, S.P.; Slagboom, P.E.; Westendorp, R.G.; Pijl, H.; van Heemst, D.; Leiden Longevity Study Group. Familial longevity is marked by enhanced insulin sensitivity. Aging Cell 2011, 10, 114–121. [Google Scholar] [CrossRef]
- Blüher, M.; Kahn, B.B.; Kahn, C.R. Extended longevity in mice lacking the insulin receptor in adipose tissue. Science 2003, 299, 572–574. [Google Scholar] [CrossRef]
- Munro, D.; Pamenter, M.E. Comparative studies of mitochondrial reactive oxygen species in animal longevity: Technical pitfalls and possibilities. Aging Cell 2019, 18, e13009. [Google Scholar] [CrossRef]
- Ranieri, S.C.; Fusco, S.; Panieri, E.; Labate, V.; Mele, M.; Tesori, V.; Ferrara, A.M.; Maulucci, G.; De Spirito, M.; Martorana, G.E.; et al. Mammalian life-span determinant p66shcA mediates obesity-induced insulin resistance. Proc. Natl. Acad. Sci. USA 2010, 107, 13420–13425. [Google Scholar] [CrossRef]
- Yu, C.; Xiao, J.H. The Keap1-Nrf2 System: A Mediator between Oxidative Stress and Aging. Oxid. Med. Cell Longev. 2021, 2021, 6635460. [Google Scholar] [CrossRef]
- Szechyńska-Hebda, M.; Ghalami, R.Z.; Kamran, M.; Van Breusegem, F.; Karpiński, S. To Be or Not to Be? Are Reactive Oxygen Species, Antioxidants, and Stress Signalling Universal Determinants of Life or Death? Cells 2022, 11, 4105. [Google Scholar] [CrossRef]
- Milligan, G.; Ulven, T.; Murdoch, H.; Hudson, B.D. G-protein-coupled receptors for free fatty acids: Nutritional and therapeutic targets. Br J. Nutr. 2014, 111, S3–S7. [Google Scholar] [CrossRef] [PubMed]
- Lappano, R.; Maggiolini, M. G protein-coupled receptors: Novel targets for drug discovery in cancer. Nat. Rev. Drug Discov. 2011, 10, 47–60. [Google Scholar] [CrossRef] [PubMed]
- Nieto Gutierrez, A.; McDonald, P.H. GPCRs: Emerging anti-cancer drug targets. Cell. Signal. 2018, 41, 65–74. [Google Scholar] [CrossRef] [PubMed]
- Nieto, A.; Hara, M.R.; Quereda, V.; Grant, W.; Saunders, V.; Xiao, K.; McDonald, P.H.; Duckett, D.R. βarrestin-1 regulates DNA repair by acting as an E3-ubiquitin ligase adaptor for 53BP1. Cell Death Differ. 2020, 27, 1200–1213. [Google Scholar] [CrossRef]
- Zhang, X.; Pan, L.; Yang, K.; Fu, Y.; Liu, Y.; Chen, W.; Ma, X.; Yin, X. Alterations of relaxin and its receptor system components in experimental diabetic cardiomyopathy rats. Cell Tissue Res. 2017, 370, 297–304. [Google Scholar] [CrossRef]
- Calvez, J.; de Ávila, C.; Matte, L.O.; Guèvremont, G.; Gundlach, A.L.; Timofeeva, E. Role of relaxin-3/RXFP3 system in stress-induced binge-like eating in female rats. Neuropharmacology 2016, 102, 207–215. [Google Scholar] [CrossRef]
- Walker, A.W.; Smith, C.M.; Gundlach, A.L.; Lawrence, A.J. Relaxin-3 receptor (Rxfp3) gene deletion reduces operant sucrose- but not alcohol-responding in mice. Genes Brain Behav. 2015, 14, 625–634. [Google Scholar] [CrossRef]
- Ryan, P.J.; Krstew, E.V.; Sarwar, M.; Gundlach, A.L.; Lawrence, A.J. Relaxin-3 mRNA levels in nucleus incertus correlate with alcohol and sucrose intake in rats. Drug Alcohol Depend. 2014, 140, 8–16. [Google Scholar] [CrossRef]
- Jasaszwili, M.; Wojciechowicz, T.; Billert, M.; Strowski, M.Z.; Nowak, K.W.; Skrzypski, M. Effects of adropin on proliferation and differentiation of 3T3-L1 cells and rat primary preadipocytes. Mol. Cell Endocrinol. 2019, 496, 110532. [Google Scholar] [CrossRef]
- Stelcer, E.; Milecka, P.; Komarowska, H.; Jopek, K.; Tyczewska, M.; Szyszka, M.; Lesniczak, M.; Suchorska, W.; Bekova, K.; Szczepaniak, B.; et al. Adropin Stimulates Proliferation and Inhibits Adrenocortical Steroidogenesis in the Human Adrenal Carcinoma (HAC15) Cell Line. Front. Endocrinol. 2020, 11, 561370. [Google Scholar] [CrossRef]
- Yamamoto, H.; Shimokawa, H.; Haga, T.; Fukui, Y.; Iguchi, K.; Unno, K.; Hoshino, M.; Takeda, A. The expression of relaxin-3 in adipose tissue and its effects on adipogenesis. Protein Pept. Lett. 2014, 21, 517–522. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Qiu, Y.; Gu, H.; Gan, M.; Zhu, Y.; Zhu, K.; Chen, L.; Zhao, Y.; Niu, L.; Zhang, S.; et al. Regulation of Adipose Thermogenesis and its Critical Role in Glucose and Lipid Metabolism. Int. J. Biol. Sci. 2022, 18, 4950–4962. [Google Scholar] [CrossRef]
- Wang, X.; Xu, M.; Li, Y. Adipose Tissue Aging and Metabolic Disorder, and the Impact of Nutritional Interventions. Nutrients 2022, 14, 3134. [Google Scholar] [CrossRef] [PubMed]
- Brun, J.F.; Myzia, J.; Varlet-Marie, E.; Raynaud de Mauverger, E.; Mercier, J. Beyond the Calorie Paradigm: Taking into Account in Practice the Balance of Fat and Carbohydrate Oxidation during Exercise? Nutrients 2022, 14, 1605. [Google Scholar] [CrossRef] [PubMed]
- Whitehall, J.C.; Smith, A.L.M.; Greaves, L.C. Mitochondrial DNA Mutations and Ageing. Subcell. Biohhem. 2023, 102, 77–98. [Google Scholar] [CrossRef]
- Mercer, J.R.; Cheng, K.K.; Figg, N.; Gorenne, I.; Mahmoudi, M.; Griffin, J.; Vidal-Puig, A.; Logan, A.; Murphy, M.P.; Bennett, M. DNA damage links mitochondrial dysfunction to atherosclerosis and the metabolic syndrome. Circ. Res. 2010, 107, 1021–1031, Erratum in Circ. Res. 2011, 108, e2. [Google Scholar] [CrossRef]
- Gureev, A.P.; Khorolskaya, V.G.; Sadovnikova, I.S.; Shaforostova, E.A.; Cherednichenko, V.R.; Burakova, I.Y.; Plotnikov, E.Y.; Popov, V.N. Age-Related Decline in Nrf2/ARE Signaling Is Associated with the Mitochondrial DNA Damage and Cognitive Impairments. Int. J. Mol. Sci. 2022, 23, 15197. [Google Scholar] [CrossRef]
- Golubev, A.G.; Anisimov, V.N. Aging and cancer: Is glucose a mediator between them? Oncotarget 2019, 10, 6758–6767. [Google Scholar] [CrossRef]
- Anton, S.D.; Moehl, K.; Donahoo, W.T.; Marosi, K.; Lee, S.A.; Mainous, A.G., 3rd; Leeuwenburgh, C.; Mattson, M.P. Flipping the Metabolic Switch: Understanding and Applying the Health Benefits of Fasting. Obesity 2018, 26, 254–268. [Google Scholar] [CrossRef]
- Puchalska, P.; Crawford, P.A. Multi-dimensional Roles of Ketone Bodies in Fuel Metabolism, Signaling, and Therapeutics. Cell Metab. 2017, 25, 262–284. [Google Scholar] [CrossRef]
- Volek, J.S.; Noakes, T.; Phinney, S.D. Rethinking fat as a fuel for endurance exercise. Eur. J. Sport Sci. 2015, 15, 13–20. [Google Scholar] [CrossRef] [PubMed]
- Shi, Y.; Shu, Z.J.; Xue, X.; Yeh, C.K.; Katz, M.S.; Kamat, A. β2-Adrenergic receptor ablation modulates hepatic lipid accumulation and glucose tolerance in aging mice. Exp. Gerontol. 2016, 78, 32–38. [Google Scholar] [CrossRef] [PubMed]
- Rogers, R.P.; Rogina, B. The role of INDY in metabolism, health, and longevity. Front. Genet. 2015, 6, 204. [Google Scholar] [CrossRef]
- Tjahjono, E.; Kirienko, D.R.; Kirienko, N.V. The emergent role of mitochondrial surveillance in cellular health. Aging Cell 2022, 21, e13710. [Google Scholar] [CrossRef] [PubMed]
- Sligar, J.; DeBruin, D.A.; Saner, N.J.; Philp, A.M.; Philp, A. The importance of mitochondrial quality control for maintaining skeletal muscle function across health span. Am. J. Physiol. Cell Physiol. 2022, 322, C461–C467. [Google Scholar] [CrossRef] [PubMed]
- Vaddavalli, P.L.; Schumacher, B. The p53 network: Cellular and systemic DNA damage responses in cancer and aging. Trends Genet. 2022, 38, 598–612. [Google Scholar] [CrossRef] [PubMed]
- Mijit, M.; Caracciolo, V.; Melillo, A.; Amicarelli, F.; Giordano, A. Role of p53 in the Regulation of Cellular Senescence. Biomolecules 2020, 10, 420. [Google Scholar] [CrossRef] [PubMed]
- Chrzanowska-Lightowlers, Z.M.; Pajak, A.; Lightowlers, R.N. Termination of protein synthesis in mammalian mitochondria. J. Biol. Chem. 2011, 286, 34479–34485. [Google Scholar] [CrossRef]
- Sengupta, D.; Sengupta, K. Lamin A and telomere maintenance in aging: Two to Tango. Mutat. Res. 2022, 825, 111788. [Google Scholar] [CrossRef]
- Romero-Bueno, R.; de la Cruz Ruiz, P.; Artal-Sanz, M.; Askjaer, P.; Dobrzynska, A. Nuclear Organization in Stress and Aging. Cells 2019, 8, 664. [Google Scholar] [CrossRef]
- Han, Y.; Micklem, G.; Kim, S.Y. Transcriptional landscape of oncogene-induced senescence: A machine learning based meta analytic approach. Ageing Res. Rev. 2023, 85, 101849. [Google Scholar] [CrossRef] [PubMed]
- Domen, A.; Deben, C.; Verswyvel, J.; Flieswasser, T.; Prenen, H.; Peeters, M.; Lardon, F.; Wouters, A. Cellular senescence in cancer: Clinical detection and prognostic implications. J. Exp. Clin. Cancer Res. 2022, 41, 360. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.H.; Park, S.J.; Jeong, S.Y.; Kim, M.J.; Jun, S.; Lee, H.S.; Chang, I.Y.; Lim, S.C.; Yoon, S.P.; Yong, J.; et al. MicroRNA-22 Suppresses DNA Repair and Promotes Genomic Instability through Targeting of MDC1. Cancer Res. 2015, 75, 1298–1310, Erratum in Cancer Res. 2018, 78, 6023. [Google Scholar] [CrossRef] [PubMed]
- Bencokova, Z.; Kaufmann, M.R.; Pires, I.M.; Lecane, P.S.; Giaccia, A.J.; Hammond, E.M. ATM activation and signaling under hypoxic conditions. Mol. Cell Biol. 2009, 29, 526–537. [Google Scholar] [CrossRef]
- Stubbs, T.; Bingman, J.I.; Besse, J.; Mykytyn, K. Ciliary signaling proteins are mislocalized in the brains of Bardet-Biedl syndrome 1-null mice. Front. Cell Dev. Biol. 2023, 10, 1092161. [Google Scholar] [CrossRef]
- Hossain, M.S.; Mineno, K.; Katafuchi, T. Neuronal Orphan G-Protein Coupled Receptor Proteins Mediate Plasmalogens-Induced Activation of ERK and Akt Signaling. PLoS ONE 2016, 11, e0150846. [Google Scholar] [CrossRef]
- Jin, X.; Zhu, L.; Cui, Z.; Tang, J.; Xie, M.; Ren, G. Elevated expression of GNAS promotes breast cancer cell proliferation and migration via the PI3K/AKT/Snail1/E-cadherin axis. Clin. Transl. Oncol. 2019, 21, 1207–1219. [Google Scholar] [CrossRef]
- Folk, W.P.; Kumari, A.; Iwasaki, T.; Pyndiah, S.; Johnson, J.C.; Cassimere, E.K.; Abdulovic-Cui, A.L.; Sakamuro, D. Loss of the tumor suppressor BIN1 enables ATM Ser/Thr kinase activation by the nuclear protein E2F1 and renders cancer cells resistant to cisplatin. J. Biol. Chem. 2019, 294, 5700–5719. [Google Scholar] [CrossRef]
- Zhou, H.; Qin, Y.; Ji, S.; Ling, J.; Fu, J.; Zhuang, Z.; Fan, X.; Song, L.; Yu, X.; Chiao, P.J. SOX9 activity is induced by oncogenic Kras to affect MDC1 and MCMs expression in pancreatic cancer. Oncogene 2018, 37, 912–923. [Google Scholar] [CrossRef]
- Barry, S.P.; Townsend, P.A.; Knight, R.A.; Scarabelli, T.M.; Latchman, D.S.; Stephanou, A. STAT3 modulates the DNA damage response pathway. Int. J. Exp. Pathol. 2010, 91, 506–514. [Google Scholar] [CrossRef]
- Peter, M.R.; Bilenky, M.; Davies, A.; Isserlin, R.; Bader, G.D.; Fleshner, N.E.; Hirst, M.; Zoubeidi, A.; Bapat, B. Distinct DNA methylation patterns associated with treatment resistance in metastatic castration resistant prostate cancer. Sci. Rep. 2021, 11, 6630. [Google Scholar] [CrossRef] [PubMed]
- Bannister, A.J.; Kouzarides, T. Regulation of chromatin by histone modifications. Cell Res. 2011, 21, 381–395. [Google Scholar] [CrossRef] [PubMed]
- Liu, H.; Fan, H.; He, P.; Zhuang, H.; Liu, X.; Chen, M.; Zhong, W.; Zhang, Y.; Zhen, C.; Li, Y.; et al. Prohibitin 1 regulates mtDNA release and downstream inflammatory responses. EMBO J. 2022, 41, e111173. [Google Scholar] [CrossRef] [PubMed]
- Saura-Esteller, J.; Sánchez-Vera, I.; Núñez-Vázquez, S.; Cosialls, A.M.; Gama-Pérez, P.; Bhosale, G.; Mendive-Tapia, L.; Lavilla, R.; Pons, G.; Garcia-Roves, P.M.; et al. Activation of the Integrated Stress Response and ER Stress Protect from Fluorizoline-Induced Apoptosis in HEK293T and U2OS Cell Lines. Int. J. Mol. Sci. 2021, 22, 6117. [Google Scholar] [CrossRef]
- Hernando-Rodríguez, B.; Artal-Sanz, M. Mitochondrial Quality Control Mechanisms and the PHB (Prohibitin) Complex. Cells 2018, 7, 238. [Google Scholar] [CrossRef]
- Silva, L.E.; Souza, R.C.; Kitano, E.S.; Monteiro, L.F.; Iwai, L.K.; Forti, F.L. Proteomic and Interactome Approaches Reveal PAK4, PHB-2, and 14-3-3η as Targets of Overactivated Cdc42 in Cellular Responses to Genomic Instability. J. Proteome Res. 2019, 18, 3597–3614. [Google Scholar] [CrossRef]
- Bang, J.I.; Bae, D.W.; Lee, H.S.; Deb, G.K.; Kim, M.O.; Sohn, S.H.; Han, C.H.; Kong, I.K. Proteomic analysis of placentas from cloned cat embryos identifies a set of differentially expressed proteins related to oxidative damage, senescence, and apoptosis. Proteomics 2011, 11, 4454–4467. [Google Scholar] [CrossRef]
- Oyang, L.; Li, J.; Jiang, X.; Lin, J.; Xia, L.; Yang, L.; Tan, S.; Wu, N.; Han, Y.; Yang, Y.; et al. The function of prohibitins in mitochondria and the clinical potentials. Cancer Cell Int. 2022, 22, 343. [Google Scholar] [CrossRef]
- Ren, L.; Meng, L.; Gao, J.; Lu, M.; Guo, C.; Li, Y.; Rong, Z.; Ye, Y. PHB2 promotes colorectal cancer cell proliferation and tumorigenesis through NDUFS1-mediated oxidative phosphorylation. Cell Death Dis. 2023, 14, 44. [Google Scholar] [CrossRef]
- Hong, J.; Kim, Y.H. Fatty Liver/Adipose Tissue Dual-Targeting Nanoparticles with Heme Oxygenase-1 Inducer for Amelioration of Obesity, Obesity-Induced Type 2 Diabetes, and Steatohepatitis. Adv. Sci. 2022, 9, e2203286. [Google Scholar] [CrossRef]
- Mao, J.; Zhang, J.; Cai, L.; Cui, Y.; Liu, J.; Mao, Y. Elevated prohibitin 1 expression mitigates glucose metabolism defects in granulosa cells of infertile patients with endometriosis. Mol. Hum. Reprod. 2022, 28, gaac018. [Google Scholar] [CrossRef] [PubMed]
- Lourenço, A.B.; Artal-Sanz, M. The Mitochondrial Prohibitin (PHB) Complex in C. elegans Metabolism and Ageing Regulation. Metabolites 2021, 11, 636. [Google Scholar] [CrossRef] [PubMed]
- Gao, Z.; Daquinag, A.C.; Fussell, C.; Djehal, A.; Désaubry, L.; Kolonin, M.G. Prohibitin Inactivation in Adipocytes Results in Reduced Lipid Metabolism and Adaptive Thermogenesis Impairment. Diabetes 2021, 70, 2204–2212. [Google Scholar] [CrossRef] [PubMed]
- Rashid, M.A.; Lee, S.; Tak, E.; Lee, J.; Choi, T.G.; Lee, J.W.; Kim, J.B.; Youn, J.H.; Kang, I.; Ha, J.; et al. Carbonyl reductase 1 protects pancreatic β-cells against oxidative stress-induced apoptosis in glucotoxicity and glucolipotoxicity. Free Radic. Biol. Med. 2010, 49, 1522–1533, Erratum in Free Radic. Biol. Med. 2018, 129, 614–617. [Google Scholar] [CrossRef]
- Petersen, D.R.; Saba, L.M.; Sayin, V.I.; Papagiannakopoulos, T.; Schmidt, E.E.; Merrill, G.F.; Orlicky, D.J.; Shearn, C.T. Elevated Nrf-2 responses are insufficient to mitigate protein carbonylation in hepatospecific PTEN deletion mice. PLoS ONE 2018, 13, e0198139. [Google Scholar] [CrossRef]
- Tecza, K.; Pamula-Pilat, J.; Lanuszewska, J.; Butkiewicz, D.; Grzybowska, E. Pharmacogenetics of toxicity of 5-fluorouracil, doxorubicin, and cyclophosphamide chemotherapy in breast cancer patients. Oncotarget 2018, 9, 9114–9136. [Google Scholar] [CrossRef]
- Jo, A.; Choi, T.G.; Jo, Y.H.; Jyothi, K.R.; Nguyen, M.N.; Kim, J.H.; Lim, S.; Shahid, M.; Akter, S.; Lee, S.; et al. Inhibition of Carbonyl Reductase 1 Safely Improves the Efficacy of Doxorubicin in Breast Cancer Treatment. Antioxid. Redox Signal. 2017, 26, 70–83. [Google Scholar] [CrossRef]
- Kannan, K.; Rogina, B. The Role of Citrate Transporter INDY in Metabolism and Stem Cell Homeostasis. Metabolites 2021, 11, 705. [Google Scholar] [CrossRef]
- LaMoia, T.E.; Butrico, G.M.; Kalpage, H.A.; Goedeke, L.; Hubbard, B.T.; Vatner, D.F.; Gaspar, R.C.; Zhang, X.M.; Cline, G.W.; Nakahara, K.; et al. Metformin, phenformin, and galegine inhibit complex IV activity and reduce glycerol-derived gluconeogenesis. Proc. Natl. Acad. Sci. USA 2022, 119, e2122287119. [Google Scholar] [CrossRef]
- Marshall, R.N.; McKendry, J.; Smeuninx, B.; Seabright, A.P.; Morgan, P.T.; Greig, C.; Breen, L. Acute resistance exercise training does not augment mitochondrial remodelling in master athletes or untrained older adults. Front. Physiol. 2023, 13, 1097988. [Google Scholar] [CrossRef]
- Klecker, T.; Westermann, B. Pathways shaping the mitochondrial inner membrane. Open Biol. 2021, 11, 210238. [Google Scholar] [CrossRef] [PubMed]
- Jezek, P.; Jaburek, M.; Holendova, B.; Engstová, H.; Dlasková, A. Mitochondrial cristae morphology reflecting metabolism, superoxide formation, redox homeostasis, and pathology. Antioxid. Redox Signal. 2023; ahead of print. [Google Scholar] [CrossRef]
- Hwang, M.S.; Rohlena, J.; Dong, L.F.; Neuzil, J.; Grimm, S. Powerhouse down: Complex II dissociation in the respiratory chain. Mitochondrion 2014, 19, 20–28. [Google Scholar] [CrossRef] [PubMed]
- Dumbali, S.P.; Wenzel, P.L. Mitochondrial Permeability Transition in Stem Cells, Development, and Disease. Adv. Exp. Med. Biol. 2023, 1409, 1–22. [Google Scholar] [CrossRef] [PubMed]
- Eisenberg-Bord, M.; Schuldiner, M. Ground control to major TOM: Mitochondria-nucleus communication. FEBS J. 2017, 284, 196–210. [Google Scholar] [CrossRef]
- Kastner, S.; Voss, T.; Keuerleber, S.; Glöckel, C.; Freissmuth, M.; Sommergruber, W. Expression of G protein-coupled receptor 19 in human lung cancer cells is triggered by entry into S-phase and supports G(2)-M cell-cycle progression. Mol. Cancer Res. 2012, 10, 1343–1358. [Google Scholar] [CrossRef]
- Semizarov, D.; Kroeger, P.; Fesik, S. siRNA-mediated gene silencing: A global genome view. Nucleic Acids Res. 2004, 32, 3836–3845. [Google Scholar] [CrossRef]
- Salvador-Barbero, B.; Álvarez-Fernández, M.; Zapatero-Solana, E.; El Bakkali, A.; Menéndez, M.D.C.; López-Casas, P.P.; Di Domenico, T.; Xie, T.; VanArsdale, T.; Shields, D.J.; et al. CDK4/6 Inhibitors Impair Recovery from Cytotoxic Chemotherapy in Pancreatic Adenocarcinoma. Cancer Cell 2020, 37, 340–353.e6, Erratum in Cancer Cell 2020, 38, 584. [Google Scholar] [CrossRef]
- Mintz, R.L.; Lao, Y.H.; Chi, C.W.; He, S.; Li, M.; Quek, C.H.; Shao, D.; Chen, B.; Han, J.; Wang, S.; et al. CRISPR/Cas9-mediated mutagenesis to validate the synergy between PARP1 inhibition and chemotherapy in BRCA1-mutated breast cancer cells. Bioeng. Transl. Med. 2020, 5, e10152. [Google Scholar] [CrossRef]
- Mecham, B.H.; Klus, G.T.; Strovel, J.; Augustus, M.; Byrne, D.; Bozso, P.; Wetmore, D.Z.; Mariani, T.J.; Kohane, I.S.; Szallasi, Z. Sequence-matched probes produce increased cross-platform consistency and more reproducible biological results in microarray-based gene expression measurements. Nucleic Acids Res. 2004, 32, e74. [Google Scholar] [CrossRef]
- Tyanova, S.; Temu, T.; Cox, J. The MaxQuant computational platform for mass spectrometry-based shotgun proteomics. Nat. Protoc. 2016, 11, 2301–2319. [Google Scholar] [CrossRef]
- Cox, J.; Neuhauser, N.; Michalski, A.; Scheltema, R.A.; Olsen, J.V.; Mann, M. Andromeda: A peptide search engine integrated into the MaxQuant environment. J. Proteome Res. 2011, 10, 1794–1805. [Google Scholar] [CrossRef] [PubMed]
- Martin, B.; Chadwick, W.; Yi, T.; Park, S.S.; Lu, D.; Ni, B.; Gadkaree, S.; Farhang, K.; Becker, K.G.; Maudsley, S. VENNTURE—A novel Venn diagram investigational tool for multiple pharmacological dataset analysis. PLoS ONE 2012, 7, e36911, Erratum in PLoS ONE 2012, 7. [Google Scholar] [CrossRef] [PubMed]
- Cai, H.; Chen, H.; Yi, T.; Daimon, C.M.; Boyle, J.P.; Peers, C.; Maudsley, S.; Martin, B. VennPlex—A novel Venn diagram program for comparing and visualizing datasets with differentially regulated datapoints. PLoS ONE 2013, 8, e53388. [Google Scholar] [CrossRef] [PubMed]
- Gerstner, N.; Kehl, T.; Lenhof, K.; Müller, A.; Mayer, C.; Eckhart, L.; Grammes, N.L.; Diener, C.; Hart, M.; Hahn, O.; et al. GeneTrail 3: Advanced high-throughput enrichment analysis. Nucleic Acids Res. 2020, 48, W515–W520. [Google Scholar] [CrossRef] [PubMed]










Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Maudsley, S.; Schrauwen, C.; Harputluoğlu, İ.; Walter, D.; Leysen, H.; McDonald, P. GPR19 Coordinates Multiple Molecular Aspects of Stress Responses Associated with the Aging Process. Int. J. Mol. Sci. 2023, 24, 8499. https://doi.org/10.3390/ijms24108499
Maudsley S, Schrauwen C, Harputluoğlu İ, Walter D, Leysen H, McDonald P. GPR19 Coordinates Multiple Molecular Aspects of Stress Responses Associated with the Aging Process. International Journal of Molecular Sciences. 2023; 24(10):8499. https://doi.org/10.3390/ijms24108499
Chicago/Turabian StyleMaudsley, Stuart, Claudia Schrauwen, İrem Harputluoğlu, Deborah Walter, Hanne Leysen, and Patricia McDonald. 2023. "GPR19 Coordinates Multiple Molecular Aspects of Stress Responses Associated with the Aging Process" International Journal of Molecular Sciences 24, no. 10: 8499. https://doi.org/10.3390/ijms24108499
APA StyleMaudsley, S., Schrauwen, C., Harputluoğlu, İ., Walter, D., Leysen, H., & McDonald, P. (2023). GPR19 Coordinates Multiple Molecular Aspects of Stress Responses Associated with the Aging Process. International Journal of Molecular Sciences, 24(10), 8499. https://doi.org/10.3390/ijms24108499





