The Influence of Spirodi(Iminohydantoin) on Charge Transfer through ds-DNA Containing 8-OXO-dG: A Theoretical Approach
Abstract
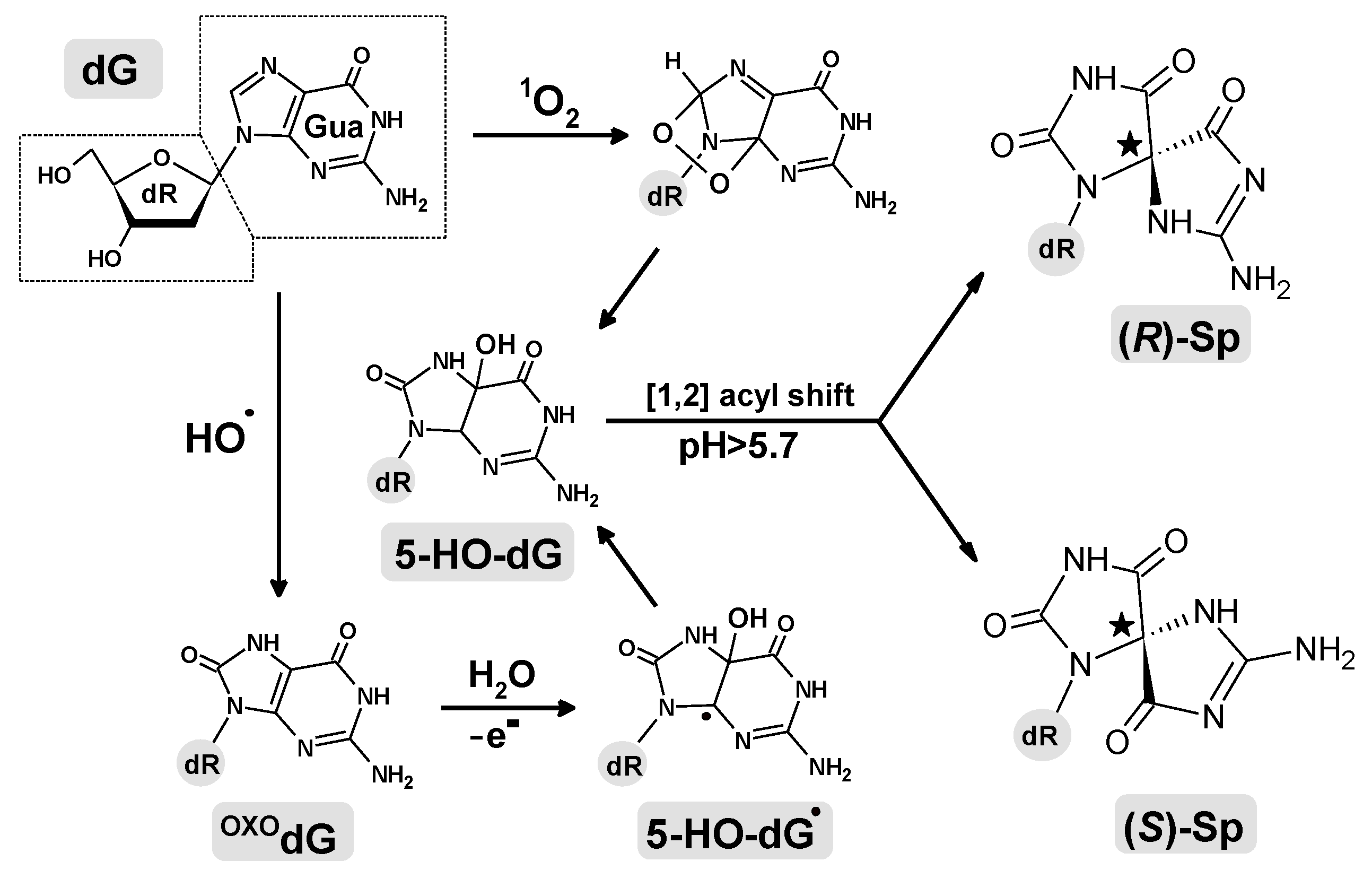
:1. Introduction
2. Results and Discussion
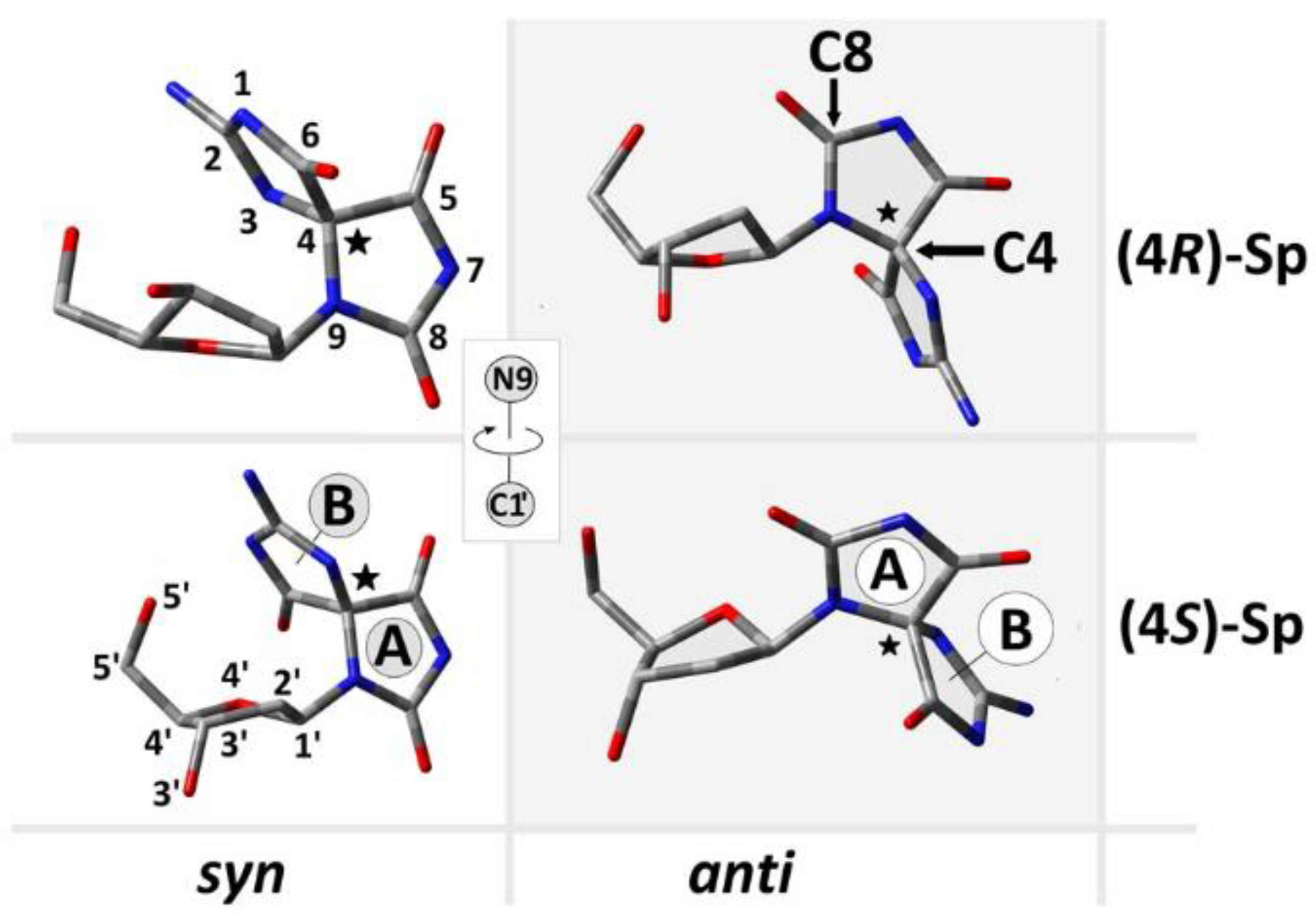
2.1. Analysis of ds-Oligos Spatial Geometries
2.2. The Electronic Properties of ds-Oligonucleotides
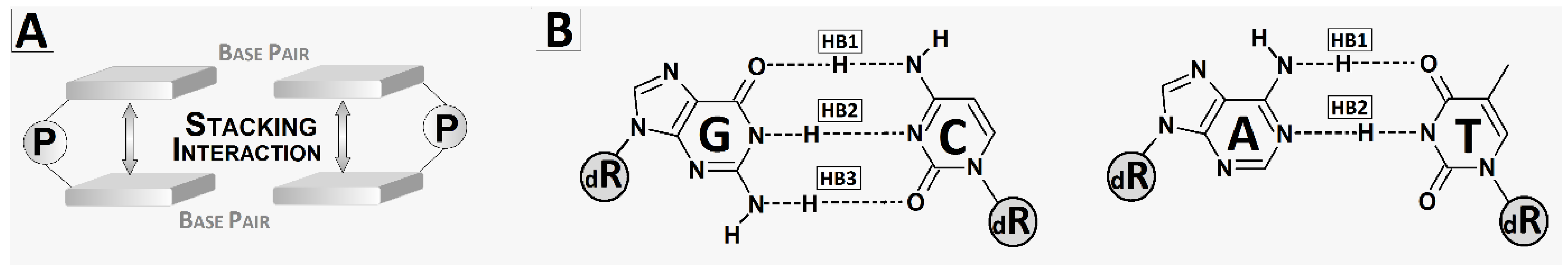
2.3. The Electronic Properties of Isolated Base Pair
2.4. The Charge and Spin Distribution
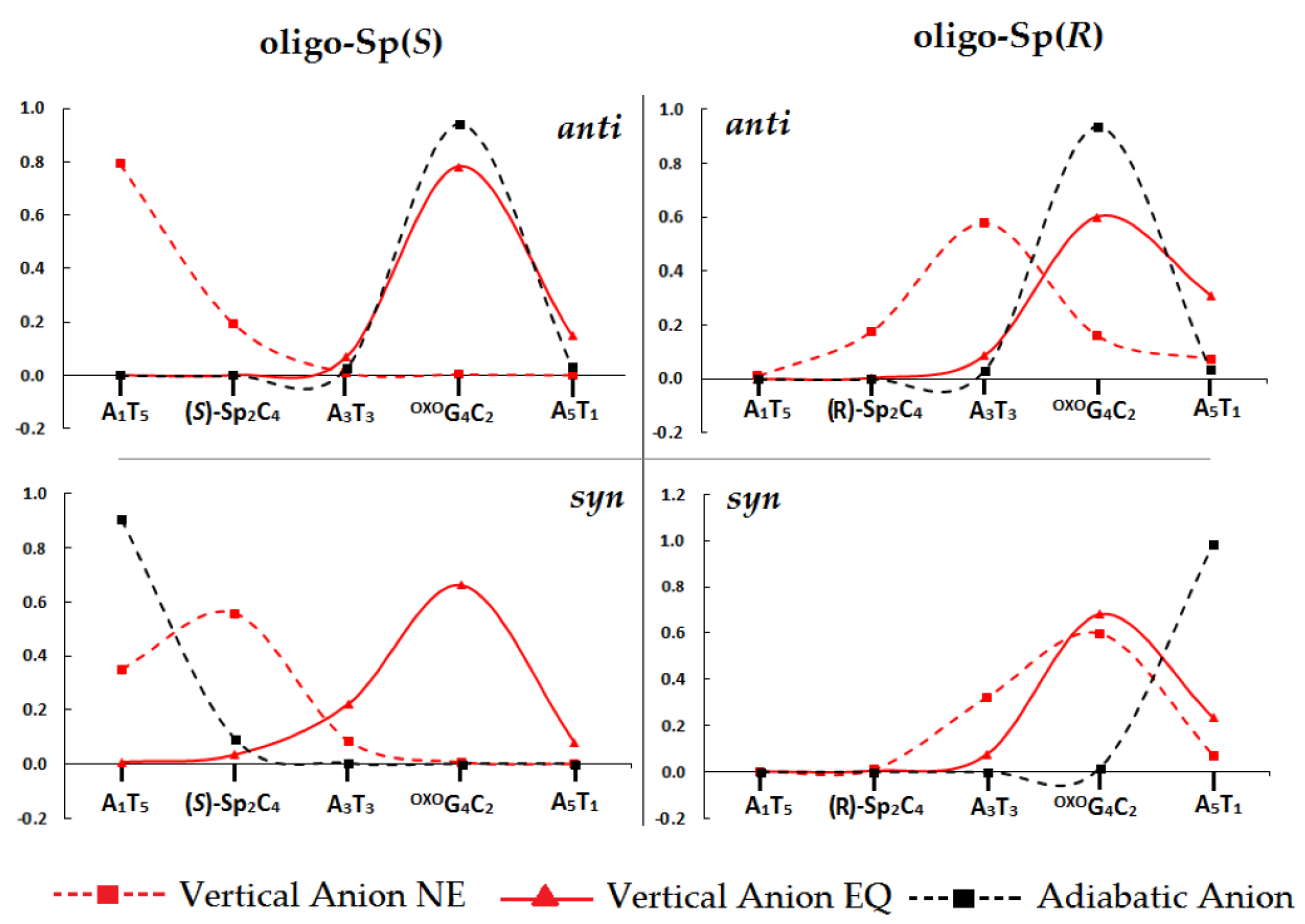
2.5. The DNA Charge Transfer
2.6. Final Remarks and the Outlook for the Future
3. Materials and Methods
3.1. Computation Methodology of ONIOM Studies
3.2. Computation Methodology of DFT Study [32]
4. Conclusions
- A structural analysis revealed the following stabilities of base pairs formed by spirodi(iminohydantoin): syn(4S)-Sp > anti(4S)-Sp > anti(4R)-Sp > syn(4R)-Sp. Additionally, the presence of an anti conformer of (4R)-Sp causes purine strand elongation with subsequent hydrogen bond elimination in the 5’-end adjected base pair.
- The electronic properties of base pairs isolated from ds-DNA show that in all cases, OXOGC adopted the lowest ionization potential, irrespective of the spirodi(iminohydantoin) form. Conversely, a higher adiabatic electron affinity was found for OXOG4C2 of oligo-(R)SpANTI and oligo-(S)SpANTI than for oligo-(R)SpSYN and oligo-(S)SpSYN for A5T1 and A1T5.
- The above strongly concurs with the charge and spin distribution analysis: for all the ds-oligo radical cations, a higher spin density was noted for the OXOGC moiety, irrespective of the Sp form. For the ds-oligo radical anion, the localization of an unpaired electron is strictly dependent on Sp conformation and its diastereomeric form. A higher density was found on OXOGC of oligo-Sp(R)ANTI and oligo-Sp(S)ANTI, while in the case of oligo-Sp(S)SYN and oligo-Sp(R)SYN, a higher density was located on the opposite ends of the double helix, i.e., A1T5 and A5T1 respectively.
- The influence of spirodi(iminohydantoin) on charge transfer was investigated according to Marcus’ theory. In most cases, Sp did not become a significant dam for hole migration, except for anti (4S)-Sp, which affected the transfer towards the base pair adjected to its 3’-end (A3T3). Furthermore, the radical cation transfer towards the OXOG4C2 base pair from proximal base pairs was found at a 107–1012 s−1 level of rate constant. The situation was different in the case of excess electron migration and a significant decrease in the charge transfer rate was noted for A1T5←(4R)-Sp2C4 of oligo-Sp(R)ANTI and OXOG4C2←A5T1oligo-Sp(S)ANTI.
Supplementary Materials
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Brown, T.A. Genomes, 2nd ed.; Wiley-Liss: Oxford, UK, 2002; ISBN 0-471-25046-5. [Google Scholar]
- Dizdaroglu, M. Oxidatively induced DNA damage and its repair in cancer. Mutat. Res.—Rev. Mutat. Res. 2015, 763, 212–245. [Google Scholar] [CrossRef] [PubMed]
- Tubbs, A.; Nussenzweig, A. Endogenous DNA Damage as a Source of Genomic Instability in Cancer. Cell 2017, 168, 644–656. [Google Scholar] [CrossRef] [PubMed]
- Wallace, S.S. Biological consequences of free radical-damaged DNA bases1,2. Free. Radic. Biol. Med. 2002, 33, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Sudhir Ambekar, S. DNA: Damage and Repair Mechanisms in Humans. Glob. J. Pharm. Pharm. Sci. 2017, 3. [Google Scholar] [CrossRef]
- Olinski, R.; Siomek, A.; Rozalski, R.; Gackowski, D.; Foksinski, M.; Guz, J.; Dziaman, T.; Szpila, A.; Tudek, B. Oxidative damage to DNA and antioxidant status in aging and age-related diseases. Acta Biochim. Pol. 2007, 54, 11–26. [Google Scholar] [CrossRef]
- Evans, M.D.; Dizdaroglu, M.; Cooke, M.S. Oxidative DNA Damage and Disease: Induction, Repair and Significance. Mutat. Res. 2004, 567, 1–61. [Google Scholar] [CrossRef]
- Jaruga, P. Mechanisms of free radical-induced damage to DNA. Free Radic. Res. 2012, 46, 382–419. [Google Scholar] [CrossRef]
- Bauer, N.C.; Corbett, A.H.; Doetsch, P.W. The current state of eukaryotic DNA base damage and repair. Nucleic Acids Res. 2015, 43, 10083–10101. [Google Scholar] [CrossRef]
- Bukowska, B.; Karwowski, B.T. Actual state of knowledge in the field of diseases related with defective nucleotide excision repair. Life Sci. 2018, 195, 6–18. [Google Scholar] [CrossRef]
- Steenken, S.; Jovanovic, S.V. How easily oxidizable is DNA? One-electron reduction potentials of adenosine and guanosine radicals in aqueous solution. J. Am. Chem. Soc. 1997, 119, 617–618. [Google Scholar] [CrossRef]
- Tudek, B.; Winczura, A.; Janik, J.; Siomek, A.; Foksinski, M.; Oliński, R. Involvement of oxidatively damaged DNA and repair in cancer development and aging. Am. J. Transl. Res. 2010, 2, 254–284. [Google Scholar]
- Fleming, A.M.; Burrows, C.J. Formation and processing of DNA damage substrates for the hNEIL enzymes. Free Radic. Biol. Med. 2017, 107, 35–52. [Google Scholar] [CrossRef]
- White, B.; Tarun, M.C.; Gathergood, N.; Rusling, J.F.; Smyth, M.R. Oxidisedguanidinohydantoin (Ghox) and spiroiminodihydantoin (Sp) are major products of iron- and copper-mediated 8-oxo-7,8-dihydroguanine and 8-oxo-7,8-dihydro-2′-deoxyguanosine oxidation. Mol. Biosyst. 2005, 1, 373–381. [Google Scholar] [CrossRef]
- Chinyengetere, F.; Jamieson, E.R. Impact of the oxidized guanine lesion spiroiminodihydantoin on the conformation and thermodynamic stability of a 15-mer DNA duplex. Biochemistry 2008, 47, 2584–2591. [Google Scholar] [CrossRef]
- Boal, A.K.; Yavin, E.; Barton, J.K. DNA repair glycosylases with a [4Fe-4S] cluster: A redox cofactor for DNA-mediated charge transport? J. Inorg. Biochem. 2007, 101, 1913–1921. [Google Scholar] [CrossRef]
- White, M.F.; Dillingham, M.S. Iron-sulphur clusters in nucleic acid processing enzymes. Curr. Opin. Struct. Biol. 2012, 22, 94–100. [Google Scholar] [CrossRef]
- Karwowski, B.T. The Influence of Single, Tandem, and Clustered DNA Damage on the Electronic Properties of the Double Helix: A Theoretical Study. Molecules 2020, 25, 3126. [Google Scholar] [CrossRef]
- Lomax, M.E.; Folkes, L.K.; Neill, P.O. Biological Consequences of Radiation-induced DNA Damage: Relevance to Radiotherapy Statement of Search Strategies Used and Sources of Information Why Radiation Damage is More Effective than Endogenous Damage at Killing Cells Ionising Radiation-induced Do. Clin. Oncol. 2013, 25, 578–585. [Google Scholar] [CrossRef]
- Huang, R.; Zhou, P. DNA damage response signaling pathways and targets for radiotherapy sensitization in cancer. Signal Transduct. Target. Ther. 2020, 5, 60. [Google Scholar] [CrossRef]
- Gong, L.; Zhang, Y.; Liu, C.; Zhang, M.; Han, S. Application of radiosensitizers in cancer radiotherapy. Int. J. Nanomed. 2021, 16, 1083–1102. [Google Scholar] [CrossRef]
- Roginskaya, M.; Moore, T.J.; Ampadu-Boateng, D.; Razskazovskiy, Y. Efficacy and site specificity of hydrogen abstraction from DNA 2-deoxyribose by carbonate radicals. Free Radic. Res. 2015, 49, 1431–1437. [Google Scholar] [CrossRef] [PubMed]
- Steenken, S.; Jovanovic, S.V.; Bietti, M.; Bernhard, K. The trap depth (in DNA) of 8-oxo-7,8-dihydro-2′deoxyguanosine as derived from electron-transfer equilibria in aqueous solution. J. Am. Chem. Soc. 2000, 122, 2373–2374. [Google Scholar] [CrossRef]
- Burrows, C.J.; Muller, J.G.; Kornyushyna, O.; Luo, W.; Duarte, V.; Michael, D.; David, S.S. Structure and Potential Mutagenicity of New Hydantoin Products from Guanosine and 8-Oxo-7, 8-Dihydroguanine Oxidation by Transition Metals. Environ. Health Perspect. 2002, 110, 713–717. [Google Scholar] [CrossRef] [PubMed]
- Fleming, A.M.; Muller, J.G.; Ji, I.; Burrows, C.J. Characterization of 2′-deoxyguanosine oxidation products observed in the Fenton-like system Cu(ii)/H2O2/reductant in nucleoside and oligodeoxynucleotide contexts. Org. Biomol. Chem. 2011, 9, 3338–3348. [Google Scholar] [CrossRef] [PubMed]
- Fleming, A.M.; Armentrout, E.I.; Zhu, J.; Muller, J.G.; Burrows, C.J. Spirodi(iminohydantoin) products from oxidation of 2′-deoxyguanosine in the presence of NH4Cl in nucleoside and oligodeoxynucleotide contexts. J. Org. Chem. 2015, 80, 711–721. [Google Scholar] [CrossRef]
- Onyango, A.N. Endogenous generation of singlet oxygen and ozone in human and animal tissues: Mechanisms, biological significance, and influence of dietary components. Oxid. Med. Cell. Longev. 2016, 2016, 2398573. [Google Scholar] [CrossRef]
- Thapa, B.; Munk, B.H.; Burrows, C.J.; Schlegel, H.B. Computational Study of the Radical Mediated Mechanism of the Formation of C8, C5, and C4 Guanine:Lysine Adducts in the Presence of the Benzophenone Photosensitizer. Chem. Res. Toxicol. 2016, 29, 1396–1409. [Google Scholar] [CrossRef]
- Thapa, B.; Munk, B.H.; Burrows, C.J.; Schlegel, H.B. Computational Study of Oxidation of Guanine by Singlet Oxygen (1Δg) and Formation of Guanine:Lysine Cross-Links. Chem.-A Eur. J. 2017, 23, 5804–5813. [Google Scholar] [CrossRef]
- Fleming, A.M.; Burrows, C.J. 8-Oxo-7,8-dihydro-2′-deoxyguanosine and abasic site tandem lesions are oxidation prone yielding hydantoin products that strongly destabilize duplex DNA. Org. Biomol. Chem. 2017, 15, 8341–8353. [Google Scholar] [CrossRef]
- Altona, C.; Sundaralingam, M. Conformational analysis of the sugar ring in nucleosides and nucleotides. Improved method for the interpretation of proton magnetic resonance coupling constants. J. Am. Chem. Soc. 1973, 95, 2333–2344. [Google Scholar] [CrossRef]
- Karwowski, B.T. The AT Interstrand Cross-Link: Structure, Electronic Properties, and Influence on Charge Transfer in dsDNA. Mol. Ther.—Nucleic Acids 2018, 13, 665–685. [Google Scholar] [CrossRef]
- Mignon, P.; Loverix, S.; Steyaert, J.; Geerlings, P. Influence of the π-π interaction on the hydrogen bonding capacity of stacked DNA/RNA bases. Nucleic Acids Res. 2005, 33, 1779–1789. [Google Scholar] [CrossRef]
- Pucci, F.; Rooman, M. Relation between DNA ionization potentials, single base substitutions and pathogenic variants. BMC Genomics 2019, 20, 551. [Google Scholar] [CrossRef]
- Douki, T.; Dimitar, J.R.; Richard, A.J. Effects of Duplex Stability on Charge-Transfer Efficiency within DNA. In Long-Range Charge Transfer in DNA I. Topics in Current Chemistry; Springer: Berlin/Heidelberg, Gernamy, 2004; pp. 1–25. [Google Scholar] [CrossRef]
- Sugiyama, H.; Saito, I. Theoretical studies of GG-specific photocleavage of DNA via electron transfer: Significant lowering of ionization potential and 5′-localization of HOMO of stacked GG bases in B-form DNA. J. Am. Chem. Soc. 1996, 118, 7063–7068. [Google Scholar] [CrossRef]
- Senthilkumar, K.; Grozema, F.C.; Guerra, C.F.; Bickelhaupt, F.M.; Siebbeles, L.D.A. Mapping the Sites for Selective Oxidation of Guanines in DNA. J. Am. Chem. Soc. 2003, 125, 13658–13659. [Google Scholar] [CrossRef]
- Voityuk, A.A.; Jortner, J.; Bixon, M.; Rösch, N. Energetics of hole transfer in DNA. Chem. Phys. Lett. 2000, 324, 430–434. [Google Scholar] [CrossRef]
- Hirshfeld, F.L. Bonded-atom fragments for describing molecular charge densities. Theor. Chim. Acta 1977, 44, 129–138. [Google Scholar] [CrossRef]
- Wagenknecht, H. Electron transfer processes in DNA: Mechanisms, biological relevance and applications in DNA analytics. Nat. Prod. Rep. 2006, 23, 973–1006. [Google Scholar] [CrossRef]
- Boal, A.K.; Yavin, E.; Lukianova, O.A.; O’Shea, V.L.; David, S.S.; Barton, J.K. DNA-bound redox activity of DNA repair glycosylases containing [4Fe-4S] clusters. Biochemistry 2005, 44, 8397–8407. [Google Scholar] [CrossRef]
- Fromme, J.C.; Banerjee, A.; Huang, S.J.; Verdine, G.L. Structural basis for removal of adenine mispaired with 8-oxoguanine by MutY adenine DNA glycosylase. Nat. Mater. 2004, 427, 652–656. [Google Scholar] [CrossRef]
- Jortner, J.; Bixon, M.; Langenbacher, T.; Michel-Beyerle, M.E. Charge transfer and transport in DNA. Proc. Natl. Acad. Sci. USA 1998, 95, 12759–12765. [Google Scholar] [CrossRef] [PubMed]
- Lewis, F.D.; Liu, J.; Weigel, W.; Rettig, W.; Kurnikov, I.V.; Beratan, D.N. Donor-bridge-acceptor energetics determine the distance dependence of electron tunneling in DNA. Proc. Natl. Acad. Sci. USA 2002, 99, 12536–12541. [Google Scholar] [CrossRef] [PubMed]
- Genereux, J.C.; Barton, J.K. Mechanisms for DNA charge transport. Chem. Rev. 2010, 110, 1642–1662. [Google Scholar] [CrossRef] [PubMed]
- Marcus, R.A. Electron Transfer Reactions in Chemistry: Theory and Experiment (Nobel Lecture). Angew. Chemie Int. Ed. English 1993, 32, 1111–1121. [Google Scholar] [CrossRef]
- Rawtani, D.; Kuntmal, B.; Agrawal, Y. Charge transfer in DNA and its diverse modelling approaches. Front. Life Sci. 2016, 9, 214–225. [Google Scholar] [CrossRef]
- Bolton, J.R.; Archer, M.D. Basic electron-transfer theory. In Electron Transfer in Inorganic, Organic and Biological Systems; American Chemical Society: Washington, DC, USA, 1991. [Google Scholar]
- Cave, R.J.; Newton, M.D. Generalization of the Mulliken-Hush treatment for the calculation of electron transfer matrix elements. Chem. Phys. Lett. 1996, 249, 15–19. [Google Scholar] [CrossRef]
- Khan, A. Reorganization energy, activation energy, and mechanism of hole transfer process in DNA: A theoretical study. J. Chem. Phys. 2008, 128, 075101. [Google Scholar] [CrossRef]
- Voityuk, A.A. Are radical cation states delocalized over GG and GGG hole traps in DNA? J. Phys. Chem. B 2005, 109, 10793–10796. [Google Scholar] [CrossRef]
- Voityuk, A.A. Computational modeling of charge transfer in DNA. In Computational Studies of RNA and DNA; Sponer, J., Lanka, F., Eds.; Springer: Dordrecht, The Netherlands, 2006; pp. 485–511. [Google Scholar]
- Dapprich, S.; Komáromi, I.; Byun, K.S.; Morokuma, K.; Frisch, M.J. A new ONIOM implementation in Gaussian98. Part I. The calculation of energies, gradients, vibrational frequencies and electric field derivatives. J. Mol. Struct. THEOCHEM 1999, 461–462, 1–21. [Google Scholar] [CrossRef]
- Gu, J.; Xie, Y.; Schaefer, H.F. Electron attachment to nucleotides in aqueous solution. ChemPhysChem 2006, 7, 1885–1887. [Google Scholar] [CrossRef]
- Gu, J.; Xie, Y.; Schaefer, H.F. Electron attachment to DNA single strands: Gas phase and aqueous solution. Nucleic Acids Res. 2007, 35, 5165–5172. [Google Scholar] [CrossRef]
- Gu, J.; Wang, J.; Leszczynski, J. Electron Attachment-Induced DNA Single Strand Breaks: C3′–O3′ σ-Bond Breaking of Pyrimidine Nucleotides Predominates. J. Am. Chem. Soc. 2006, 128, 9322–9323. [Google Scholar] [CrossRef]
- Hehre, W.J.; Radom, L.; Schleyer, P.R.; Pople, J.; Wiley, J.; Wiberg, K.B.; Clark, T.; York, N. Ab Initio Molecular Orbital Theory by A Handbook of Computational Chem-istry: A Practical Guide to Chemical Structure and Energy Calculations, by. J. Comput. Chem. 1986, 7, 379–383. [Google Scholar]
- Zhao, Y.; Pu, J.; Lynch, B.J.; Truhlar, D.G. Tests of second-generation and third-generation density functionals for thermochemical kineticsElectronic supplementary information (ESI) available: Mean errors for pure and hybrid DFT methods. Phys. Chem. Chem. Phys. 2004, 6, 673. [Google Scholar] [CrossRef]
- Li, T.C.; Tong, P.Q. Time-dependent density-functional theory for multicomponent systems. Phys. Rev. A 1986, 34, 529–532. [Google Scholar] [CrossRef]
- Lange, A.W.; Herbert, J.M. Both intra- And interstrand charge-transfer excited states in aqueous B-DNA are present at energies comparable to, or just above, the 1ππ * excitonic bright states. J. Am. Chem. Soc. 2009, 131, 3913–3922. [Google Scholar] [CrossRef]
- Miertus̃, S.; Tomasi, J. Approximate evaluations of the electrostatic free energy and internal energy changes in solution processes. Chem. Phys. 1982, 65, 239–245. [Google Scholar] [CrossRef]
- Kumar, A.; Adhikary, A.; Sevilla, M.D.; Close, D.M. One-electron oxidation of ds(5′-GGG-3′) and ds(5′-G(8OG)G-3′) and the nature of hole distribution: A density functional theory (DFT) study. Phys. Chem. Chem. Phys. 2020, 22, 5078–5089. [Google Scholar] [CrossRef]
- Karwowski, B.T. The influence of phosphorothioate on charge migration in single and double stranded DNA: A theoretical approach †. Phys. Chem. Chem. Phys. 2015, 17, 21507–21516. [Google Scholar] [CrossRef]
- Frisch, M.J.; Trucks, G.W.; Schlegel, H.B.; Scuseria, G.E.; Robb, M.A.; Cheeseman, J.R.; Scalmani, G.; Barone, V.; Mennucci, B.; Petersson, G.A.; et al. Gaussian 16, Revision C.01; Gaussian Inc.: Wallingford, CT, USA, 2019. [Google Scholar]

| Ds-Oligonucleotide Sequence | Notation |
|---|---|
| d[A1(S)Sp2SYNA3OXOG4A5] * d[T5C4T3C2T1] | oligo-(S)SpSYN |
| d[A1(S)Sp2ANTIA3OXOG4A5] * d[T5C4T3C2T1] | oligo-(S)SpANTI |
| d[A1(R)Sp2SYNA3OXOG4A5] * d[T5C4T3C2T1] | oligo-(R)SpSYN |
| d[A1(R)Sp2ANTIA3OXOG4A5] * d[T5C4T3C2T1] | oligo-(R)SpANTI |
| System | Oligo-Sp(S) | Oligo-Sp(R) | System | Oligo-Sp(S) | Oligo-Sp(R) | ||||
|---|---|---|---|---|---|---|---|---|---|
| Anti | Syn | Anti | Syn | Anti | Syn | Anti | Syn | ||
| BP Dimer | Stacking Energy | Base Pair | Hydrogen Bond Energy | ||||||
| A1T5|Sp2C4 | 15.32 | 10.30 | 17.77 | 9.89 | A1T5 | 10.07 | 9.16 | 3.36 | 10.62 |
| Sp2C4|A3T3 | 13.68 | 17.19 | 10.95 | 14.97 | Sp2C4 | 2.44 | 10.95 | 3.17 | 3.25 |
| A3T3|OG4C2 | 16.55 | 15.81 | 17.21 | 17.75 | A3T3 | 9.85 | 10.88 | 10.77 | 9.22 |
| OG4C2|A5T1 | 16.25 | 15.82 | 15.22 | 15.46 | OXOG4C2 | 18.13 | 18.08 | 17.79 | 17.97 |
| A5T1 | 10.60 | 10.63 | 10.63 | 10.66 | |||||
 | |||||||||
| Conformer | VIPNE | VIPEQ | AIP | VEANE | VEAEQ | AEA |
|---|---|---|---|---|---|---|
| oligo—Sp(S) | ||||||
| anti | 6.35 | 5.88 | 5.37 | −0.68 | −1.44 | −1.97 |
| syn | 6.66 | 5.90 | 5.48 | −0.67 | −1.34 | −1.34 |
| oligo—Sp(R) | ||||||
| anti | 6.39 | 5.81 | 5.38 | −0.62 | −1.37 | −1.94 |
| syn | 6.64 | 5.88 | 5.43 | −0.69 | −1.38 | −1.87 |
| Electronic Properties In [eV] | ||||||||
|---|---|---|---|---|---|---|---|---|
| oligo-Sp(R) | oligo-Sp(S) | |||||||
| Base Pair | anti | syn | anti | syn | ||||
| VIP | AIP | VIP | AIP | VIP | AIP | VIP | AIP | |
| A1T5 | 6.76 | 6.74 | 6.75 | 6.76 | 6.55 | 6.52 | 6.62 | 6.65 |
| Sp2C4 | 7.66 | 7.64 | 6.93 | 6.91 | 6.91 | 6.90 | 7.69 | 7.72 |
| A3T3 | 6.62 | 6.61 | 6.84 | 6.56 | 6.80 | 6.50 | 6.64 | 6.61 |
| OXOG4C2 | 5.93 | 5.53 | 5.91 | 5.54 | 5.91 | 5.56 | 5.90 | 5.56 |
| A5T1 | 6.73 | 6.68 | 6.71 | 6.67 | 6.72 | 6.68 | 6.72 | 6.67 |
| oligo-Sp(R) | oligo-Sp(S) | |||||||
| Base Pair | anti | syn | anti | syn | ||||
| VEA | AEA | VEA | AEA | VEA | AEA | VEA | AEA | |
| A1T5 | −1.51 | −1.51 | −1.40 | −1.39 | −1.57 | −1.60 | −1.42 | −1.85 |
| Sp2C4 | −1.32 | −1.33 | −1.33 | −1.32 | −1.29 | −1.34 | −1.14 | −1.49 |
| A3T3 | −1.44 | −1.34 | −1.43 | −1.42 | −1.41 | −1.42 | −1.30 | −1.44 |
| OXOG4C2 | −1.52 | −1.97 | −1.53 | −1.53 | −1.53 | −1.96 | −1.50 | −1.48 |
| A5T1 | −1.43 | −1.41 | −1.43 | −1.83 | −1.42 | −1.38 | −1.44 | −1.44 |
| Excess-Electron Transfer | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| System | λ | ΔG | Ea | V12 | kHT | System | λ | ΔG | Ea | V12 | kHT |
| X:syn (S)-Sp | oligo-Sp(S)SYN | X:syn (R)-Sp | oligo-Sp(R)SYN | ||||||||
| A1T5←X2C4 | 0.51 | −0.36 | 0.01 | 0.11 | 1.9 × 1014 | A1T5←X2C4 | 0.03 | −0.07 | 0.01 | 0.01 | 6.6 × 1012 |
| X2C4←A3T3 | 0.34 | −0.05 | 0.06 | 0.07 | 3.4 × 1013 | X2C4→A3T3 | 0.03 | −0.09 | 0.04 | 0.02 | 6.6 × 1012 |
| A3T3→OXOG4C2 | −0.02 | −0.04 | −0.04 | 0.01 | N.D | A3T3→OXOG4C2 | 0.01 | −0.11 | 0.36 | 0.03 | 1.2 × 108 |
| OXOG4C2←A5T1 | −0.02 | −0.04 | −0.05 | 0.05 | N.D | OXOG4C2→A5T1 | 0.47 | −0.30 | 0.01 | 0.05 | 4.2 × 1013 |
| A1T5←A3T3 | 0.43 | −0.41 | 0.00 | 0.07 | 1.3 × 1014 | A1T5→A3T3 | 0.01 | −0.03 | 0.03 | 0.06 | 2.2 × 1014 |
| X2C4←OXOG4C2 | 0.30 | −0.01 | 0.07 | 0.06 | 4.0 × 1012 | X2C4→OXOG4C2 | 0.04 | −0.21 | 0.18 | 0.06 | 3.3 × 1011 |
| A3T3←A5T1 | 0.14 | −0.00 | 0.03 | 0.08 | 8.0 × 1013 | A3T3→A5T1 | 0.41 | −0.42 | 0.00 | 0.08 | 1.7 × 1014 |
| X:anti (S)-Sp | oligo-Sp(S)ANTI | X:anti (R)-Sp | oligo-Sp(R)ANTI | ||||||||
| A1T5←X2C4 | 0.03 | −0.27 | 0.41 | 0.05 | 2.9 × 107 | A1T5←X2C4 | 0.01 | −0.17 | 1.35 | 0.03 | 2.3 × 10−9 |
| X2C4→A3T3 | 0.00 | −0.08 | 0.33 | 0.04 | 7.0 × 1010 | X2C4→A3T3 | −0.10 | −0.01 | −0.03 | 0.05 | N.D |
| A3T3→OXOG4C2 | 0.44 | −0.55 | 0.01 | 0.06 | 6.2 × 1013 | A3T3→OXOG4C2 | 0.52 | −0.63 | 0.01 | 0.02 | 6.3 × 1012 |
| OXOG4C2←A5T1 | 0.03 | −0.58 | 2.75 | 0.05 | 8 × 10−33 | OXOG4C2←A5T1 | 0.47 | −0.55 | 0.00 | 0.05 | 6.1 × 1013 |
| A1T5←A3T3 | 0.02 | −0.19 | 0.30 | 0.06 | 4.4 × 109 | A1T5←A3T3 | −0.08 | −0.17 | −0.20 | 0.08 | N.D |
| X2C4→OXOG4C2 | 0.43 | −0.62 | 0.02 | 0.07 | 5.5 × 1013 | X2C4→OXOG4C2 | 0.45 | −0.64 | 0.02 | 0.05 | 3.1 × 1013 |
| A3T3←A5T1 | −0.02 | −0.03 | −0.04 | 0.08 | N.D | A3T3→A5T1 | 0.06 | −0.08 | 0.00 | 0.08 | 4.1 × 1014 |
| Hole-Electron Transfer | |||||||||||
| System | λ | ΔG | Ea | V12 | kHT | System | λ | ΔG | Ea | V12 | kHT |
| X: syn (S)-Sp | oligo-Sp(S)SYN | X: syn (R)-Sp | oligo-Sp(R)SYN | ||||||||
| A1T5←X2C4 | −0.02 | −1.06 | −19.3 | 0.28 | N.D | A1T5←X2C4 | −0.04 | −0.15 | −0.24 | 0.14 | N.D |
| X2C4→A3T3 | 0.04 | −1.10 | 6.78 | 0.13 | 3.5 × 10−100 | X2C4→A3T3 | 0.26 | −0.35 | 0.01 | 0.04 | 3.6 × 1013 |
| A3T3→OXOG4C2 | 0.33 | −1.05 | 0.40 | 0.38 | 7.3 × 108 | A3T3→OXOG4C2 | 0.29 | −1.02 | 0.47 | 0.48 | 8.2 × 107 |
| OXOG4C2←A5T1 | 0.34 | −1.11 | 0.44 | 0.40 | 1.7 × 1012 | OXOG4C2←A5T1 | 0.37 | −1.13 | 0.38 | 0.40 | 1.7 × 109 |
| A1T5→A3T3 | 0.01 | −0.04 | 0.02 | 0.07 | 4.4 × 1014 | A1T5→A3T3 | 0.29 | −0.20 | 0.01 | 0.05 | 5.9 × 1013 |
| X2C4→OXOG4C2 | 0.36 | −2.16 | 2.26 | 0.61 | 7.4 × 10−23 | X2C4→OXOG4C2 | 0.34 | −1.38 | 0.77 | 0.56 | 8.7 × 102 |
| A3T3←A5T1 | 0.03 | −0.06 | 0.01 | 0.04 | 1.1 × 1014 | A3T3←A5T1 | 0.28 | −0.11 | 0.03 | 0.07 | 5.4 × 1013 |
| X: anti (S)-Sp | oligo-Sp(S)ANTI | X: anti (R)-Sp | oligo-Sp(R)ANTI | ||||||||
| A1T5←X2C4 | 0.15 | −0.39 | 0.10 | 0.09 | 7.2 × 1012 | A1T5←X2C4 | −0.01 | −0.89 | −15.9 | 0.14 | N.D |
| X2C4→A3T3 | 0.42 | −0.40 | 0.00 | 0.08 | 1.7 × 1014 | X2C4→A3T3 | −0.02 | −1.02 | −11.7 | 0.22 | N.D |
| A3T3→OXOG4C2 | 0.34 | −0.94 | 0.26 | 0.46 | 2.5 × 1011 | A3T3→OXOG4C2 | 0.41 | −1.09 | 0.29 | 0.42 | 5.8 × 1010 |
| OXOG4C2←A5T1 | 0.38 | −1.12 | 0.36 | 0.41 | 3.8 × 109 | OXOG4C2←A5T1 | 0.41 | −1.15 | 0.34 | 0.40 | 7.5 × 109 |
| A1T5→A3T3 | 0.32 | −0.02 | 0.07 | 0.13 | 3.0 × 1013 | A1T5→A3T3 | 0.00 | −0.13 | −1.08 | 0.05 | 2.1 × 1033 |
| X2C4→OXOG4C2 | 0.47 | −1.34 | 0.41 | 0.54 | 8.1 × 108 | X2C4→OXOG4C2 | 0.38 | −2.11 | 1.95 | 0.10 | 3.6 × 10−19 |
| A3T3←A5T1 | 0.28 | −0.18 | 0.01 | 0.04 | 5.6 × 1013 | A3T3←A5T1 | 0.00 | −0.07 | 1.04 | 0.07 | 2.2 × 10−2 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the author. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Karwowski, B.T. The Influence of Spirodi(Iminohydantoin) on Charge Transfer through ds-DNA Containing 8-OXO-dG: A Theoretical Approach. Int. J. Mol. Sci. 2023, 24, 8570. https://doi.org/10.3390/ijms24108570
Karwowski BT. The Influence of Spirodi(Iminohydantoin) on Charge Transfer through ds-DNA Containing 8-OXO-dG: A Theoretical Approach. International Journal of Molecular Sciences. 2023; 24(10):8570. https://doi.org/10.3390/ijms24108570
Chicago/Turabian StyleKarwowski, Boleslaw T. 2023. "The Influence of Spirodi(Iminohydantoin) on Charge Transfer through ds-DNA Containing 8-OXO-dG: A Theoretical Approach" International Journal of Molecular Sciences 24, no. 10: 8570. https://doi.org/10.3390/ijms24108570
APA StyleKarwowski, B. T. (2023). The Influence of Spirodi(Iminohydantoin) on Charge Transfer through ds-DNA Containing 8-OXO-dG: A Theoretical Approach. International Journal of Molecular Sciences, 24(10), 8570. https://doi.org/10.3390/ijms24108570






