Dysfunction of Foxp3+ Regulatory T Cells Induces Dysbiosis of Gut Microbiota via Aberrant Binding of Immunoglobulins to Microbes in the Intestinal Lumen
Abstract
1. Introduction
2. Results
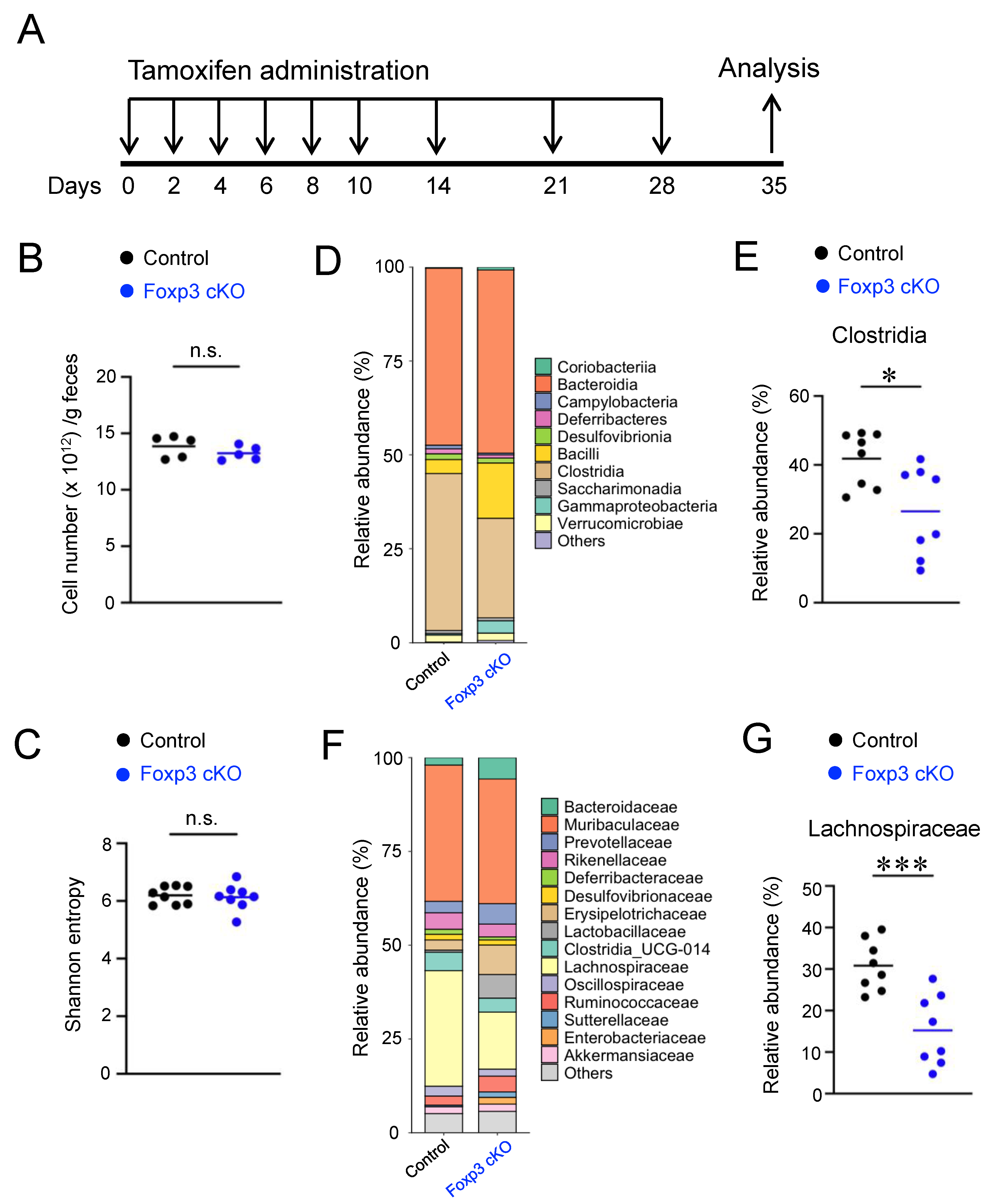
2.1. Conditional Knockout of Foxp3 Changed the Composition of Gut Microbiota
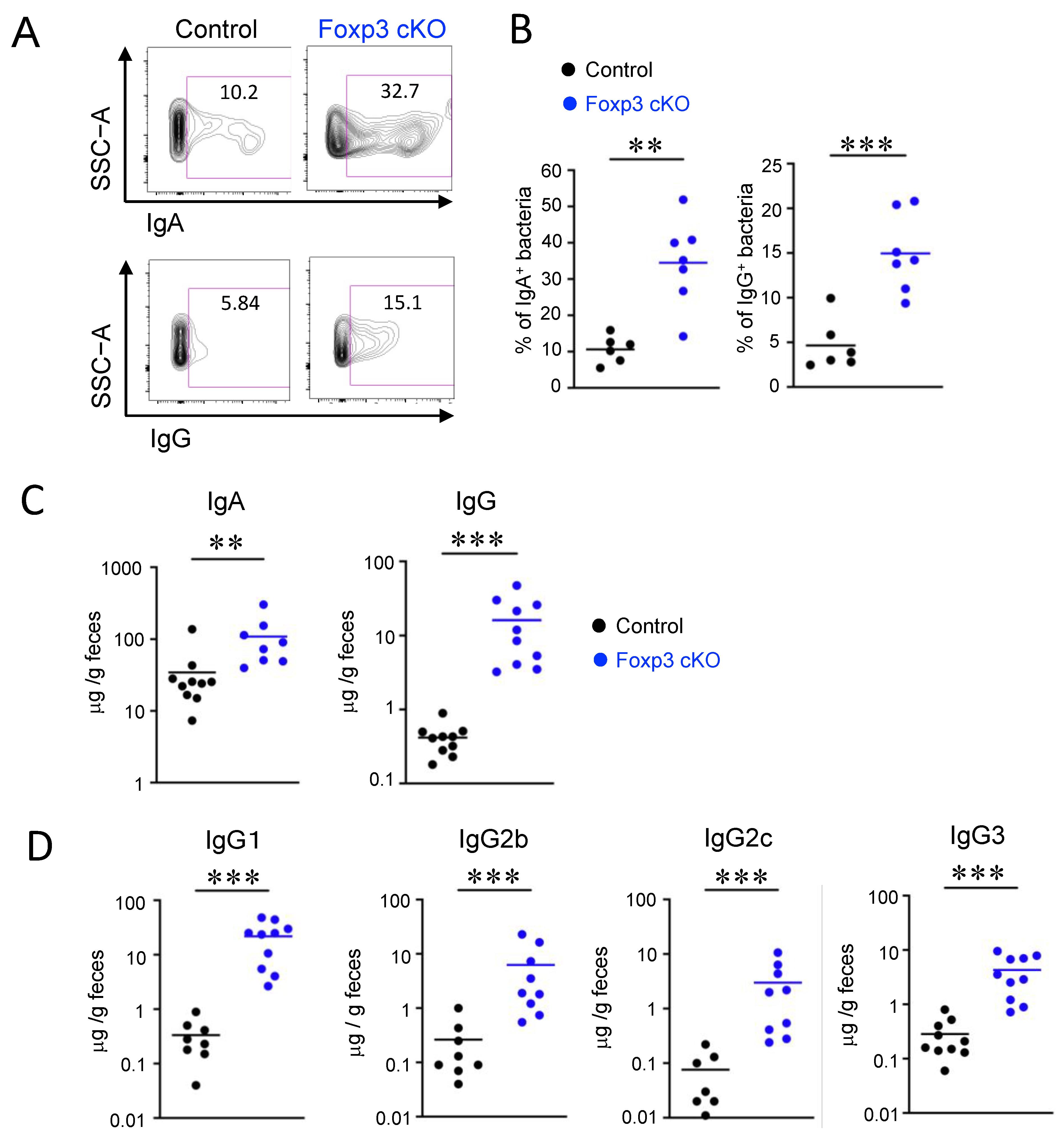
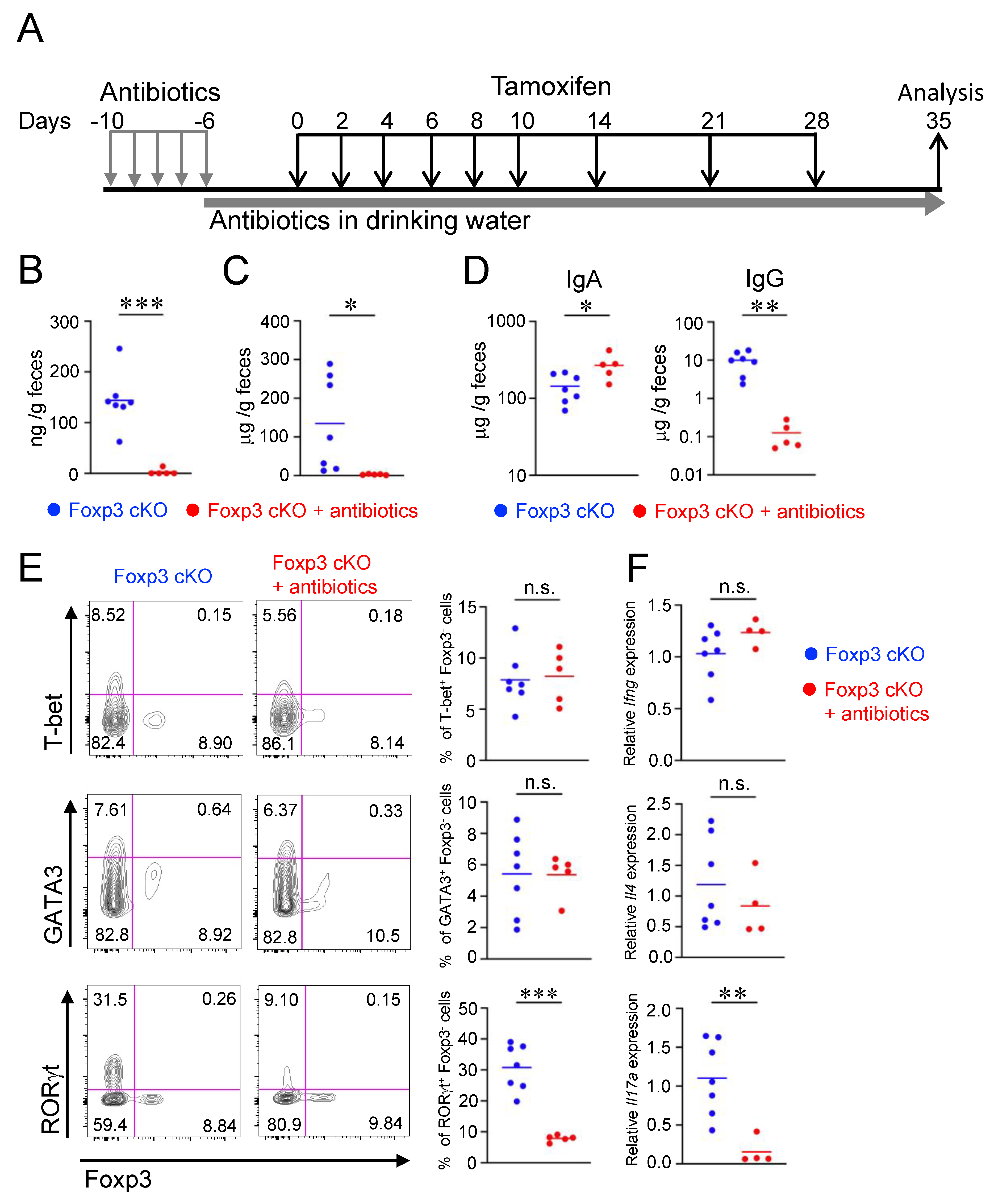
2.2. Dysfunction of Treg Cells Increased Ig-Coated Bacteria
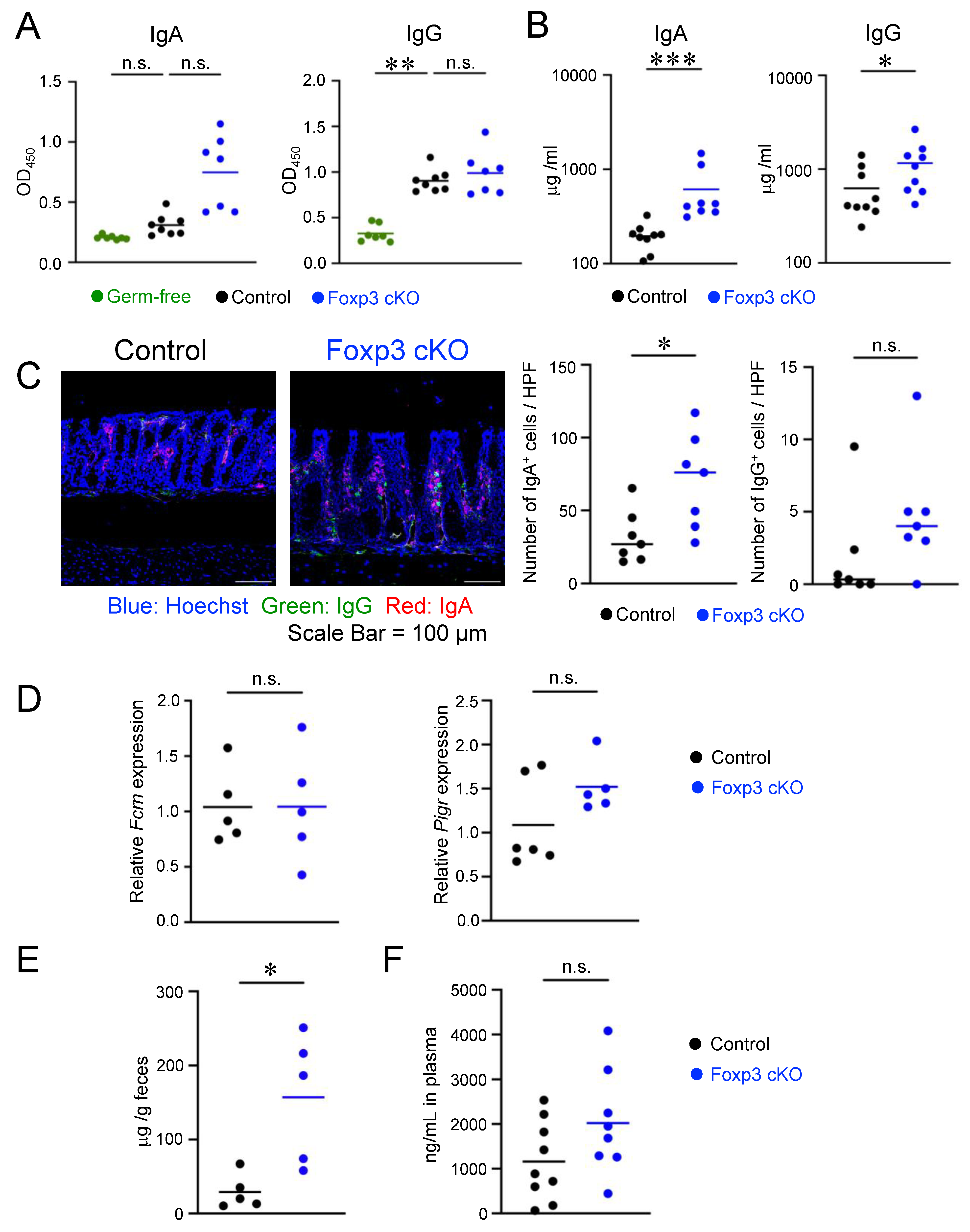
2.3. Deletion of Foxp3 Promoted Intestinal Permeability but Did Not Increase the Production of Bacteria-Specific IgG
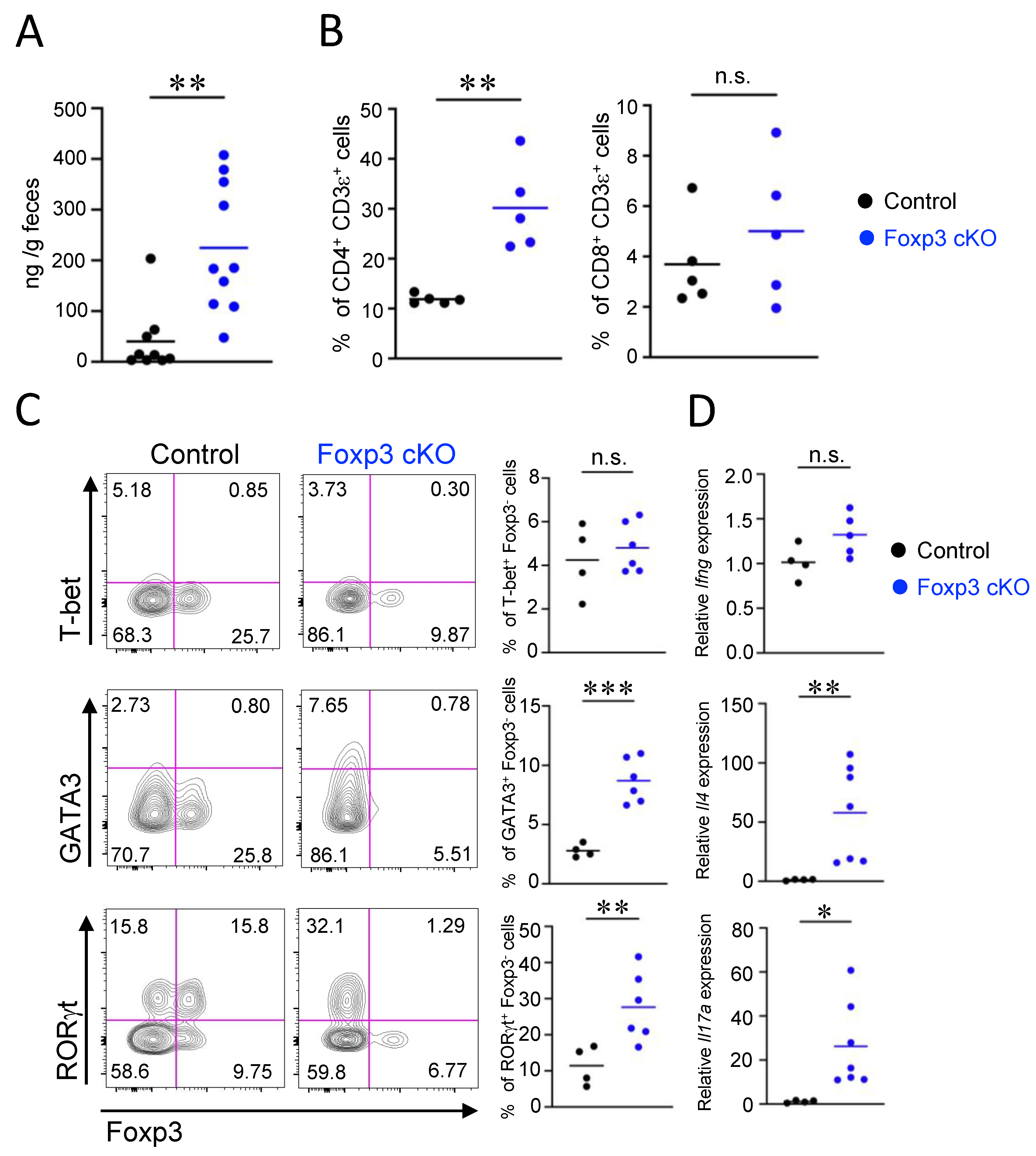
2.4. Th2- and Th17-Associated Intestinal Inflammatory Responses Were Induced by Dysfunction of Treg Cells
2.5. Impairment of Intestinal Barrier by Dysfunction of Treg Cells Depends on Gut Microbiota
3. Discussion
4. Materials and Methods
4.1. Mice
4.2. Tamoxifen Administration
4.3. Antibiotic Treatments
4.4. Isolation and Stimulation of Cells from Lymphoid Tissues
4.5. Flow Cytometry
4.6. Enzyme-Linked Immunosorbent Assay
4.7. Histological Analysis
4.8. Fluorescent Immunohistochemistry
4.9. Quantitative Reverse Transcription Polymerase Chain Reaction (RT-qPCR)
4.10. 16S rRNA Gene Sequence
4.11. Quantification of Fecal Bacteria
4.12. In Vivo Intestinal Permeability Assay
4.13. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Fontenot, J.D.; Gavin, M.A.; Rudensky, A.Y. Foxp3 programs the development and function of CD4+CD25+ regulatory T cells. Nat. Immunol. 2003, 4, 330–336. [Google Scholar] [CrossRef] [PubMed]
- Hori, S.; Nomura, T.; Sakaguchi, S. Control of regulatory T cell development by the transcription factor Foxp3. Science 2003, 299, 1057–1061. [Google Scholar] [CrossRef] [PubMed]
- Rajendeeran, A.; Tenbrock, K. Regulatory T cell function in autoimmune disease. J. Transl. Autoimmun. 2021, 4, 100130. [Google Scholar] [CrossRef] [PubMed]
- Grover, P.; Goel, P.N.; Greene, M.I. Regulatory T Cells: Regulation of Identity and Function. Front. Immunol. 2021, 12, 750542. [Google Scholar] [CrossRef] [PubMed]
- Ramsdell, F.; Ziegler, S.F. FOXP3 and scurfy: How it all began. Nat. Rev. Immunol. 2014, 14, 343–349. [Google Scholar] [CrossRef]
- Bennett, C.L.; Christie, J.; Ramsdell, F.; Brunkow, M.E.; Ferguson, P.J.; Whitesell, L.; Kelly, T.E.; Saulsbury, F.T.; Chance, P.F.; Ochs, H.D. The immune dysregulation, polyendocrinopathy, enteropathy, X-linked syndrome (IPEX) is caused by mutations of FOXP3. Nat. Genet. 2001, 27, 20–21. [Google Scholar] [CrossRef] [PubMed]
- Bacchetta, R.; Barzaghi, F.; Roncarolo, M.G. From IPEX syndrome to FOXP3 mutation: A lesson on immune dysregulation. Ann. N. Y. Acad. Sci. 2018, 1417, 5–22. [Google Scholar] [CrossRef]
- Consonni, F.; Ciullini Mannurita, S.; Gambineri, E. Atypical Presentations of IPEX: Expect the Unexpected. Front. Pediatr. 2021, 9, 643094. [Google Scholar] [CrossRef]
- Bilate, A.M.; Lafaille, J.J. Induced CD4+Foxp3+ regulatory T cells in immune tolerance. Annu. Rev. Immunol. 2012, 30, 733–758. [Google Scholar] [CrossRef]
- Ai, T.L.; Solomon, B.D.; Hsieh, C.S. T-cell selection and intestinal homeostasis. Immunol. Rev. 2014, 259, 60–74. [Google Scholar] [CrossRef]
- Kim, K.S. Regulation of T cell repertoires by commensal microbiota. Front. Cell Infect. Microbiol. 2022, 12, 1004339. [Google Scholar] [CrossRef] [PubMed]
- Jacobse, J.; Li, J.; Rings, E.; Samsom, J.N.; Goettel, J.A. Intestinal Regulatory T Cells as Specialized Tissue-Restricted Immune Cells in Intestinal Immune Homeostasis and Disease. Front. Immunol. 2021, 12, 716499. [Google Scholar] [CrossRef]
- Macpherson, A.J.; Yilmaz, B.; Limenitakis, J.P.; Ganal-Vonarburg, S.C. IgA Function in Relation to the Intestinal Microbiota. Annu. Rev. Immunol. 2018, 36, 359–381. [Google Scholar] [CrossRef] [PubMed]
- Leon, E.D.; Francino, M.P. Roles of Secretory Immunoglobulin A in Host-Microbiota Interactions in the Gut Ecosystem. Front. Microbiol. 2022, 13, 880484. [Google Scholar] [CrossRef] [PubMed]
- Kamada, N.; Sakamoto, K.; Seo, S.U.; Zeng, M.Y.; Kim, Y.G.; Cascalho, M.; Vallance, B.A.; Puente, J.L.; Nunez, G. Humoral Immunity in the Gut Selectively Targets Phenotypically Virulent Attaching-and-Effacing Bacteria for Intraluminal Elimination. Cell Host Microbe 2015, 17, 617–627. [Google Scholar] [CrossRef]
- Caballero-Flores, G.; Sakamoto, K.; Zeng, M.Y.; Wang, Y.; Hakim, J.; Matus-Acuna, V.; Inohara, N.; Nunez, G. Maternal Immunization Confers Protection to the Offspring against an Attaching and Effacing Pathogen through Delivery of IgG in Breast Milk. Cell Host Microbe 2019, 25, 313–323.e4. [Google Scholar] [CrossRef]
- Sanidad, K.Z.; Amir, M.; Ananthanarayanan, A.; Singaraju, A.; Shiland, N.B.; Hong, H.S.; Kamada, N.; Inohara, N.; Nunez, G.; Zeng, M.Y. Maternal gut microbiome-induced IgG regulates neonatal gut microbiome and immunity. Sci. Immunol. 2022, 7, eabh3816. [Google Scholar] [CrossRef]
- Chen, K.; Magri, G.; Grasset, E.K.; Cerutti, A. Rethinking mucosal antibody responses: IgM, IgG and IgD join IgA. Nat. Rev. Immunol. 2020, 20, 427–441. [Google Scholar] [CrossRef]
- Caballero-Flores, G.; Pickard, J.M.; Nunez, G. Regulation of Citrobacter rodentium colonization: Virulence, immune response and microbiota interactions. Curr. Opin. Microbiol. 2021, 63, 142–149. [Google Scholar] [CrossRef]
- Wu, H.J.; Wu, E. The role of gut microbiota in immune homeostasis and autoimmunity. Gut Microbes 2012, 3, 4–14. [Google Scholar] [CrossRef]
- McLean, M.H.; Dieguez, D., Jr.; Miller, L.M.; Young, H.A. Does the microbiota play a role in the pathogenesis of autoimmune diseases? Gut 2015, 64, 332–341. [Google Scholar] [CrossRef] [PubMed]
- Honda, K.; Littman, D.R. The microbiota in adaptive immune homeostasis and disease. Nature 2016, 535, 75–84. [Google Scholar] [CrossRef] [PubMed]
- He, B.; Hoang, T.K.; Wang, T.; Ferris, M.; Taylor, C.M.; Tian, X.; Luo, M.; Tran, D.Q.; Zhou, J.; Tatevian, N.; et al. Resetting microbiota by Lactobacillus reuteri inhibits T reg deficiency-induced autoimmunity via adenosine A2A receptors. J. Exp. Med. 2017, 214, 107–123. [Google Scholar] [CrossRef] [PubMed]
- He, B.; Liu, Y.; Hoang, T.K.; Tian, X.; Taylor, C.M.; Luo, M.; Tran, D.Q.; Tatevian, N.; Rhoads, J.M. Antibiotic-modulated microbiome suppresses lethal inflammation and prolongs lifespan in Treg-deficient mice. Microbiome 2019, 7, 145. [Google Scholar] [CrossRef]
- Kinashi, Y.; Hase, K. Partners in Leaky Gut Syndrome: Intestinal Dysbiosis and Autoimmunity. Front. Immunol. 2021, 12, 673708. [Google Scholar] [CrossRef]
- Josefowicz, S.Z.; Niec, R.E.; Kim, H.Y.; Treuting, P.; Chinen, T.; Zheng, Y.; Umetsu, D.T.; Rudensky, A.Y. Extrathymically generated regulatory T cells control mucosal TH2 inflammation. Nature 2012, 482, 395–399. [Google Scholar] [CrossRef]
- Campbell, C.; Dikiy, S.; Bhattarai, S.K.; Chinen, T.; Matheis, F.; Calafiore, M.; Hoyos, B.; Hanash, A.; Mucida, D.; Bucci, V.; et al. Extrathymically Generated Regulatory T Cells Establish a Niche for Intestinal Border-Dwelling Bacteria and Affect Physiologic Metabolite Balance. Immunity 2018, 48, 1245–1257.e9. [Google Scholar] [CrossRef]
- Kawamoto, S.; Maruya, M.; Kato, L.M.; Suda, W.; Atarashi, K.; Doi, Y.; Tsutsui, Y.; Qin, H.; Honda, K.; Okada, T.; et al. Foxp3(+) T cells regulate immunoglobulin a selection and facilitate diversification of bacterial species responsible for immune homeostasis. Immunity 2014, 41, 152–165. [Google Scholar] [CrossRef]
- Atarashi, K.; Tanoue, T.; Shima, T.; Imaoka, A.; Kuwahara, T.; Momose, Y.; Cheng, G.; Yamasaki, S.; Saito, T.; Ohba, Y.; et al. Induction of colonic regulatory T cells by indigenous Clostridium species. Science 2011, 331, 337–341. [Google Scholar] [CrossRef]
- Furusawa, Y.; Obata, Y.; Fukuda, S.; Endo, T.A.; Nakato, G.; Takahashi, D.; Nakanishi, Y.; Uetake, C.; Kato, K.; Kato, T.; et al. Commensal microbe-derived butyrate induces the differentiation of colonic regulatory T cells. Nature 2013, 504, 446–450. [Google Scholar] [CrossRef]
- Atarashi, K.; Tanoue, T.; Oshima, K.; Suda, W.; Nagano, Y.; Nishikawa, H.; Fukuda, S.; Saito, T.; Narushima, S.; Hase, K.; et al. Treg induction by a rationally selected mixture of Clostridia strains from the human microbiota. Nature 2013, 500, 232–236. [Google Scholar] [CrossRef]
- Arpaia, N.; Campbell, C.; Fan, X.; Dikiy, S.; van der Veeken, J.; deRoos, P.; Liu, H.; Cross, J.R.; Pfeffer, K.; Coffer, P.J.; et al. Metabolites produced by commensal bacteria promote peripheral regulatory T-cell generation. Nature 2013, 504, 451–455. [Google Scholar] [CrossRef] [PubMed]
- Calvo-Barreiro, L.; Zhang, L.; Abdel-Rahman, S.A.; Naik, S.P.; Gabr, M. Gut Microbial-Derived Metabolites as Immune Modulators of T Helper 17 and Regulatory T Cells. Int. J. Mol. Sci. 2023, 24, 1806. [Google Scholar] [CrossRef]
- Hang, S.; Paik, D.; Yao, L.; Kim, E.; Trinath, J.; Lu, J.; Ha, S.; Nelson, B.N.; Kelly, S.P.; Wu, L.; et al. Bile acid metabolites control TH17 and Treg cell differentiation. Nature 2019, 576, 143–148. [Google Scholar] [CrossRef] [PubMed]
- Zeng, M.Y.; Cisalpino, D.; Varadarajan, S.; Hellman, J.; Warren, H.S.; Cascalho, M.; Inohara, N.; Nunez, G. Gut Microbiota-Induced Immunoglobulin G Controls Systemic Infection by Symbiotic Bacteria and Pathogens. Immunity 2016, 44, 647–658. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Ye, L.; Christianson, G.J.; Yang, J.Q.; Roopenian, D.C.; Zhu, X. NF-kappaB signaling regulates functional expression of the MHC class I-related neonatal Fc receptor for IgG via intronic binding sequences. J. Immunol. 2007, 179, 2999–3011. [Google Scholar] [CrossRef] [PubMed]
- Qian, S.; Gao, Z.; Cao, R.; Yang, K.; Cui, Y.; Li, S.; Meng, X.; He, Q.; Li, Z. Transmissible Gastroenteritis Virus Infection Up-Regulates FcRn Expression via Nucleocapsid Protein and Secretion of TGF-beta in Porcine Intestinal Epithelial Cells. Front. Microbiol. 2019, 10, 3085. [Google Scholar] [CrossRef]
- Wang, L.; Llorente, C.; Hartmann, P.; Yang, A.M.; Chen, P.; Schnabl, B. Methods to determine intestinal permeability and bacterial translocation during liver disease. J. Immunol. Methods 2015, 421, 44–53. [Google Scholar] [CrossRef]
- Bednar, K.J.; Lee, J.H.; Ort, T. Tregs in Autoimmunity: Insights into Intrinsic Brake Mechanism Driving Pathogenesis and Immune Homeostasis. Front. Immunol. 2022, 13, 932485. [Google Scholar] [CrossRef]
- Wohlfert, E.A.; Grainger, J.R.; Bouladoux, N.; Konkel, J.E.; Oldenhove, G.; Ribeiro, C.H.; Hall, J.A.; Yagi, R.; Naik, S.; Bhairavabhotla, R.; et al. GATA3 controls Foxp3(+) regulatory T cell fate during inflammation in mice. J. Clin. Investig. 2011, 121, 4503–4515. [Google Scholar] [CrossRef]
- Schiering, C.; Krausgruber, T.; Chomka, A.; Frohlich, A.; Adelmann, K.; Wohlfert, E.A.; Pott, J.; Griseri, T.; Bollrath, J.; Hegazy, A.N.; et al. The alarmin IL-33 promotes regulatory T-cell function in the intestine. Nature 2014, 513, 564–568. [Google Scholar] [CrossRef] [PubMed]
- Lochner, M.; Peduto, L.; Cherrier, M.; Sawa, S.; Langa, F.; Varona, R.; Riethmacher, D.; Si-Tahar, M.; Di Santo, J.P.; Eberl, G. In vivo equilibrium of proinflammatory IL-17+ and regulatory IL-10+ Foxp3+ RORgamma t+ T cells. J. Exp. Med. 2008, 205, 1381–1393. [Google Scholar] [CrossRef] [PubMed]
- Ohnmacht, C.; Park, J.H.; Cording, S.; Wing, J.B.; Atarashi, K.; Obata, Y.; Gaboriau-Routhiau, V.; Marques, R.; Dulauroy, S.; Fedoseeva, M.; et al. The microbiota regulates type 2 immunity through RORgammat(+) T cells. Science 2015, 349, 989–993. [Google Scholar] [CrossRef] [PubMed]
- Russler-Germain, E.V.; Rengarajan, S.; Hsieh, C.S. Antigen-specific regulatory T-cell responses to intestinal microbiota. Mucosal. Immunol. 2017, 10, 1375–1386. [Google Scholar] [CrossRef] [PubMed]
- Geuking, M.B.; Cahenzli, J.; Lawson, M.A.; Ng, D.C.; Slack, E.; Hapfelmeier, S.; McCoy, K.D.; Macpherson, A.J. Intestinal bacterial colonization induces mutualistic regulatory T cell responses. Immunity 2011, 34, 794–806. [Google Scholar] [CrossRef] [PubMed]
- Tsuji, M.; Komatsu, N.; Kawamoto, S.; Suzuki, K.; Kanagawa, O.; Honjo, T.; Hori, S.; Fagarasan, S. Preferential generation of follicular B helper T cells from Foxp3+ T cells in gut Peyer’s patches. Science 2009, 323, 1488–1492. [Google Scholar] [CrossRef] [PubMed]
- Rath, T.; Kuo, T.T.; Baker, K.; Qiao, S.W.; Kobayashi, K.; Yoshida, M.; Roopenian, D.; Fiebiger, E.; Lencer, W.I.; Blumberg, R.S. The immunologic functions of the neonatal Fc receptor for IgG. J. Clin. Immunol. 2013, 33, S9–S17. [Google Scholar] [CrossRef] [PubMed]
- van der Waaij, L.A.; Kroese, F.G.; Visser, A.; Nelis, G.F.; Westerveld, B.D.; Jansen, P.L.; Hunter, J.O. Immunoglobulin coating of faecal bacteria in inflammatory bowel disease. Eur. J. Gastroenterol. Hepatol. 2004, 16, 669–674. [Google Scholar] [CrossRef]
- Harmsen, H.J.; Pouwels, S.D.; Funke, A.; Bos, N.A.; Dijkstra, G. Crohn’s disease patients have more IgG-binding fecal bacteria than controls. Clin. Vaccine Immunol. 2012, 19, 515–521. [Google Scholar] [CrossRef]
- Lin, R.; Chen, H.; Shu, W.; Sun, M.; Fang, L.; Shi, Y.; Pang, Z.; Wu, W.; Liu, Z. Clinical significance of soluble immunoglobulins A and G and their coated bacteria in feces of patients with inflammatory bowel disease. J. Transl. Med. 2018, 16, 359. [Google Scholar] [CrossRef]
- Sheehan, D.; Moran, C.; Shanahan, F. The microbiota in inflammatory bowel disease. J. Gastroenterol. 2015, 50, 495–507. [Google Scholar] [CrossRef] [PubMed]
- Nishida, A.; Inoue, R.; Inatomi, O.; Bamba, S.; Naito, Y.; Andoh, A. Gut microbiota in the pathogenesis of inflammatory bowel disease. Clin. J. Gastroenterol. 2018, 11, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Sellon, R.K.; Tonkonogy, S.; Schultz, M.; Dieleman, L.A.; Grenther, W.; Balish, E.; Rennick, D.M.; Sartor, R.B. Resident enteric bacteria are necessary for development of spontaneous colitis and immune system activation in interleukin-10-deficient mice. Infect Immun. 1998, 66, 5224–5231. [Google Scholar] [CrossRef] [PubMed]
- Madsen, K.L.; Doyle, J.S.; Tavernini, M.M.; Jewell, L.D.; Rennie, R.P.; Fedorak, R.N. Antibiotic therapy attenuates colitis in interleukin 10 gene-deficient mice. Gastroenterology 2000, 118, 1094–1105. [Google Scholar] [CrossRef]
- Panwala, C.M.; Jones, J.C.; Viney, J.L. A novel model of inflammatory bowel disease: Mice deficient for the multiple drug resistance gene, mdr1a, spontaneously develop colitis. J. Immunol. 1998, 161, 5733–5744. [Google Scholar] [CrossRef]
- Hans, W.; Scholmerich, J.; Gross, V.; Falk, W. The role of the resident intestinal flora in acute and chronic dextran sulfate sodium-induced colitis in mice. Eur. J. Gastroenterol. Hepatol. 2000, 12, 267–273. [Google Scholar] [CrossRef]
- Chinen, T.; Volchkov, P.Y.; Chervonsky, A.V.; Rudensky, A.Y. A critical role for regulatory T cell-mediated control of inflammation in the absence of commensal microbiota. J. Exp. Med. 2010, 207, 2323–2330. [Google Scholar] [CrossRef]
- Fujino, S.; Andoh, A.; Bamba, S.; Ogawa, A.; Hata, K.; Araki, Y.; Bamba, T.; Fujiyama, Y. Increased expression of interleukin 17 in inflammatory bowel disease. Gut 2003, 52, 65–70. [Google Scholar] [CrossRef]
- Andoh, A.; Zhang, Z.; Inatomi, O.; Fujino, S.; Deguchi, Y.; Araki, Y.; Tsujikawa, T.; Kitoh, K.; Kim-Mitsuyama, S.; Takayanagi, A.; et al. Interleukin-22, a member of the IL-10 subfamily, induces inflammatory responses in colonic subepithelial myofibroblasts. Gastroenterology 2005, 129, 969–984. [Google Scholar] [CrossRef]
- Rovedatti, L.; Kudo, T.; Biancheri, P.; Sarra, M.; Knowles, C.H.; Rampton, D.S.; Corazza, G.R.; Monteleone, G.; Di Sabatino, A.; Macdonald, T.T. Differential regulation of interleukin 17 and interferon gamma production in inflammatory bowel disease. Gut 2009, 58, 1629–1636. [Google Scholar] [CrossRef]
- Ito, R.; Kita, M.; Shin-Ya, M.; Kishida, T.; Urano, A.; Takada, R.; Sakagami, J.; Imanishi, J.; Iwakura, Y.; Okanoue, T.; et al. Involvement of IL-17A in the pathogenesis of DSS-induced colitis in mice. Biochem. Biophys. Res. Commun. 2008, 377, 12–16. [Google Scholar] [CrossRef]
- Zhang, Z.; Zheng, M.; Bindas, J.; Schwarzenberger, P.; Kolls, J.K. Critical role of IL-17 receptor signaling in acute TNBS-induced colitis. Inflamm. Bowel. Dis. 2006, 12, 382–388. [Google Scholar] [CrossRef] [PubMed]
- Ogawa, A.; Andoh, A.; Araki, Y.; Bamba, T.; Fujiyama, Y. Neutralization of interleukin-17 aggravates dextran sulfate sodium-induced colitis in mice. Clin. Immunol. 2004, 110, 55–62. [Google Scholar] [CrossRef] [PubMed]
- Maxwell, J.R.; Zhang, Y.; Brown, W.A.; Smith, C.L.; Byrne, F.R.; Fiorino, M.; Stevens, E.; Bigler, J.; Davis, J.A.; Rottman, J.B.; et al. Differential Roles for Interleukin-23 and Interleukin-17 in Intestinal Immunoregulation. Immunity 2015, 43, 739–750. [Google Scholar] [CrossRef] [PubMed]
- Leppkes, M.; Becker, C.; Ivanov, I.I.; Hirth, S.; Wirtz, S.; Neufert, C.; Pouly, S.; Murphy, A.J.; Valenzuela, D.M.; Yancopoulos, G.D.; et al. RORgamma-expressing Th17 cells induce murine chronic intestinal inflammation via redundant effects of IL-17A and IL-17F. Gastroenterology 2009, 136, 257–267. [Google Scholar] [CrossRef]
- Tai, Y.; Sakamoto, K.; Takano, A.; Haga, K.; Harada, Y. Dysregulation of humoral immunity in Foxp3 conditional-knockout mice. Biochem. Biophys. Res. Commun. 2019, 513, 787–793. [Google Scholar] [CrossRef]
- Bolyen, E.; Rideout, J.R.; Dillon, M.R.; Bokulich, N.A.; Abnet, C.C.; Al-Ghalith, G.A.; Alexander, H.; Alm, E.J.; Arumugam, M.; Asnicar, F.; et al. Reproducible, interactive, scalable and extensible microbiome data science using QIIME 2. Nat. Biotechnol. 2019, 37, 852–857. [Google Scholar] [CrossRef] [PubMed]
- Martin, M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet J. 2011, 17, 10–12. [Google Scholar] [CrossRef]
- Callahan, B.J.; McMurdie, P.J.; Rosen, M.J.; Han, A.W.; Johnson, A.J.; Holmes, S.P. DADA2: High-resolution sample inference from Illumina amplicon data. Nat. Methods 2016, 13, 581–583. [Google Scholar] [CrossRef]
- Camacho, C.; Coulouris, G.; Avagyan, V.; Ma, N.; Papadopoulos, J.; Bealer, K.; Madden, T.L. BLAST+: Architecture and applications. BMC Bioinform. 2009, 10, 421. [Google Scholar] [CrossRef]
- Pruesse, E.; Quast, C.; Knittel, K.; Fuchs, B.M.; Ludwig, W.; Peplies, J.; Glockner, F.O. SILVA: A comprehensive online resource for quality checked and aligned ribosomal RNA sequence data compatible with ARB. Nucleic Acids Res. 2007, 35, 7188–7196. [Google Scholar] [CrossRef] [PubMed]
- Weiss, S.; Xu, Z.Z.; Peddada, S.; Amir, A.; Bittinger, K.; Gonzalez, A.; Lozupone, C.; Zaneveld, J.R.; Vazquez-Baeza, Y.; Birmingham, A.; et al. Normalization and microbial differential abundance strategies depend upon data characteristics. Microbiome 2017, 5, 27. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Koshida, K.; Ito, M.; Yakabe, K.; Takahashi, Y.; Tai, Y.; Akasako, R.; Kimizuka, T.; Takano, S.; Sakamoto, N.; Haniuda, K.; et al. Dysfunction of Foxp3+ Regulatory T Cells Induces Dysbiosis of Gut Microbiota via Aberrant Binding of Immunoglobulins to Microbes in the Intestinal Lumen. Int. J. Mol. Sci. 2023, 24, 8549. https://doi.org/10.3390/ijms24108549
Koshida K, Ito M, Yakabe K, Takahashi Y, Tai Y, Akasako R, Kimizuka T, Takano S, Sakamoto N, Haniuda K, et al. Dysfunction of Foxp3+ Regulatory T Cells Induces Dysbiosis of Gut Microbiota via Aberrant Binding of Immunoglobulins to Microbes in the Intestinal Lumen. International Journal of Molecular Sciences. 2023; 24(10):8549. https://doi.org/10.3390/ijms24108549
Chicago/Turabian StyleKoshida, Kouhei, Mitsuki Ito, Kyosuke Yakabe, Yoshimitsu Takahashi, Yuki Tai, Ryouhei Akasako, Tatsuki Kimizuka, Shunsuke Takano, Natsumi Sakamoto, Kei Haniuda, and et al. 2023. "Dysfunction of Foxp3+ Regulatory T Cells Induces Dysbiosis of Gut Microbiota via Aberrant Binding of Immunoglobulins to Microbes in the Intestinal Lumen" International Journal of Molecular Sciences 24, no. 10: 8549. https://doi.org/10.3390/ijms24108549
APA StyleKoshida, K., Ito, M., Yakabe, K., Takahashi, Y., Tai, Y., Akasako, R., Kimizuka, T., Takano, S., Sakamoto, N., Haniuda, K., Ogawa, S., Kimura, S., Kim, Y.-G., Hase, K., & Harada, Y. (2023). Dysfunction of Foxp3+ Regulatory T Cells Induces Dysbiosis of Gut Microbiota via Aberrant Binding of Immunoglobulins to Microbes in the Intestinal Lumen. International Journal of Molecular Sciences, 24(10), 8549. https://doi.org/10.3390/ijms24108549






