Perfluoroalkyl Substances (PFAS) Affect Inflammation in Lung Cells and Tissues
Abstract
1. Introduction
2. Results
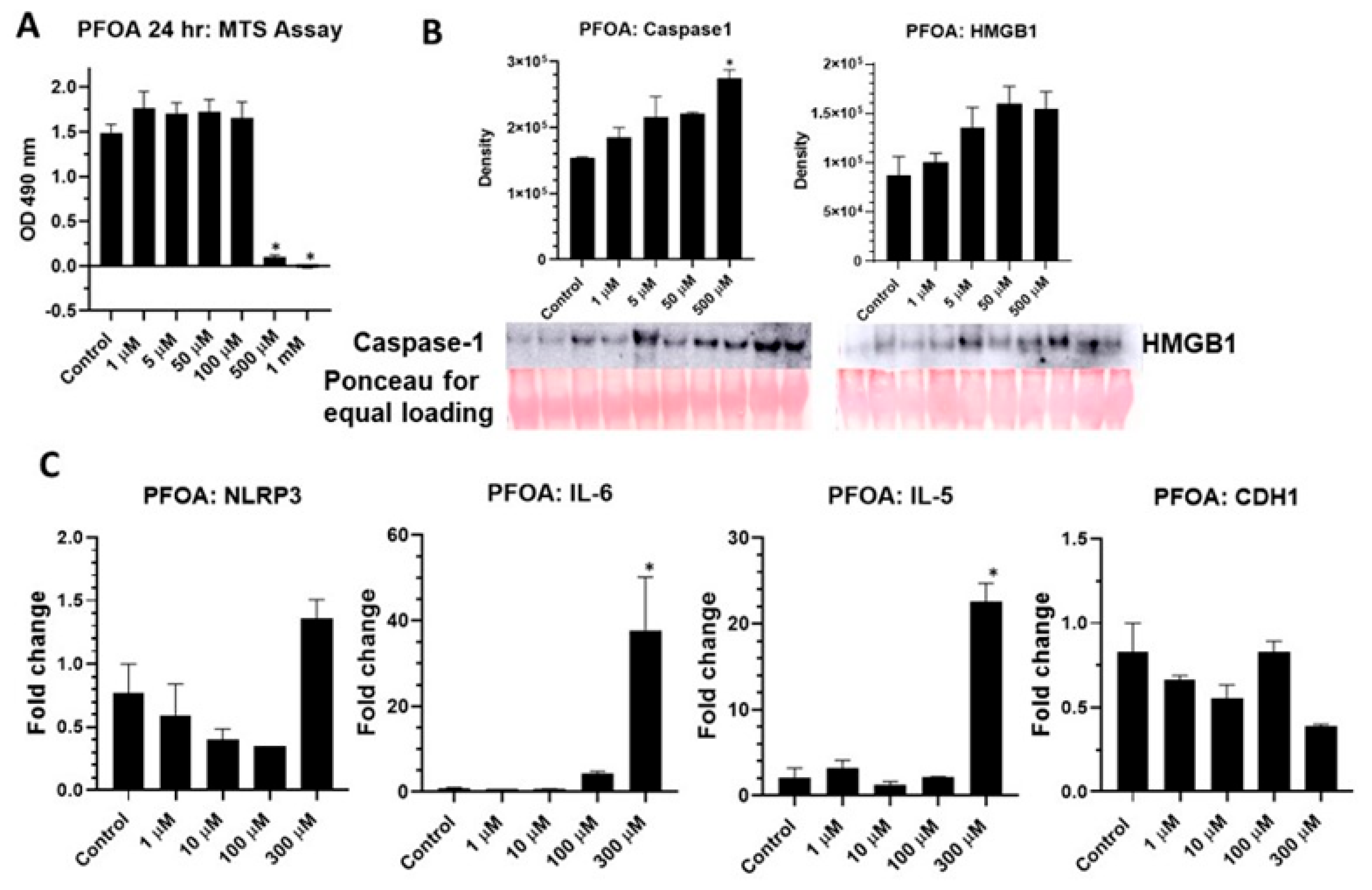
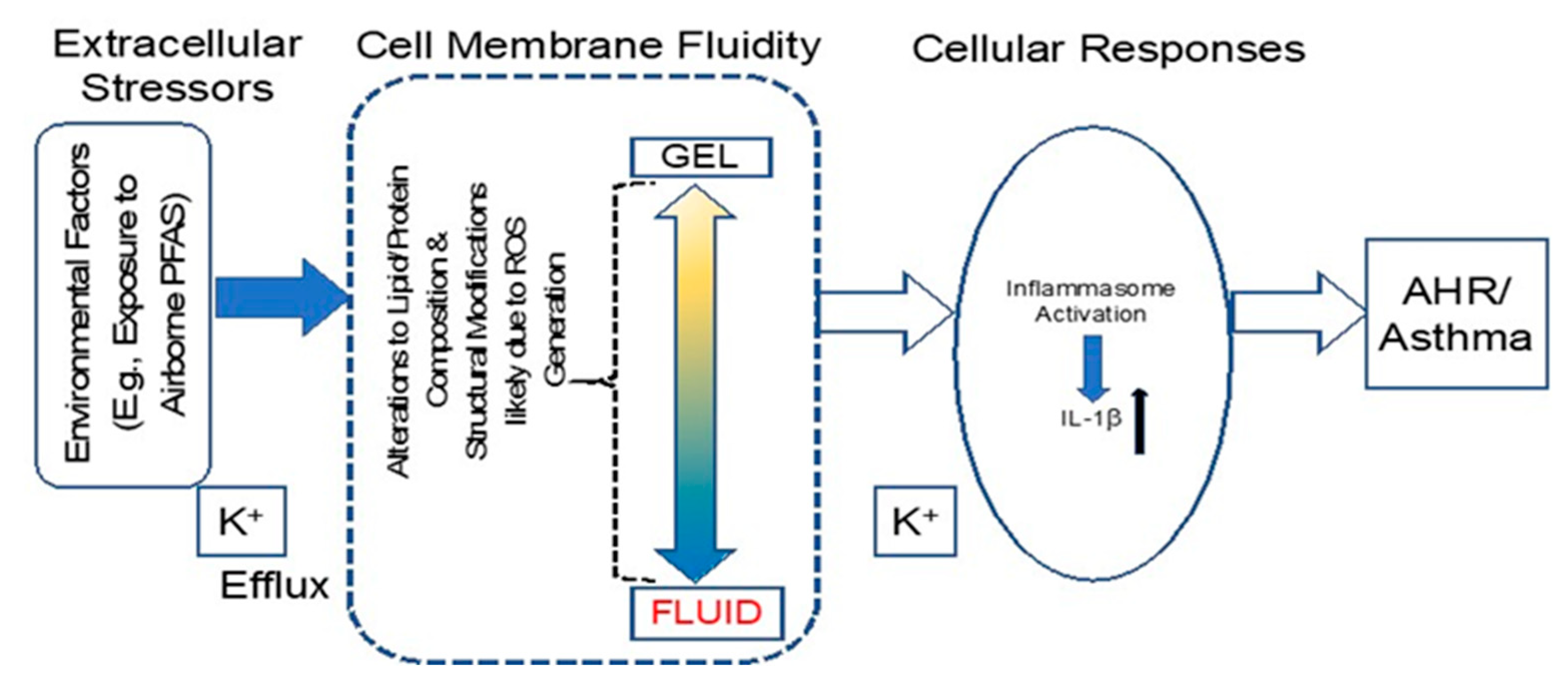
2.1. PFOA Significantly Altered Membrane Properties in BEAS2B Cells
2.2. PFOA Activates NLRP3 Inflammasome in BEAS2B Cells
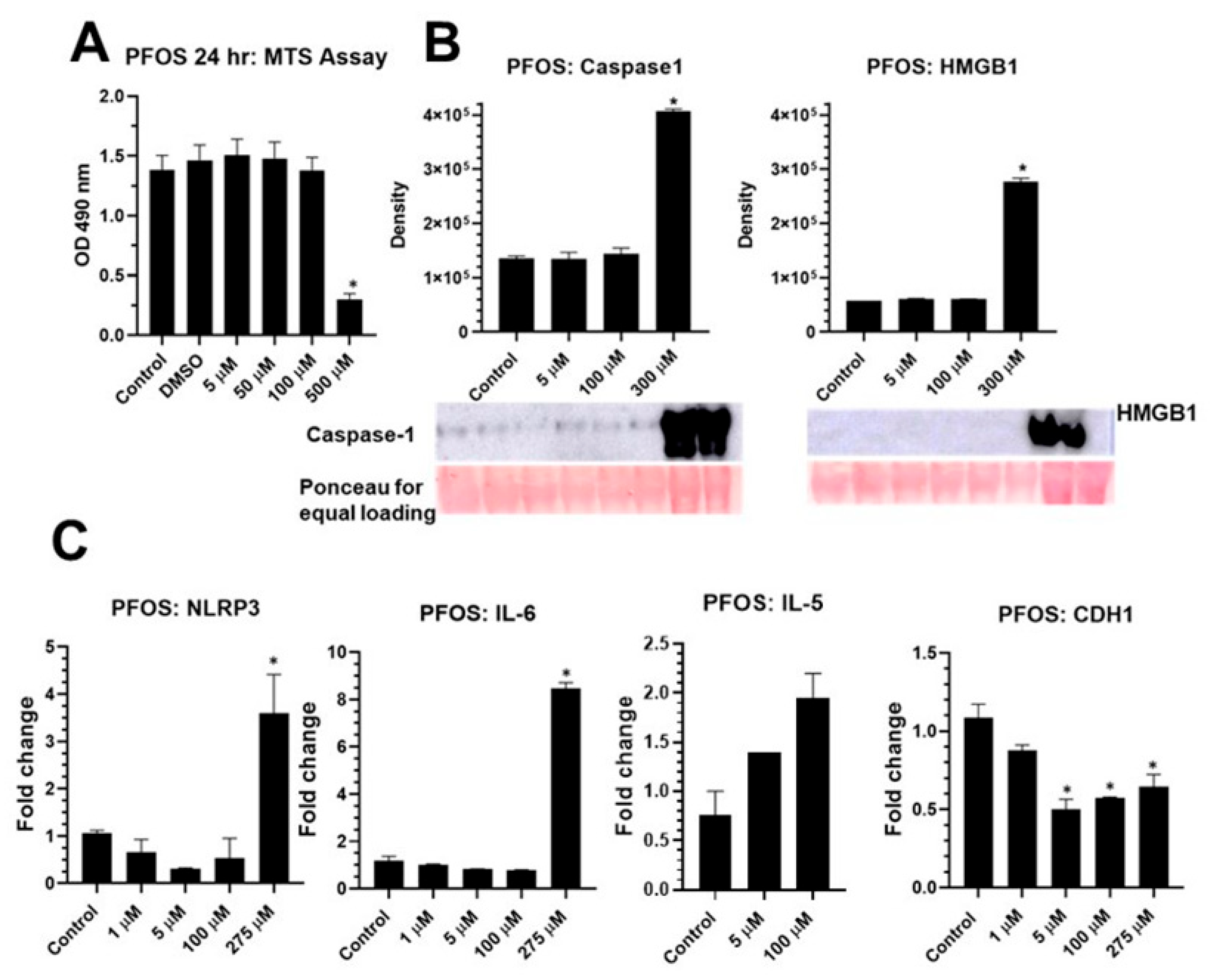
2.3. PFOS Activates NLRP3 Inflammasome in BEAS2B Cells
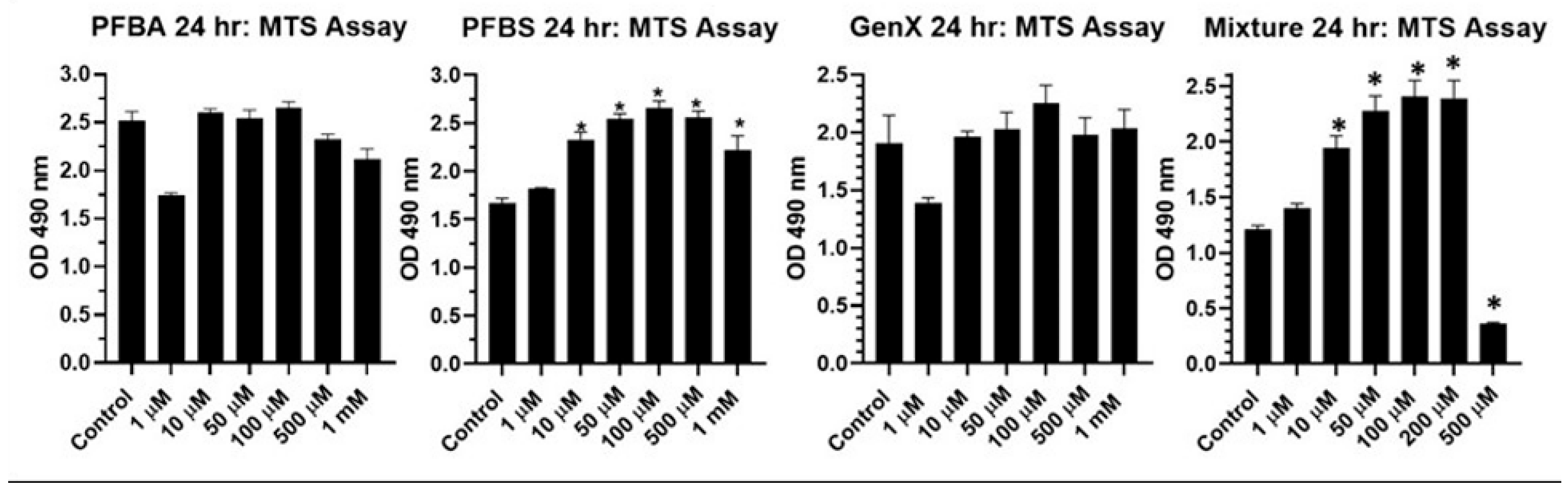
2.4. Effect of Short Chain and a Mixture of Long and Short Chain PFAS on BEAS2B Cell Viability
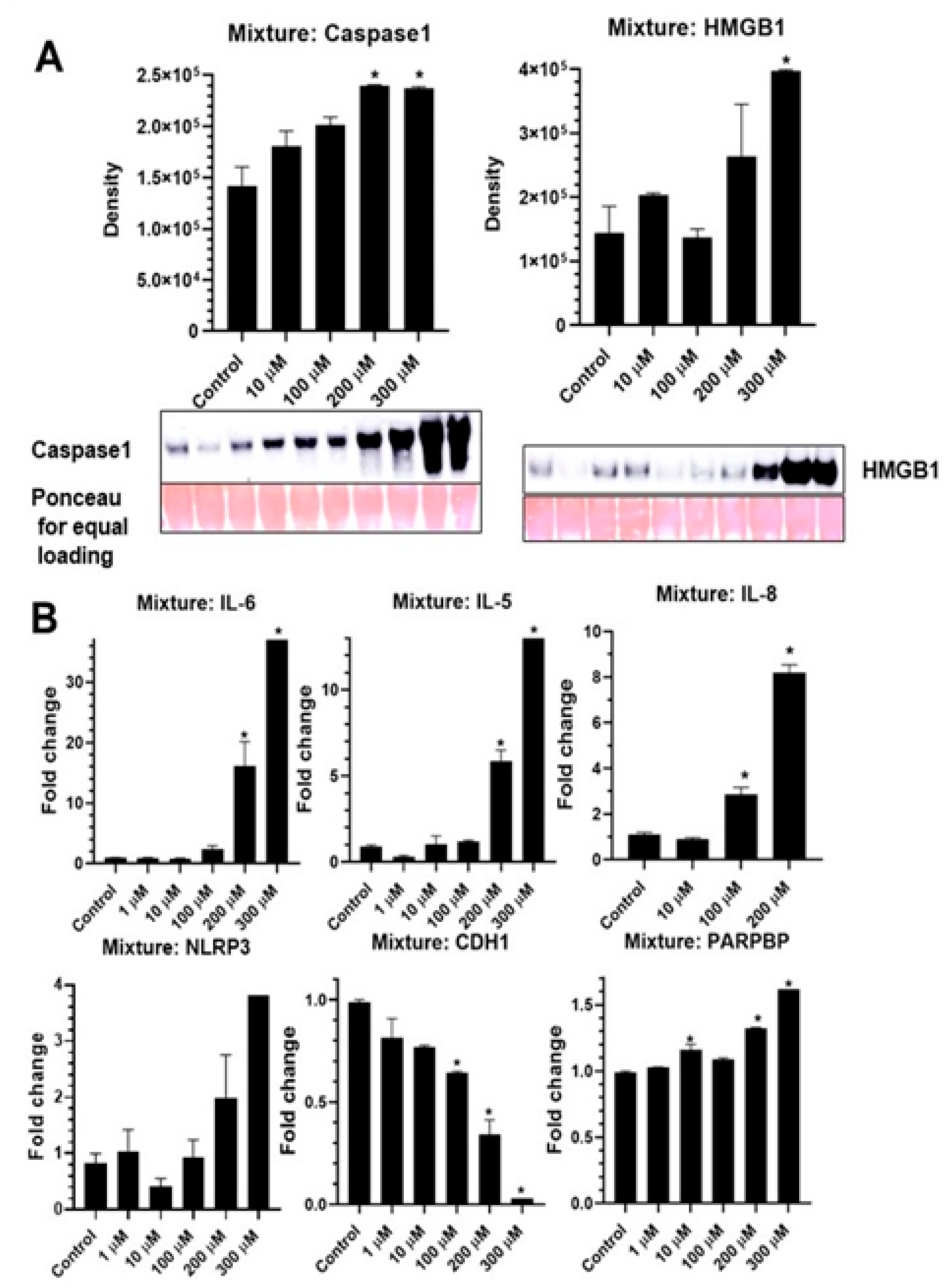
2.5. PFAS Mixture Activates NLRP3 Inflammasome and Pro-Inflammatory Cytokines in Lung Epithelial Cells
2.6. In Vivo Studies
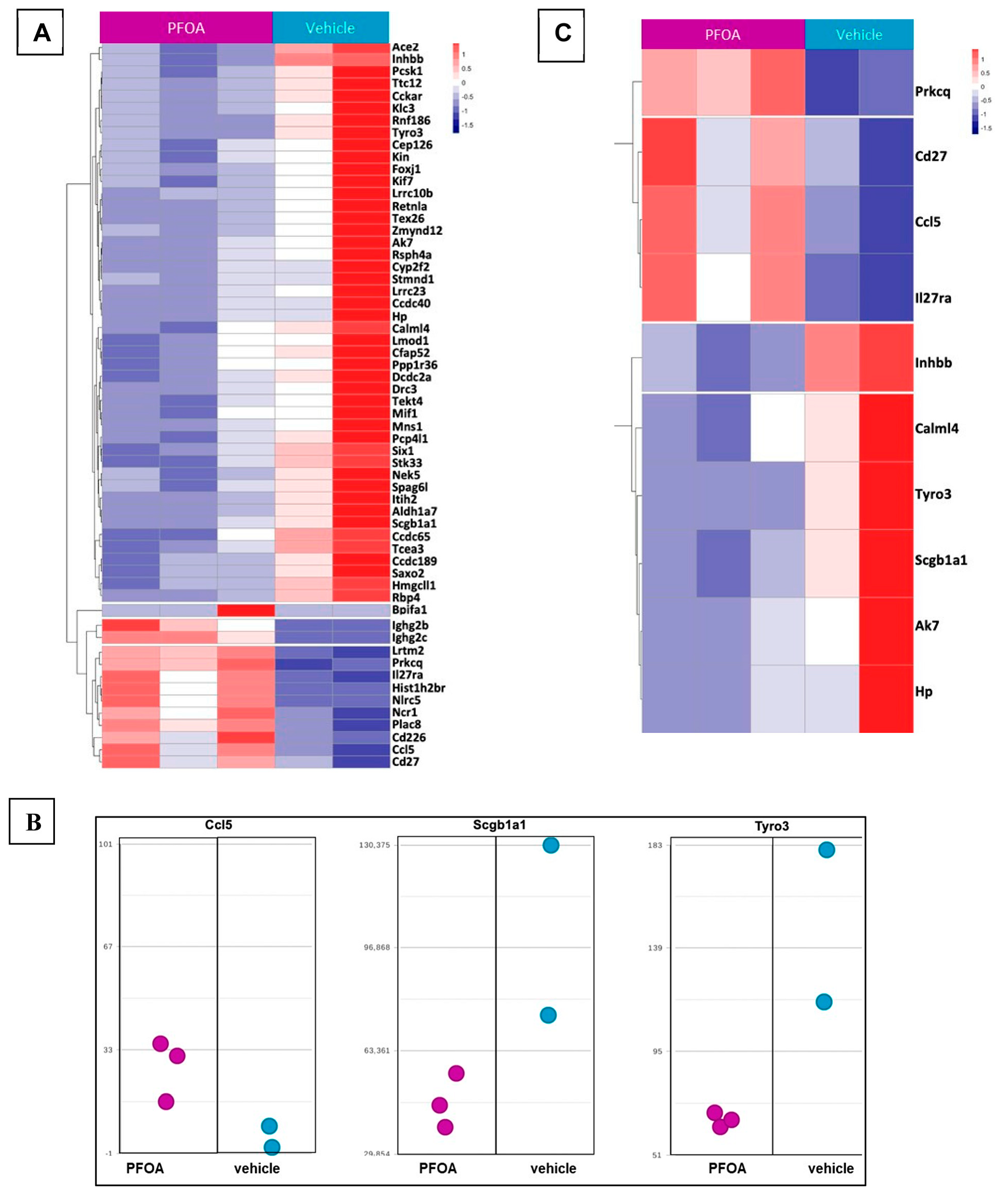
2.7. RNA Seq-Gene Expression on Mouse Lung Tissues Exposed to PFOA
3. Discussion
4. Materials and Methods
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Zhu, W.; Khan, K.; Roakes, H.; Maker, E.; Underwood, K.L.; Zemba, S.; Badireddy, A.R. Vermont-wide assessment of anthropogenic background concentrations of perfluoroalkyl substances in surface soils. J. Hazard. Mater. 2022, 438, 129479. [Google Scholar] [CrossRef]
- Kurwadkar, S.; Dane, J.; Kanel, S.R.; Nadagouda, M.N.; Cawdrey, R.W.; Ambade, B.; Struckhoff, G.C.; Wilkin, R. Per- and polyfluoroalkyl substances in water and wastewater: A critical review of their global occurrence and distribution. Sci. Total Environ. 2022, 809, 151003. [Google Scholar] [CrossRef]
- Sznajder-Katarzyńska, K.; Surma, M.; Cieślik, I. A Review of Perfluoroalkyl Acids (PFAAs) in terms of Sources, Applications, Human Exposure, Dietary Intake, Toxicity, Legal Regulation, and Methods of Determination. J. Chem. 2019, 2019, 2717528. [Google Scholar] [CrossRef]
- Kjølholt, J.; Jensen, A.A.; Warming, M. Short-Chain Polyfluoroalkyl Substances (PFAS). A Literature Review of Information on Human Health Effects and Environmental Fate and Effect Aspects of Short-Chain PFAS; Danish Environmental Protection Agency: Copenhagen, Denmark, 2015. [Google Scholar]
- Bruton, T.A.; Blum, A. Proposal for coordinated health research in PFAS-contaminated communities in the United States. Environ. Health 2017, 16, 120. [Google Scholar] [CrossRef]
- Daly, E.R.; Chan, B.P.; Talbot, E.A.; Nassif, J.; Bean, C.; Cavallo, S.J.; Metcalf, E.; Simone, K.; Woolf, A.D. Per- and polyfluoroalkyl substance (PFAS) exposure assessment in a community exposed to contaminated drinking water, New Hampshire, 2015. Int. J. Hyg. Environ. Health 2018, 221, 569–577. [Google Scholar] [CrossRef]
- Zemba, S.G.; Pope, S.; Badireddy, A.R. Ecological risk and bioremediation of per- and polyfluoroalkyl substances. Sci. J. Biol. Life Sci.-SJBLS 2020, 1, 1–5. [Google Scholar] [CrossRef]
- Schecter, A.; Colacino, J.; Haffner, D.; Patel, K.; Opel, M.; Papke, O.; Birnbaum, L. Perfluorinated compounds, polychlorinated biphenyls, and organochlorine pesticide contamination in composite food samples from Dallas, Texas, USA. Environ. Health Perspect. 2010, 118, 796–802. [Google Scholar] [CrossRef]
- Begley, T.H.; White, K.; Honigfort, P.; Twaroski, M.L.; Neches, R.; Walker, R.A. Perfluorochemicals: Potential sources of and migration from food packaging. Food Addit. Contam. 2005, 22, 1023–1031. [Google Scholar] [CrossRef]
- Hu, X.C.; Andrews, D.Q.; Lindstrom, A.B.; Bruton, T.A.; Schaider, L.A.; Grandjean, P.; Lohmann, R.; Carignan, C.C.; Blum, A.; Balan, S.A.; et al. Detection of Poly- and Perfluoroalkyl Substances (PFASs) in U.S. Drinking Water Linked to Industrial Sites, Military Fire Training Areas, and Wastewater Treatment Plants. Environ. Sci. Technol. Lett. 2016, 3, 344–350. [Google Scholar] [CrossRef]
- Calafat, A.M.; Wong, L.Y.; Kuklenyik, Z.; Reidy, J.A.; Needham, L.L. Polyfluoroalkyl chemicals in the U.S. population: Data from the National Health and Nutrition Examination Survey (NHANES) 2003-2004 and comparisons with NHANES 1999-2000. Environ. Health Perspect. 2007, 115, 1596–1602. [Google Scholar] [CrossRef]
- Hussain, A.; Hameed, S.; Stoeckli-Evans, H. 1-(4-Chloro-phenyl-sulfon-yl)-5-(4-fluoro-phen-yl)-5-methyl-imidazolidine-2,4-dio ne. Acta Crystallogr. Sect. E Struct. Rep. Online 2009, 65 Pt 4, o858–o859. [Google Scholar] [CrossRef]
- Taniyasu, S.; Yamashita, N.; Moon, H.B.; Kwok, K.Y.; Lam, P.K.; Horii, Y.; Petrick, G.; Kannan, K. Does wet precipitation represent local and regional atmospheric transportation by perfluorinated alkyl substances? Environ. Int. 2013, 55, 25–32. [Google Scholar] [CrossRef]
- Dreyer, A.; Matthias, V.; Weinberg, I.; Ebinghaus, R. Wet deposition of poly- and perfluorinated compounds in Northern Germany. Environ. Pollut. 2010, 158, 1221–1227. [Google Scholar] [CrossRef]
- Shoeib, M.; Vlahos, P.; Hamer, T.; Peters, A.; Graustein, M.; Narayan, J. Survey of polyfluorinated chemicals (PFCs) in the atmosphere over the northeast Atlantic Ocean. Atmos. Environ. 2010, 44, 2887–2893. [Google Scholar] [CrossRef]
- Koskela, A.; Koponen, J.; Lehenkari, P.; Viluksela, M.; Korkalainen, M.; Tuukkanen, J. Perfluoroalkyl substances in human bone: Concentrations in bones and effects on bone cell differentiation. Sci. Rep. 2017, 7, 6841. [Google Scholar] [CrossRef]
- Ahmad, S.; Wen, Y.; Irudayaraj, J.M.K. PFOA induces alteration in DNA methylation regulators and SARS-CoV-2 targets Ace2 and Tmprss2 in mouse lung tissues. Toxicol. Rep. 2021, 8, 1892–1898. [Google Scholar] [CrossRef]
- Olsen, G.W.; Burris, J.M.; Ehresman, D.J.; Froehlich, J.W.; Seacat, A.M.; Butenhoff, J.L.; Zobel, L.R. Half-life of serum elimination of perfluorooctanesulfonate, perfluorohexanesulfonate, and perfluorooctanoate in retired fluorochemical production workers. Environ. Health Perspect. 2007, 115, 1298–1305. [Google Scholar] [CrossRef]
- Morck, T.A.; Nielsen, F.; Nielsen, J.K.S.; Siersma, V.D.; Grandjean, P.; Knudsen, L.E. PFAS concentrations in plasma samples from Danish school children and their mothers. Chemosphere 2015, 129, 203–209. [Google Scholar] [CrossRef]
- Barry, V.; Winquist, A.; Steenland, K. Perfluorooctanoic acid (PFOA) exposures and incident cancers among adults living near a chemical plant. Environ. Health Perspect. 2013, 121, 1313–1318. [Google Scholar] [CrossRef]
- Melzer, D.; Rice, N.; Depledge, M.H.; Henley, W.E.; Galloway, T.S. Association between serum perfluorooctanoic acid (PFOA) and thyroid disease in the U.S. National Health and Nutrition Examination Survey. Environ. Health Perspect. 2010, 118, 686–692. [Google Scholar] [CrossRef]
- Panikkar, B.; Lemmond, B.; Allen, L.; DiPirro, C.; Kasper, S. Making the invisible visible: Results of a community-led health survey following PFAS contamination of drinking water in Merrimack, New Hampshire. Environ. Health 2019, 18, 79. [Google Scholar] [CrossRef] [PubMed]
- McGlinchey, A.; Sinioja, T.; Lamichhane, S.; Sen, P.; Bodin, J.; Siljander, H.; Dickens, A.M.; Geng, D.; Carlsson, C.; Duberg, D.; et al. Prenatal exposure to perfluoroalkyl substances modulates neonatal serum phospholipids, increasing risk of type 1 diabetes. Environ. Int. 2020, 143, 105935. [Google Scholar] [CrossRef] [PubMed]
- Sinisalu, L.; Sen, P.; Salihovic, S.; Virtanen, S.M.; Hyoty, H.; Ilonen, J.; Toppari, J.; Veijola, R.; Oresic, M.; Knip, M.; et al. Early-life exposure to perfluorinated alkyl substances modulates lipid metabolism in progression to celiac disease. Environ. Res. 2020, 188, 109864. [Google Scholar] [CrossRef] [PubMed]
- Goodrich, J.A.; Walker, D.I.; He, J.; Lin, X.; Baumert, B.O.; Hu, X.; Alderete, T.L.; Chen, Z.; Valvi, D.; Fuentes, Z.C.; et al. Metabolic Signatures of Youth Exposure to Mixtures of Per- and Polyfluoroalkyl Substances: A Multi-Cohort Study. Environ. Health Perspect. 2023, 131, 27005. [Google Scholar] [CrossRef] [PubMed]
- Kotlarz, N.; McCord, J.; Collier, D.; Lea, C.S.; Strynar, M.; Lindstrom, A.B.; Wilkie, A.A.; Islam, J.Y.; Matney, K.; Tarte, P.; et al. Measurement of Novel, Drinking Water-Associated PFAS in Blood from Adults and Children in Wilmington, North Carolina. Environ. Health Perspect. 2020, 128, 77005. [Google Scholar] [CrossRef] [PubMed]
- Sorli, J.B.; Lag, M.; Ekeren, L.; Perez-Gil, J.; Haug, L.S.; Da Silva, E.; Matrod, M.N.; Gutzkow, K.B.; Lindeman, B. Per- and polyfluoroalkyl substances (PFASs) modify lung surfactant function and pro-inflammatory responses in human bronchial epithelial cells. Toxicol. Vitr. Int. J. Publ. Assoc. BIBRA 2019, 62, 104656. [Google Scholar] [CrossRef]
- Wang, L.Q.; Liu, T.; Yang, S.; Sun, L.; Zhao, Z.Y.; Li, L.Y.; She, Y.C.; Zheng, Y.Y.; Ye, X.Y.; Bao, Q.; et al. Author Correction: Perfluoroalkyl substance pollutants activate the innate immune system through the AIM2 inflammasome. Nat. Commun. 2022, 13, 5667. [Google Scholar] [CrossRef]
- Zhou, X.; Dong, T.; Fan, Z.; Peng, Y.; Zhou, R.; Wang, X.; Song, N.; Han, M.; Fan, B.; Jia, J.; et al. Perfluorodecanoic acid stimulates NLRP3 inflammasome assembly in gastric cells. Sci. Rep. 2017, 7, 45468. [Google Scholar] [CrossRef]
- Shao, W.; Xu, J.; Xu, C.; Weng, Z.; Liu, Q.; Zhang, X.; Liang, J.; Li, W.; Zhang, Y.; Jiang, Z.; et al. Early-life perfluorooctanoic acid exposure induces obesity in male offspring and the intervention role of chlorogenic acid. Environ. Pollut. 2020, 272, 115974. [Google Scholar] [CrossRef]
- Zhang, H.; Lu, H.; Yu, L.; Yuan, J.; Qin, S.; Li, C.; Ge, R.S.; Chen, H.; Ye, L. Effects of gestational exposure to perfluorooctane sulfonate on the lung development of offspring rats. Environ. Pollut. 2021, 272, 115535. [Google Scholar] [CrossRef]
- Yang, Z.; Roth, K.; Ding, J.; Kassotis, C.D.; Mor, G.; Petriello, M.C. Exposure to a mixture of per-and polyfluoroalkyl substances modulates pulmonary expression of ACE2 and circulating hormones and cytokines. Toxicol. Appl. Pharmacol. 2022, 456, 116284. [Google Scholar] [CrossRef]
- Dong, G.H.; Tung, K.Y.; Tsai, C.H.; Liu, M.M.; Wang, D.; Liu, W.; Jin, Y.H.; Hsieh, W.S.; Lee, Y.L.; Chen, P.C. Serum polyfluoroalkyl concentrations, asthma outcomes, and immunological markers in a case-control study of Taiwanese children. Environ. Health Perspect. 2013, 121, 507–513. [Google Scholar] [CrossRef]
- Humblet, O.; Diaz-Ramirez, L.G.; Balmes, J.R.; Pinney, S.M.; Hiatt, R.A. Perfluoroalkyl chemicals and asthma among children 12–19 years of age: NHANES (1999–2008). Environ. Health Perspect. 2014, 122, 1129–1133. [Google Scholar] [CrossRef]
- Chang, E.T.; Adami, H.O.; Boffetta, P.; Wedner, H.J.; Mandel, J.S. A critical review of perfluorooctanoate and perfluorooctanesulfonate exposure and immunological health conditions in humans. Crit. Rev. Toxicol. 2016, 46, 279–331. [Google Scholar] [CrossRef]
- Ryu, M.H.; Jha, A.; Ojo, O.O.; Mahood, T.H.; Basu, S.; Detillieux, K.A.; Nikoobakht, N.; Wong, C.S.; Loewen, M.; Becker, A.B.; et al. Chronic exposure to perfluorinated compounds: Impact on airway hyperresponsiveness and inflammation. Am. J. Physiol. Lung Cell. Mol. Physiol. 2014, 307, L765–L774. [Google Scholar] [CrossRef]
- Impinen, A.; Nygaard, U.C.; Lodrup Carlsen, K.C.; Mowinckel, P.; Carlsen, K.H.; Haug, L.S.; Granum, B. Prenatal exposure to perfluoralkyl substances (PFASs) associated with respiratory tract infections but not allergy- and asthma-related health outcomes in childhood. Environ. Res. 2018, 160, 518–523. [Google Scholar] [CrossRef]
- Smit, L.A.; Lenters, V.; Hoyer, B.B.; Lindh, C.H.; Pedersen, H.S.; Liermontova, I.; Jonsson, B.A.; Piersma, A.H.; Bonde, J.P.; Toft, G.; et al. Prenatal exposure to environmental chemical contaminants and asthma and eczema in school-age children. Allergy 2015, 70, 653–660. [Google Scholar] [CrossRef]
- Averina, M.; Brox, J.; Huber, S.; Furberg, A.S.; Sorensen, M. Serum perfluoroalkyl substances (PFAS) and risk of asthma and various allergies in adolescents. The Tromso study Fit Futures in Northern Norway. Environ. Res. 2019, 169, 114–121. [Google Scholar] [CrossRef]
- Huang, H.; Yu, K.; Zeng, X.; Chen, Q.; Liu, Q.; Zhao, Y.; Zhang, J.; Zhang, X.; Huang, L. Association between prenatal exposure to perfluoroalkyl substances and respiratory tract infections in preschool children. Environ. Res. 2020, 191, 110256. [Google Scholar] [CrossRef]
- Sanchez Garcia, D.; Sjodin, M.; Hellstrandh, M.; Norinder, U.; Nikiforova, V.; Lindberg, J.; Wincent, E.; Bergman, A.; Cotgreave, I.; Munic Kos, V. Cellular accumulation and lipid binding of perfluorinated alkylated substances (PFASs)—A comparison with lysosomotropic drugs. Chem.-Biol. Interact. 2018, 281, 1–10. [Google Scholar] [CrossRef]
- Zhang, L.; Louie, A.; Rigutto, G.; Guo, H.; Zhao, Y.; Ahn, S.; Dahlberg, S.; Sholinbeck, M.; Smith, M.T. A systematic evidence map of chronic inflammation and immunosuppression related to per- and polyfluoroalkyl substance (PFAS) exposure. Environ. Res. 2022, 220, 115188. [Google Scholar] [CrossRef] [PubMed]
- Han, M.W.; Kim, S.H.; Oh, I.; Kim, Y.H.; Lee, J. Serum IL-1beta can be a biomarker in children with severe persistent allergic rhinitis. Allergy Asthma Clin. Immunol. 2019, 15, 58. [Google Scholar] [CrossRef] [PubMed]
- Ruggeri, P.; Caramori, G. Functional Role of Inflammasome Activation in a Subset of Obese Nonsmoking Patients with Severe Asthma. Am. J. Respir. Crit. Care Med. 2019, 199, 1045–1047. [Google Scholar] [CrossRef] [PubMed]
- Li, R.; Wang, J.; Zhu, F.; Xu, W.; Zha, G.; He, G.; Cao, H.; Wang, Y.; Yang, J. ATP/P2x7-NLRP3 axis of dendritic cells participates in the regulation of airway inflammation and hyper-responsiveness in asthma by mediating HMGB1 expression and secretion. Exp. Cell Res. 2018, 366, 1–15. [Google Scholar] [CrossRef]
- Xiao, Y.; Xu, W.; Su, W. NLRP3 inflammasome: A likely target for the treatment of allergic diseases. Clin. Exp. Allergy 2018, 48, 1080–1091. [Google Scholar] [CrossRef]
- Dostert, C.; Petrilli, V.; Van Bruggen, R.; Steele, C.; Mossman, B.T.; Tschopp, J. Innate immune activation through Nalp3 inflammasome sensing of asbestos and silica. Science 2008, 320, 674–677. [Google Scholar] [CrossRef]
- Hillegass, J.M.; Miller, J.M.; MacPherson, M.B.; Westbom, C.M.; Sayan, M.; Thompson, J.K.; Macura, S.L.; Perkins, T.N.; Beuschel, S.L.; Alexeeva, V.; et al. Asbestos and erionite prime and activate the NLRP3 inflammasome that stimulates autocrine cytokine release in human mesothelial cells. Part. Fibre Toxicol. 2013, 10, 39. [Google Scholar] [CrossRef]
- Marinello, W.P.; Mohseni, Z.S.; Cunningham, S.J.; Crute, C.; Huang, R.; Zhang, J.J.; Feng, L. Perfluorobutane sulfonate exposure disrupted human placental cytotrophoblast cell proliferation and invasion involving in dysregulating preeclampsia related genes. FASEB J. Off. Publ. Fed. Am. Soc. Exp. Biol. 2020, 34, 14182–14199. [Google Scholar] [CrossRef]
- Alcorn, J.F.; Guala, A.S.; van der Velden, J.; McElhinney, B.; Irvin, C.G.; Davis, R.J.; Janssen-Heininger, Y.M. Jun N-terminal kinase 1 regulates epithelial-to-mesenchymal transition induced by TGF-beta1. J. Cell Sci. 2008, 121, 1036–1045. [Google Scholar] [CrossRef]
- Thompson, J.K.; MacPherson, M.B.; Beuschel, S.L.; Shukla, A. Asbestos-Induced Mesothelial to Fibroblastic Transition Is Modulated by the Inflammasome. Am. J. Pathol. 2017, 187, 665–678. [Google Scholar] [CrossRef]
- Ferrero, M.R.; Tavares, L.P.; Garcia, C.C. The Dual Role of CCR5 in the Course of Influenza Infection: Exploring Treatment Opportunities. Front. Immunol. 2021, 12, 826621. [Google Scholar] [CrossRef]
- Habib, N.; Pasha, M.A.; Tang, D.D. Current Understanding of Asthma Pathogenesis and Biomarkers. Cells 2022, 11, 2764. [Google Scholar] [CrossRef]
- Khan, P.; Fatima, M.; Khan, M.A.; Batra, S.K.; Nasser, M.W. Emerging role of chemokines in small cell lung cancer: Road signs for metastasis, heterogeneity, and immune response. Semin. Cancer Biol. 2022, 87, 117126. [Google Scholar] [CrossRef]
- Mootz, M.; Jakwerth, C.A.; Schmidt-Weber, C.B.; Zissler, U.M. Secretoglobins in the big picture of immunoregulation in airway diseases. Allergy 2022, 77, 767–777. [Google Scholar] [CrossRef]
- Aehnlich, P.; Powell, R.M.; Peeters, M.J.W.; Rahbech, A.; Thor Straten, P. TAM Receptor Inhibition-Implications for Cancer and the Immune System. Cancers 2021, 13, 1195. [Google Scholar] [CrossRef]
- Phelps, D.W.; Palekar, A.I.; Conley, H.E.; Ferrero, G.; Driggers, J.H.; Linder, K.E.; Kullman, S.W.; Reif, D.M.; Sheats, M.K.; DeWitt, J.C.; et al. Legacy and emerging per- and polyfluoroalkyl substances suppress the neutrophil respiratory burst. J. Immunotoxicol. 2023, 20, 2176953. [Google Scholar] [CrossRef]
- Wang, C.; Fu, H.; Yang, J.; Liu, L.; Zhang, F.; Yang, C.; Li, H.; Chen, J.; Li, Q.; Wang, X.; et al. PFO5DoDA disrupts hepatic homeostasis primarily through glucocorticoid signaling inhibition. J. Hazard. Mater. 2023, 447, 130831. [Google Scholar] [CrossRef]
- Schlezinger, J.J.; Puckett, H.; Oliver, J.; Nielsen, G.; Heiger-Bernays, W.; Webster, T.F. Perfluorooctanoic acid activates multiple nuclear receptor pathways and skews expression of genes regulating cholesterol homeostasis in liver of humanized PPARalpha mice fed an American diet. Toxicol. Appl. Pharm. 2020, 405, 115204. [Google Scholar] [CrossRef]
- Munson, P.; Lam, Y.W.; Dragon, J.; MacPherson, M.; Shukla, A. Exosomes from asbestos-exposed cells modulate gene expression in mesothelial cells. FASEB J. 2018, 32, 4328–4342. [Google Scholar] [CrossRef]
- Perkins, T.N.; Peeters, P.M.; Shukla, A.; Arijs, I.; Dragon, J.; Wouters, E.F.; Reynaert, N.L.; Mossman, B.T. Indications for distinct pathogenic mechanisms of asbestos and silica through gene expression profiling of the response of lung epithelial cells. Hum. Mol. Genet. 2015, 24, 1374–1389. [Google Scholar] [CrossRef]
- Frederix, P.L.; Bosshart, P.D.; Engel, A. Atomic force microscopy of biological membranes. Biophys. J. 2009, 96, 329–338. [Google Scholar] [CrossRef] [PubMed]
- Al-Rekabi, Z.; Contera, S. Multifrequency AFM reveals lipid membrane mechanical properties and the effect of cholesterol in modulating viscoelasticity. Proc. Natl. Acad. Sci. USA 2018, 115, 2658–2663. [Google Scholar] [CrossRef] [PubMed]
- Olsen, G.W.; Zobel, L.R. Assessment of lipid, hepatic, and thyroid parameters with serum perfluorooctanoate (PFOA) concentrations in fluorochemical production workers. Int. Arch. Occup. Environ. Health 2007, 81, 231–246. [Google Scholar] [CrossRef] [PubMed]
- Schlezinger, J.J.; Hyotylainen, T.; Sinioja, T.; Boston, C.; Puckett, H.; Oliver, J.; Heiger-Bernays, W.; Webster, T.F. Perfluorooctanoic acid induces liver and serum dyslipidemia in humanized PPARalpha mice fed an American diet. Toxicol. Appl. Pharm. 2021, 426, 115644. [Google Scholar] [CrossRef]
- Yang, Q.; Nagano, T.; Shah, Y.; Cheung, C.; Ito, S.; Gonzalez, F.J. The PPAR alpha-humanized mouse: A model to investigate species differences in liver toxicity mediated by PPAR alpha. Toxicol. Sci. 2008, 101, 132–139. [Google Scholar] [CrossRef]
- USDA. What We Eat in America. 2018. Available online: https://www.ars.usda.gov/northeast-area/beltsville-md-bhnrc/beltsville-human-nutrition-research-center/food-surveys-research-group/docs/wweianhanes-overview (accessed on 15 January 2018).

| Treatment | Plasticity | Force Ratio | Elastic Modulus (Pa) | ||||||
|---|---|---|---|---|---|---|---|---|---|
| Mean | SEM | n | Mean | SEM | n | Mean | SEM | n | |
| DMSO | 0.276 | 0.0217 | 89 | 5.299 | 0.4668 | 95 | 521,803 | 41,398 | 97 |
| PFOS | 0.238 | 0.0158 | 96 | 5.013 | 0.2219 | 95 | 602,628 | 38,920 | 100 |
| PFOA | 0.305 | 0.0801 | 95 | 2.921 | 0.2770 | 95 | 699,743 | 159,913 | 96 |
| Ensembl | Chr | Total Counts | p-Value | FDR (Step-Up) | Fold Change |
|---|---|---|---|---|---|
| Bpifa1 | 2 | 946 | 5.29 × 10−9 | 9.88 × 10−5 | 2.36 × 106 |
| Ighg2c | 12 | 232 | 1.34 × 10−4 | 9.61 × 10−2 | 9.80 × 101 |
| Hist1h2br | 13 | 85 | 9.02 × 10−4 | 1.87 × 10−1 | 7.89 × 101 |
| Ighg2b | 12 | 234 | 6.84 × 10−4 | 1.75 × 10−1 | 1.46 × 101 |
| Ncr1 | 7 | 551 | 8.33 × 10−4 | 1.87 × 10−1 | 9.14 × 100 |
| Ccl5 | 11 | 663 | 2.69 × 10−4 | 1.32 × 10−1 | 5.59 × 100 |
| I830077J02Rik | 3 | 371 | 9.65 × 10−5 | 7.36 × 10−2 | 5.18 × 100 |
| Plac8 | 5 | 745 | 7.30 × 10−4 | 1.75 × 10−1 | 3.86 × 100 |
| Cd27 | 6 | 453 | 3.25 × 10−4 | 1.38 × 10−1 | 3.32 × 100 |
| Cd226 | 18 | 873 | 2.47 × 10−4 | 1.32 × 10−1 | 3.30 × 100 |
| Il27ra | 8 | 647 | 6.11 × 10−4 | 1.70 × 10−1 | 2.65 × 100 |
| Lrtm2 | 6 | 1051 | 2.07 × 10−4 | 1.21 × 10−1 | 2.41 × 100 |
| Prkcq | 2 | 1757 | 2.81 × 10−7 | 2.62 × 10−3 | 2.38 × 100 |
| Nlrc5 | 8 | 3571 | 1.09 × 10−3 | 1.96 × 10−1 | 2.01 × 100 |
| Kin | 2 | 2205 | 6.73 × 10−4 | 1.75 × 10−1 | −2.03 × 100 |
| Lmod1 | 1 | 6101 | 1.19 × 10−3 | 2.00 × 10−1 | −2.04 × 100 |
| 5330417C22Rik | 3 | 12,012 | 8.49 × 10−6 | 2.54 × 10−2 | −2.04 × 100 |
| Kif7 | 7 | 1439 | 4.81 × 10−4 | 1.66 × 10−1 | −2.07 × 100 |
| Cyp2f2 | 7 | 142,500 | 5.99 × 10−4 | 1.70 × 10−1 | −2.07 × 100 |
| Inhbb | 1 | 2105 | 9.52 × 10−6 | 2.54 × 10−2 | −2.08 × 100 |
| Rnf186 | 4 | 1163 | 8.24 × 10−4 | 1.87 × 10−1 | −2.10 × 100 |
| Hp | 8 | 44,707 | 2.96 × 10−4 | 1.38 × 10−1 | −2.14 × 100 |
| Lrrc10b | 19 | 1435 | 5.51 × 10−4 | 1.70 × 10−1 | −2.15 × 100 |
| Cckar | 5 | 3927 | 7.17 × 10−5 | 6.38 × 10−2 | −2.18 × 100 |
| Hmgcll1 | 9 | 1103 | 3.40 × 10−4 | 1.38 × 10−1 | −2.24 × 100 |
| Mns1 | 9 | 4942 | 6.98 × 10−4 | 1.75 × 10−1 | −2.24 × 100 |
| Drc3 | 11 | 2063 | 1.19 × 10−3 | 2.00 × 10−1 | −2.26 × 100 |
| Ttc12 | 9 | 2140 | 4.64 × 10−5 | 5.30 × 10−2 | −2.27 × 100 |
| Aldh1a7 | 19 | 9082 | 1.30 × 10−5 | 2.70 × 10−2 | −2.28 × 100 |
| Foxj1 | 11 | 4375 | 1.10 × 10−3 | 1.96 × 10−1 | −2.30 × 100 |
| Rbp4 | 19 | 1647 | 3.00 × 10−5 | 4.54 × 10−2 | −2.35 × 100 |
| Ccdc189 | 7 | 1386 | 6.25 × 10−5 | 5.83 × 10−2 | −2.35 × 100 |
| Pcp4l1 | 1 | 3787 | 1.92 × 10−4 | 1.17 × 10−1 | −2.39 × 100 |
| Lrrc23 | 6 | 2829 | 5.69 × 10−4 | 1.70 × 10−1 | −2.40 × 100 |
| Ak7 | 12 | 5344 | 8.81 × 10−4 | 1.87 × 10−1 | −2.42 × 100 |
| Tcea3 | 4 | 1315 | 9.63 × 10−4 | 1.88 × 10−1 | −2.43 × 100 |
| Scgb1a1 | 19 | 910,638 | 5.78 × 10−6 | 2.54 × 10−2 | −2.52 × 100 |
| Ccdc40 | 11 | 6721 | 5.28 × 10−4 | 1.70 × 10−1 | −2.53 × 100 |
| Tyro3 | 2 | 1570 | 2.54 × 10−5 | 4.32 × 10−2 | −2.55 × 100 |
| Ccdc65 | 15 | 945 | 1.10 × 10−3 | 1.96 × 10−1 | −2.56 × 100 |
| Cep126 | 9 | 3598 | 9.01 × 10−4 | 1.87 × 10−1 | −2.56 × 100 |
| Calml4 | 9 | 1519 | 3.79 × 10−4 | 1.42 × 10−1 | −2.58 × 100 |
| Klc3 | 7 | 956 | 5.11 × 10−5 | 5.30 × 10−2 | −2.59 × 100 |
| Stk33 | 7 | 1254 | 3.48 × 10−4 | 1.38 × 10−1 | −2.59 × 100 |
| Rsph4a | 10 | 4311 | 3.38 × 10−4 | 1.38 × 10−1 | −2.65 × 100 |
| Ace2 | X | 1782 | 5.94 × 10−5 | 5.83 × 10−2 | −2.69 × 100 |
| Saxo2 | 7 | 1942 | 5.28 × 10−4 | 1.70 × 10−1 | −2.71 × 100 |
| Spag6l | 16 | 2335 | 3.38 × 10−4 | 1.38 × 10−1 | −2.72 × 100 |
| Nek5 | 8 | 1539 | 4.64 × 10−5 | 5.30 × 10−2 | −2.77 × 100 |
| Ppp1r36 | 12 | 987 | 6.87 × 10−4 | 1.75 × 10−1 | −2.81 × 100 |
| Dcdc2a | 13 | 1097 | 6.02 × 10−4 | 1.70 × 10−1 | −2.81 × 100 |
| Cfap52 | 11 | 1362 | 5.18 × 10−4 | 1.70 × 10−1 | −2.98 × 100 |
| Tekt4 | 17 | 984 | 2.81 × 10−4 | 1.35 × 10−1 | −3.12 × 100 |
| Mlf1 | 3 | 1739 | 1.03 × 10−3 | 1.92 × 10−1 | −3.13 × 100 |
| Six1 | 12 | 935 | 1.03 × 10−3 | 1.92 × 10−1 | −3.45 × 100 |
| Stmnd1 | 13 | 1442 | 6.27 × 10−4 | 1.70 × 10−1 | −3.54 × 100 |
| Itih2 | 2 | 466 | 1.59 × 10−4 | 1.06 × 10−1 | −3.71 × 100 |
| Retnla | 16 | 2736 | 7.31 × 10−6 | 2.54 × 10−2 | −5.01 × 100 |
| Pcsk1 | 13 | 218 | 9.91 × 10−4 | 1.91 × 10−1 | −5.06 × 100 |
| Zmynd12 | 4 | 172 | 5.76 × 10−4 | 1.70 × 10−1 | −6.57 × 100 |
| Tex26 | 5 | 147 | 9.66 × 10−4 | 1.88 × 10−1 | −8.53 × 100 |
| Gm3417 | 17 | 384 | 1.20 × 10−5 | 2.70 × 10−2 | −2.31 × 101 |
| Gene Set | Description | Enrichment Score | p-Value | Genes in List | Genes not in List |
|---|---|---|---|---|---|
| GO: 0050727 | regulation of inflammatory response | 4.14199 | 0.0158912 | 4 | 286 |
| GO: 0050728 | negative regulation of inflammatory response | 1.06038 | 0.346326 | 1 | 126 |
| GO: 0050729 | positive regulation of inflammatory response | 2.98854 | 0.0503611 | 2 | 106 |
| GO: 0002437 | inflammatory response to antigenic stimulus | 2.52479 | 0.0800755 | 1 | 24 |
| GO: 0002526 | acute inflammatory response | 1.81842 | 0.162283 | 1 | 52 |
| GO: 0002673 | regulation of acute inflammatory response | 4.23522 | 0.0144766 | 2 | 53 |
| GO: 0002675 | positive regulation of acute inflammatory response | 5.54591 | 0.00390341 | 2 | 26 |
| GO: 0002861 | regulation of inflammatory response to antigenic stimulus | 2.41631 | 0.0892499 | 1 | 27 |
| GO: 0002863 | positive regulation of inflammatory response to antigenic stimulus | 3.01936 | 0.0488324 | 1 | 14 |
| GO: 0002864 | regulation of acute inflammatory response to antigenic stimulus | 2.95645 | 0.0520031 | 1 | 15 |
| GO: 0002866 | positive regulation of acute inflammatory response to antigenic stimulus | 3.32299 | 0.0360447 | 1 | 10 |
| GO: 0006954 | inflammatory response | 5.10092 | 0.00609113 | 5 | 344 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Dragon, J.; Hoaglund, M.; Badireddy, A.R.; Nielsen, G.; Schlezinger, J.; Shukla, A. Perfluoroalkyl Substances (PFAS) Affect Inflammation in Lung Cells and Tissues. Int. J. Mol. Sci. 2023, 24, 8539. https://doi.org/10.3390/ijms24108539
Dragon J, Hoaglund M, Badireddy AR, Nielsen G, Schlezinger J, Shukla A. Perfluoroalkyl Substances (PFAS) Affect Inflammation in Lung Cells and Tissues. International Journal of Molecular Sciences. 2023; 24(10):8539. https://doi.org/10.3390/ijms24108539
Chicago/Turabian StyleDragon, Julie, Michael Hoaglund, Appala Raju Badireddy, Greylin Nielsen, Jennifer Schlezinger, and Arti Shukla. 2023. "Perfluoroalkyl Substances (PFAS) Affect Inflammation in Lung Cells and Tissues" International Journal of Molecular Sciences 24, no. 10: 8539. https://doi.org/10.3390/ijms24108539
APA StyleDragon, J., Hoaglund, M., Badireddy, A. R., Nielsen, G., Schlezinger, J., & Shukla, A. (2023). Perfluoroalkyl Substances (PFAS) Affect Inflammation in Lung Cells and Tissues. International Journal of Molecular Sciences, 24(10), 8539. https://doi.org/10.3390/ijms24108539






