BcMYB111 Responds to BcCBF2 and Induces Flavonol Biosynthesis to Enhance Tolerance under Cold Stress in Non-Heading Chinese Cabbage
Abstract
1. Introduction
2. Results
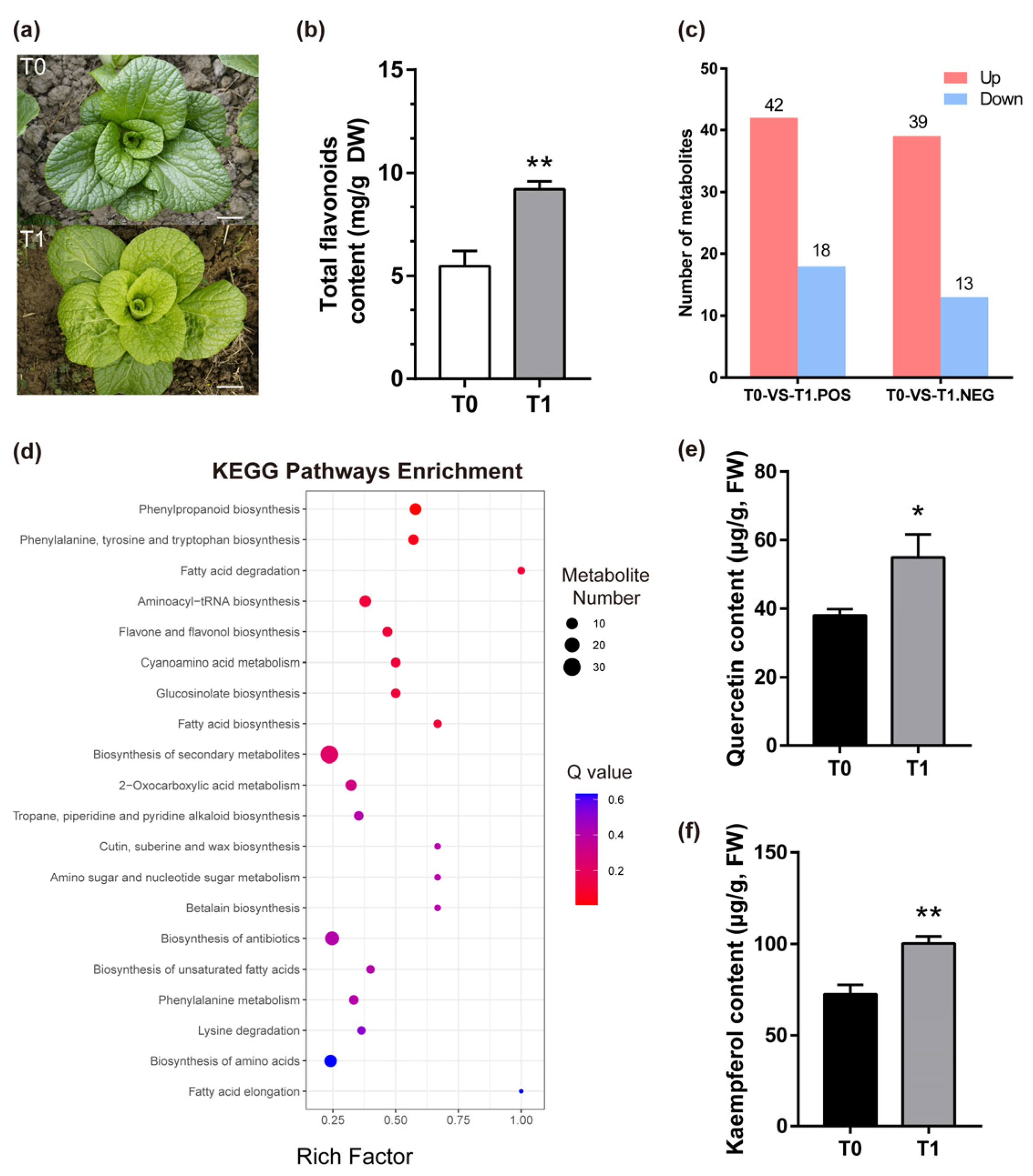
2.1. Metabolomic Analysis of NHCC in Response to Cold Stress
2.2. Characterization and Expression Profile of BcMYB111
2.3. Cold Stress Induces the Up-Regulated Expression of BcMYB111 and the Accumulation of Flavonols
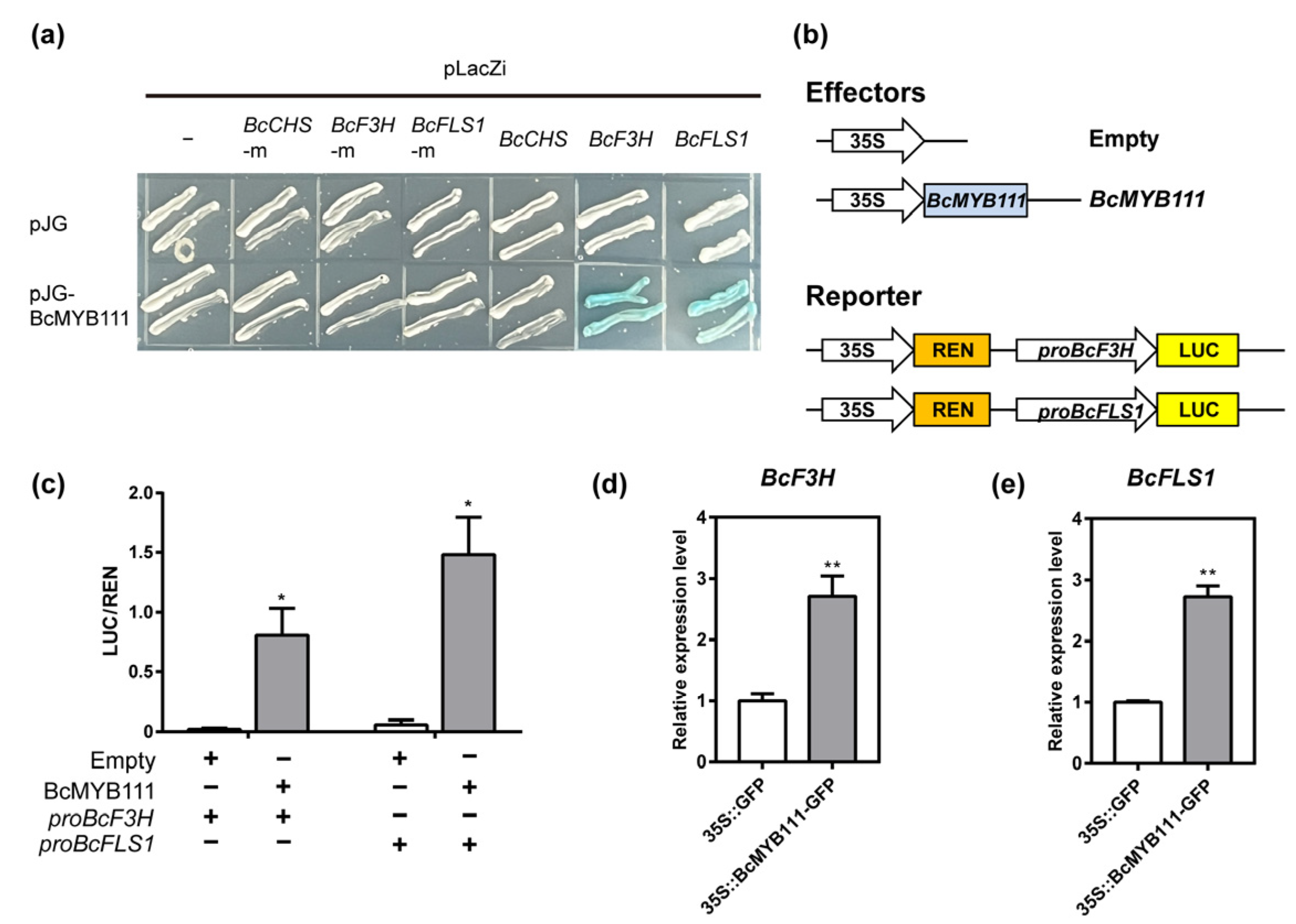
2.4. BcMYB111 Is a Transcriptional Activator of BcF3H and BcFLS1
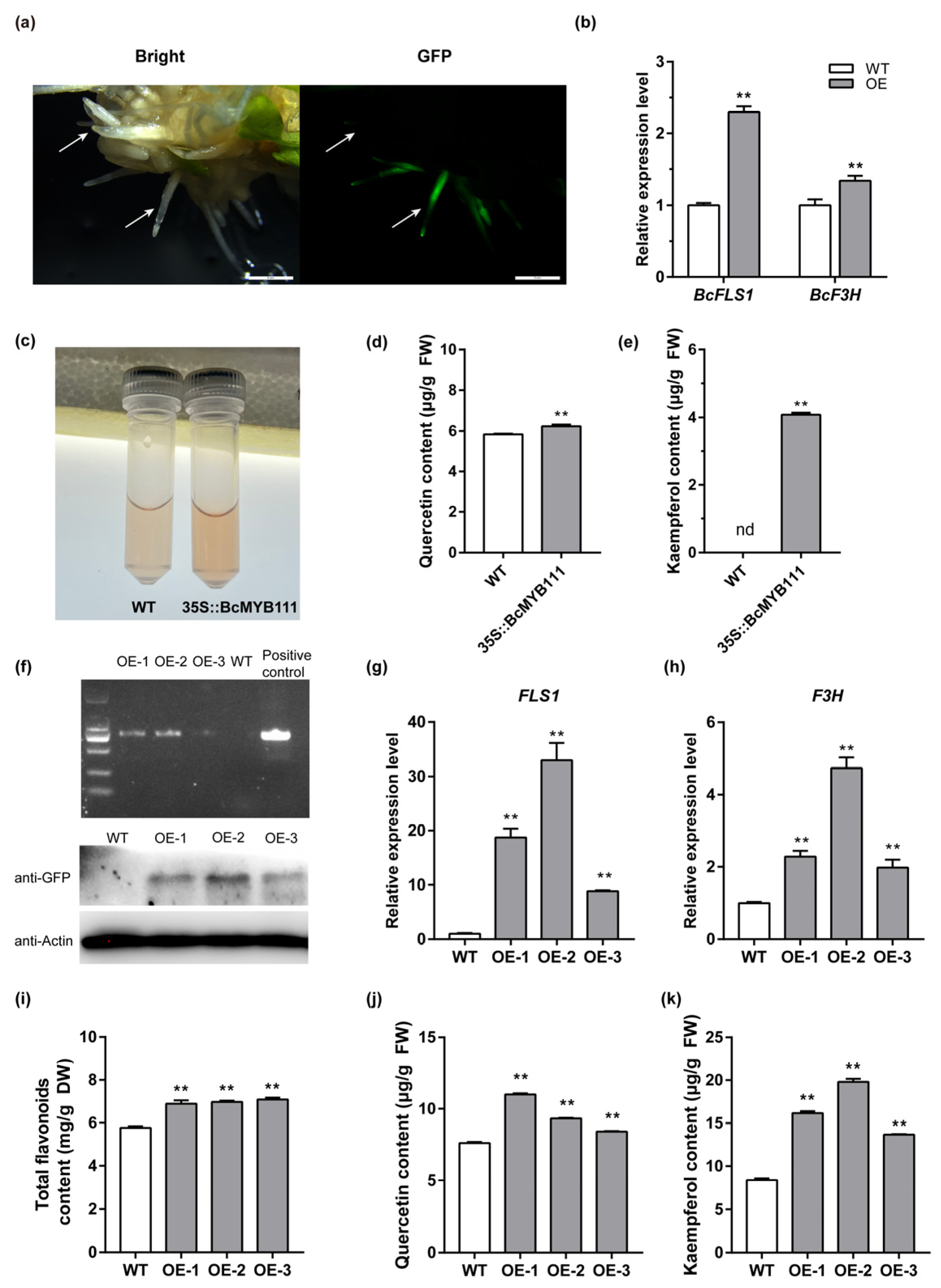
2.5. BcMYB111 Promotes the Accumulation of Flavonols
2.6. The Silencing of BcMYB111 Decreases the Synthesis of Flavonols
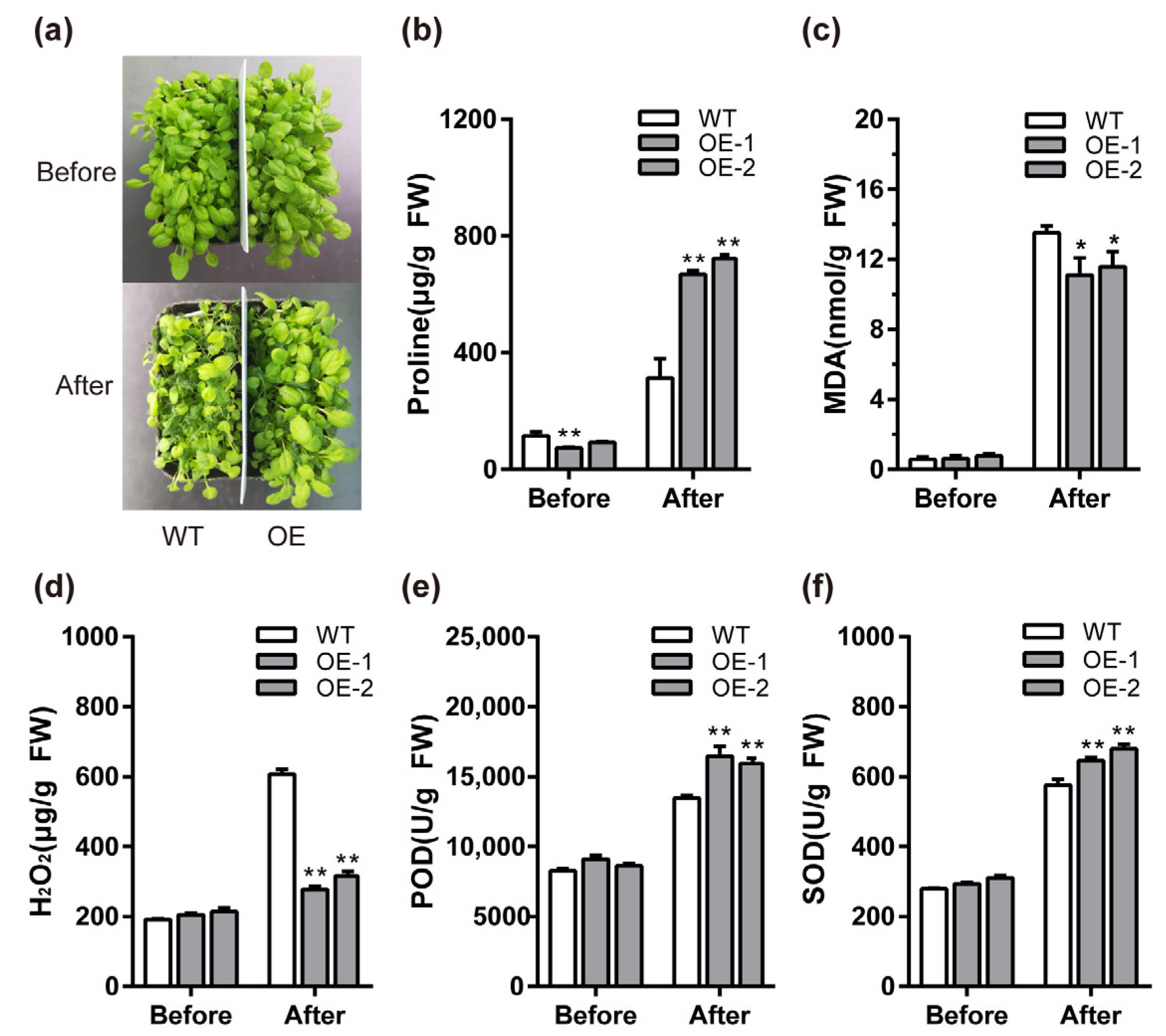
2.7. BcMYB111 Improves Cold Tolerance
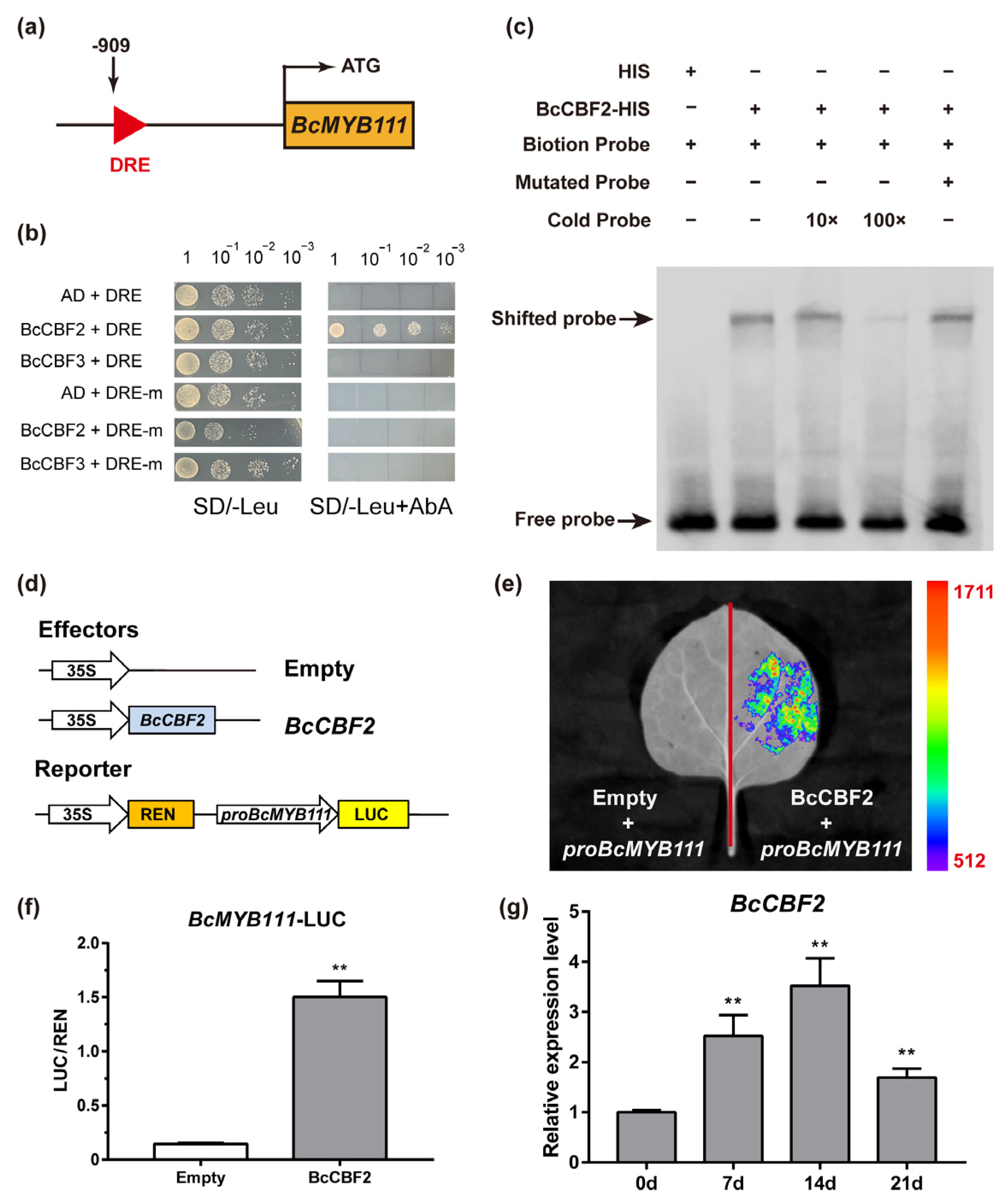
2.8. BcMYB111 Is a Direct Target Gene of BcCBF2
3. Discussion
4. Materials and Methods
4.1. Plant Materials, Growth Conditions, and Treatments
4.2. Untargeted Metabolomic Analysis
4.3. Subcellular Localization and the Assay of Transient Expression in the Cotyledon of NHCC
4.4. Quantitative Real-Time PCR Analysis
4.5. Measurement of the Total Flavonoid, Quercetin, and Kaempferol Contents
4.6. Dual Luciferase Assay
4.7. Yeast One-Hybrid Assay
4.8. Electrophoretic Mobility Shift Assay (EMSA)
4.9. Silencing of BcMYB111 in NHCC by Virus-Induced Gene Silencing (VIGS)
4.10. Generation of Transgenic Arabidopsis thaliana
4.11. Measurement of MDA, H2O2, and Enzyme Activity of POD and SOD
4.12. Agrobacterium Rhizogenes-Mediated Transformation of Hairy Roots
4.13. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Buer, C.S.; Imin, N.; Djordjevic, M.A. Flavonoids: New roles for old molecules. J. Integr. Plant Biol. 2010, 52, 98–111. [Google Scholar] [CrossRef] [PubMed]
- Tan, H.; Man, C.; Xie, Y.; Yan, J.; Chu, J.; Huang, J. A Crucial Role of GA-Regulated Flavonol Biosynthesis in Root Growth of Arabidopsis. Mol. Plant 2019, 12, 17. [Google Scholar] [CrossRef]
- Samanta, A.; Das, G.; Das, S.K. Roles of flavonoids in plants. Carbon 2011, 100, 12–35. [Google Scholar]
- Brunetti, C. Flavonoids as Antioxidants in Plants Under Abiotic Stresses; Springer: New York, NY, USA, 2012; pp. 159–179. [Google Scholar]
- Dixon, R.A.; Paiva, N.L. Stress-induced phenylpropanoid metabolism. Plant Cell 1995, 7, 1085. [Google Scholar] [CrossRef]
- Pourcel, L.; Routaboul, J.M.; Cheynier, V.; Lepiniec, L.; Debeaujon, I. Flavonoid oxidation in plants: From biochemical properties to ph ysiological functions. Trends Plant Sci. 2007, 12, 29–36. [Google Scholar] [CrossRef] [PubMed]
- Nakabayashi, R.; Yonekura-Sakakibara, K.; Urano, K.; Suzuki, M.; Yamada, Y.; Nishizawa, T.; Matsuda, F.; Kojima, M.; Sakakibara, H.; Shinozaki, K.; et al. Enhancement of oxidative and drought tolerance in Arabidopsis by overaccumulation of antioxidant flavonoids. Plant J. 2014, 77, 367–379. [Google Scholar] [CrossRef]
- Bilger, W.; Rolland, M.; Nybakken, L. UV screening in higher plants induced by low temperature in the absence of UV-B radiation. Photochem. Photobiol. Sci. 2007, 6, 190–195. [Google Scholar] [CrossRef] [PubMed]
- Korn, M.; Peterek, S.; Mock, H.P.; Heyer, A.G.; Hincha, D.K. Heterosis in the freezing tolerance, and sugar and flavonoid contents of crosses between Arabidopsis thaliana accessions of widely varying freezing tolerance. Plant Cell Environ. 2008, 31, 813–827. [Google Scholar] [CrossRef]
- Mi, A.; Ar, B.; Ai, C.; Mn, D.; Mas, E.; Iak, F.; Ai, G.; Ieo, H.; Mr, I.; Ma, J. Corrigendum to Luteolin, a flavonoid, as an anticancer agent: A review. Biomed. Pharmacother. 2019, 116, 108621. [Google Scholar]
- Selvakumar, P.; Badgeley, A.; Murphy, P.; Anwar, H.; Sharma, U.; Lawrence, K.; Lakshmikuttyamma, A. Flavonoids and Other Polyphenols Act as Epigenetic Modifiers in Breast Cancer. Nutrients 2020, 12, 761. [Google Scholar] [CrossRef]
- Corradini, E.; Foglia, P.; Giansanti, P.; Gubbiotti, R.; Samperi, R.; Lagana, A. Flavonoids: Chemical properties and analytical methodologies of identification and quantitation in foods and plants. Nat. Prod. Res. 2011, 25, 469–495. [Google Scholar] [CrossRef]
- Liu, W.; Feng, Y.; Yu, S.; Fan, Z.; Li, X.; Li, J.; Yin, H. The Flavonoid Biosynthesis Network in Plants. Int. J. Mol. Sci. 2021, 22, 12824. [Google Scholar] [CrossRef]
- Yonekura-Sakakibara, K.; Higashi, Y.; Nakabayashi, R. The Origin and Evolution of Plant Flavonoid Metabolism. Front. Plant Sci. 2019, 10, 943. [Google Scholar] [CrossRef]
- Li, H.; Li, Y.; Yu, J.; Wu, T.; Zhang, J.; Tian, J.; Yao, Y. MdMYB8 is associated with flavonol biosynthesis via the activation of the MdFLS promoter in the fruits of Malus crabapple. Hortic. Res. 2020, 7, 19. [Google Scholar] [CrossRef]
- Yan, H.; Pei, X.; Zhang, H.; Li, X.; Zhao, X. MYB-Mediated Regulation of Anthocyanin Biosynthesis. Int. J. Mol. Sci. 2021, 22, 3103. [Google Scholar] [CrossRef] [PubMed]
- Cao, Y.; Jia, H.; Xing, M.; Jin, R.; Grierson, D.; Gao, Z.; Sun, C.; Chen, K.; Xu, C.; Li, X. Genome-Wide Analysis of MYB Gene Family in Chinese Bayberry (Morella rubra) and Identification of Members Regulating Flavonoid Biosynthesis. Front. Plant Sci. 2021, 12, 691384. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Luan, Q.; Han, J.; Zhang, C.; Liu, M.; Ren, Z. CsMYB60 directly and indirectly activates structural genes to promote the biosynthesis of flavonols and proanthocyanidins in cucumber. Hortic. Res. 2020, 7, 103. [Google Scholar] [CrossRef]
- Huang, W.; Khaldun, A.B.; Chen, J.; Zhang, C.; Lv, H.; Yuan, L.; Wang, Y. A R2R3-MYB Transcription Factor Regulates the Flavonol Biosynthetic Pathway in a Traditional Chinese Medicinal Plant, Epimedium sagittatum. Front. Plant Sci. 2016, 7, 1089. [Google Scholar] [CrossRef] [PubMed]
- Wang, N.; Xu, H.; Jiang, S.; Zhang, Z.; Lu, N.; Qiu, H.; Qu, C.; Wang, Y.; Wu, S.; Chen, X. MYB12 and MYB22 play essential roles in proanthocyanidin and flavonol synthesis in red-fleshed apple (Malus sieversii f. niedzwetzkyana). Plant J. 2017, 90, 276–292. [Google Scholar] [CrossRef]
- Zhang, T.; Xu, H.; Yang, G.; Wang, N.; Zhang, J.; Wang, Y.; Jiang, S.; Fang, H.; Zhang, Z.; Chen, X. Molecular mechanism of MYB111 and WRKY40 involved in anthocyanin biosynthesis in red-fleshed apple callus. Plant Cell Tissue Organ Cult. (PCTOC) 2019, 139, 467–478. [Google Scholar] [CrossRef]
- Fornalé, S.; Lopez, E.; Salazar-Henao, J.E.; Fernández-Nohales, P.; Rigau, J.; Caparros-Ruiz, D. AtMYB7, a new player in the regulation of UV-sunscreens in Arabidopsis thaliana. Plant Cell Physiol. 2014, 55, 507–516. [Google Scholar] [CrossRef]
- Stracke, R.; Ishihara, H.; Huep, G.; Barsch, A.; Mehrtens, F.; Niehaus, K.; Weisshaar, B. Differential regulation of closely related R2R3-MYB transcription factors controls flavonol accumulation in different parts of the Arabidopsis thaliana seedling. Plant J. 2007, 50, 660–677. [Google Scholar] [CrossRef]
- Stracke, R.; Jahns, O.; Keck, M.; Tohge, T.; Niehaus, K.; Fernie, A.R.; Weisshaar, B. Analysis of PRODUCTION OF FLAVONOL GLYCOSIDES-dependent flavonol glycoside accumulation in Arabidopsis thaliana plants reveals MYB11-, MYB12- and MYB111-independent flavonol glycoside accumulation. New Phytol. 2010, 188, 985–1000. [Google Scholar] [CrossRef] [PubMed]
- Zhang, P.; Qian, D.; Luo, C.; Niu, Y.; Li, T.; Li, C.; Xiang, Y.; Wang, X.; Niu, Y. Arabidopsis ADF5 acts as a downstream target gene of CBFs in response to low-temperature stress. Front. Cell Dev. Biol. 2021, 9, 635533. [Google Scholar] [CrossRef]
- Fürtauer, L.; Weiszmann, J.; Weckwerth, W.; Nägele, T. Dynamics of Plant Metabolism during Cold Acclimation. Int. J. Mol. Sci. 2019, 20, 5411. [Google Scholar] [CrossRef]
- Schulz, E.; Tohge, T.; Zuther, E.; Fernie, A.R.; Hincha, D.K. Flavonoids are determinants of freezing tolerance and cold acclimation in Arabidopsis thaliana. Sci. Rep. 2016, 6, 34027. [Google Scholar] [CrossRef]
- Fang, J.G.; Cheng, W.M.; Wang, X.C.; Jia, H.F. Functional conservation analysis and expression modes of grape anthocyanin synthesis genes responsive to low temperature stress. Gene Int. J. Focus. Gene Cloning Gene Struct. Funct. 2015, 574, 168–177. [Google Scholar]
- Petridis, A.; Döll, S.; Nichelmann, L.; Bilger, W.; Mock, H.P. Arabidopsis thaliana G2-Like Flavonoid Regulator and Brassinosteroid Enhanced Expression1 are low-temperature regulators of flavonoid accumulation. New Phytol. 2016, 211, 912–925. [Google Scholar] [CrossRef] [PubMed]
- Guo, J.; Zhou, X.; Wang, T.; Wang, G.; Cao, F. Regulation of flavonoid metabolism in ginkgo leaves in response to different day-night temperature combinations. Plant Physiol. Biochem. 2020, 147, 133–140. [Google Scholar] [CrossRef]
- Schmidt, S.; Zietz, M.; Schreiner, M.; Rohn, S.; Kroh, L.W.; Krumbein, A. Genotypic and climatic influences on the concentration and composition of flavonoids in kale (Brassica oleracea var. sabellica). Food Chem. 2010, 119, 1293–1299. [Google Scholar] [CrossRef]
- Schulz, E.; Tohge, T.; Zuther, E.; Fernie, A.R.; Hincha, D.K. Natural variation in flavonol and anthocyanin metabolism during cold acclimation in Arabidopsis thaliana accessions. Plant Cell Environ. 2015, 38, 1658–1672. [Google Scholar] [CrossRef] [PubMed]
- Hao, S.; Lu, Y.; Peng, Z.; Wang, E.; Chao, L.; Zhong, S.; Yao, Y. McMYB4 improves temperature adaptation by regulating phenylpropanoid metabolism and hormone signaling in apple. Hortic. Res. 2021, 8, 182. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Liu, G.F.; Ma, L.M.; Liu, T.K.; Zhang, C.W.; Xiao, D.; Zheng, H.K.; Chen, F.; Hou, X.L. A chromosome-level reference genome of non-heading Chinese cabbage [Brassica campestris (syn. Brassica rapa) ssp. chinensis]. Hortic. Res. 2020, 7, 212. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Huang, F.; Tao, Y.; Zhou, Y.; Bai, A.; Yu, Z.; Xiao, D.; Zhang, C.; Liu, T.; Hou, X. BcGR1. 1, a Cytoplasmic Localized Glutathione Reductase, Enhanced Tolerance to Copper Stress in Arabidopsis thaliana. Antioxidants 2022, 11, 389. [Google Scholar] [CrossRef]
- Zhang, Y.; Chen, G.; Dong, T.; Pan, Y.; Zhao, Z.; Tian, S.; Hu, Z. Anthocyanin accumulation and transcriptional regulation of anthocyanin biosynthesis in purple bok choy (Brassica rapa var. chinensis). J. Agric. Food Chem. 2014, 62, 12366–12376. [Google Scholar] [CrossRef]
- Wang, H.; Zong, C.; Bai, A.; Yuan, S.; Li, Y.; Yu, Z.; Tian, R.; Liu, T.; Hou, X.; Li, Y. Transcriptome sequencing and gas chromatography-mass spectrometry analyses provide insights into beta-caryophyllene biosynthesis in Brassica campestris. Food Chem. 2022, 5, 100129. [Google Scholar] [CrossRef]
- Li, B.; Fan, R.; Guo, S.; Wang, P.; Zhu, X.; Fan, Y.; Chen, Y.; He, K.; Kumar, A.; Shi, J.; et al. The Arabidopsis MYB transcription factor, MYB111 modulates salt responses by regulating flavonoid biosynthesis. Environ. Exp. Bot. 2019, 166, 103807. [Google Scholar] [CrossRef]
- Bhatia, C.; Pandey, A.; Gaddam, S.R.; Hoecker, U.; Trivedi, P.K. Low Temperature-Enhanced Flavonol Synthesis Requires Light-Associated Regulatory Components in Arabidopsis thaliana. Plant Cell Physiol. 2018, 59, 2099–2112. [Google Scholar] [CrossRef]
- Shi, Y.; Ding, Y.; Yang, S. Molecular Regulation of CBF Signaling in Cold Acclimation. Trends Plant Sci. 2018, 23, 623–637. [Google Scholar] [CrossRef]
- Kidokoro, S.; Shinozaki, K.; Yamaguchi-Shinozaki, K. Transcriptional regulatory network of plant cold-stress responses. Trends Plant Sci. 2022, 27, 922–935. [Google Scholar] [CrossRef]
- Cao, Y.; Li, K.; Li, Y.; Zhao, X.; Wang, L. MYB Transcription Factors as Regulators of Secondary Metabolism in Plants. Biology 2020, 9, 61. [Google Scholar] [CrossRef]
- Pandey, A.; Misra, P.; Trivedi, P.K. Constitutive expression of Arabidopsis MYB transcription factor, AtMYB11, in tobacco modulates flavonoid biosynthesis in favor of flavonol accumulation. Plant Cell Rep. 2015, 34, 1515–1528. [Google Scholar] [CrossRef]
- Wang, F.; Kong, W.; Wong, G.; Fu, L.; Peng, R.; Li, Z.; Yao, Q. AtMYB12 regulates flavonoids accumulation and abiotic stress tolerance in transgenic Arabidopsis thaliana. Mol. Genet. Genom. 2016, 291, 1545–1559. [Google Scholar] [CrossRef]
- Taylor, L.P.; Grotewold, E. Flavonoids as developmental regulators. Curr. Opin. Plant Biol. 2005, 8, 317–323. [Google Scholar] [CrossRef] [PubMed]
- Davies, K.M.; Schwinn, K.E.; Deroles, S.C.; Manson, D.G.; Bradley, J.M. Enhancing anthocyanin production by altering competition for substrate between flavonol synthase and dihydroflavonol 4-reductase. Euphytica 2003, 131, 259–268. [Google Scholar] [CrossRef]
- Meng, X.; Li, Y.; Zhou, T.; Sun, W.; Wang, L. Functional Differentiation of Duplicated Flavonoid 3-O-Glycosyltransferases in the Flavonol and Anthocyanin Biosynthesis of Freesia hybrida. Front. Plant Sci. 2019, 10, 1330. [Google Scholar] [CrossRef]
- Liu, C.; Su, Y.; Li, J.; Jia, B.; Cao, Z.; Qin, G. Physiological adjustment of pomegranate pericarp responding to sunburn and its underlying molecular mechanisms. BMC Plant Biol. 2022, 22, 169. [Google Scholar] [CrossRef]
- Naing, A.H.; Kim, C.K. Roles of R2R3-MYB transcription factors in transcriptional regulation of anthocyanin biosynthesis in horticultural plants. Plant Mol. Biol. 2018, 98, 1–18. [Google Scholar] [CrossRef]
- Ma, D.; Constabel, C.P. MYB repressors as regulators of phenylpropanoid metabolism in plants. Trends Plant Sci. 2019, 24, 275–289. [Google Scholar] [CrossRef]
- Heppel, S.C.; Jaffé, F.W.; Takos, A.M.; Schellmann, S.; Rausch, T.; Walker, A.R.; Bogs, J. Identification of key amino acids for the evolution of promoter target specificity of anthocyanin and proanthocyanidin regulating MYB factors. Plant Mol. Biol. 2013, 82, 457–471. [Google Scholar] [CrossRef]
- Chinnusamy, V.; Zhu, J.; Zhu, J.-K. Cold stress regulation of gene expression in plants. Trends Plant Sci. 2007, 12, 444–451. [Google Scholar] [CrossRef]
- Hwarari, D.; Guan, Y.; Ahmad, B.; Movahedi, A.; Min, T.; Hao, Z.; Lu, Y.; Chen, J.; Yang, L. ICE-CBF-COR Signaling Cascade and Its Regulation in Plants Responding to Cold Stress. Int. J. Mol. Sci. 2022, 23, 1549. [Google Scholar] [CrossRef]
- Guo, J.; Ren, Y.; Tang, Z.; Shi, W.; Zhou, M. Characterization and expression profiling of the ICE-CBF-COR genes in wheat. PeerJ 2019, 7, e8190. [Google Scholar] [CrossRef]
- Ding, Y.; Shi, Y.; Yang, S. Advances and challenges in uncovering cold tolerance regulatory mechanisms in plants. New Phytol. 2019, 222, 1690–1704. [Google Scholar] [CrossRef] [PubMed]
- Stockinger, E.J.; Gilmour, S.J.; Thomashow, M.F. Arabidopsis thaliana CBF1 encodes an AP2 domain-containing transcriptional activator that binds to the C-repeat/DRE, a cis-acting DNA regulatory element that stimulates transcription in response to low temperature and water deficit. Proc. Natl. Acad. Sci. USA 1997, 94, 1035–1040. [Google Scholar] [CrossRef]
- Liu, Q.; Kasuga, M.; Sakuma, Y.; Abe, H.; Miura, S.; Yamaguchi-Shinozaki, K.; Shinozaki, K. Two transcription factors, DREB1 and DREB2, with an EREBP/AP2 DNA binding domain separate two cellular signal transduction pathways in drought-and low-temperature-responsive gene expression, respectively, in Arabidopsis. Plant Cell 1998, 10, 1391–1406. [Google Scholar] [CrossRef] [PubMed]
- Zhao, C.; Zhang, Z.; Xie, S.; Si, T.; Li, Y.; Zhu, J.K. Mutational Evidence for the Critical Role of CBF Transcription Factors in Cold Acclimation in Arabidopsis. Plant Physiol. 2016, 171, 2744–2759. [Google Scholar] [CrossRef]
- Wang, Y.; Zhang, Y.; Fan, C.; Wei, Y.; Meng, J.; Li, Z.; Zhong, C. Genome-wide analysis of MYB transcription factors and their responses to salt stress in Casuarina equisetifolia. BMC Plant Biol. 2021, 21, 328. [Google Scholar] [CrossRef] [PubMed]
- Cominelli, E.; Galbiati, M.; Vavasseur, A.; Conti, L.; Sala, T.; Vuylsteke, M.; Leonhardt, N.; Dellaporta, S.L.; Tonelli, C. A guard-cell-specific MYB transcription factor regulates stomatal movements and plant drought tolerance. Curr. Biol. 2005, 15, 1196–1200. [Google Scholar] [CrossRef]
- Cominelli, E.; Sala, T.; Calvi, D.; Gusmaroli, G.; Tonelli, C. Over-expression of the Arabidopsis AtMYB41 gene alters cell expansion and leaf surface permeability. Plant J. 2008, 53, 53–64. [Google Scholar] [CrossRef]
- Du, Y.-T.; Zhao, M.-J.; Wang, C.-T.; Gao, Y.; Wang, Y.-X.; Liu, Y.-W.; Chen, M.; Chen, J.; Zhou, Y.-B.; Xu, Z.-S. Identification and characterization of GmMYB118 responses to drought and salt stress. BMC Plant Biol. 2018, 18, 320. [Google Scholar] [CrossRef] [PubMed]
- Wu, J.; Jiang, Y.; Liang, Y.; Chen, L.; Chen, W.; Cheng, B. Expression of the maize MYB transcription factor ZmMYB3R enhances drought and salt stress tolerance in transgenic plants. Plant Physiol. Biochem. 2019, 137, 179–188. [Google Scholar] [CrossRef] [PubMed]
- Chen, K.; Song, M.; Guo, Y.; Liu, L.; Xue, H.; Dai, H.; Zhang, Z. MdMYB46 could enhance salt and osmotic stress tolerance in apple by directly activating stress-responsive signals. Plant Biotechnol. J. 2019, 17, 2341–2355. [Google Scholar] [CrossRef] [PubMed]
- Lee, H.G.; Seo, P.J. The MYB 96–HHP module integrates cold and abscisic acid signaling to activate the CBF–COR pathway in Arabidopsis. Plant J. 2015, 82, 962–977. [Google Scholar] [CrossRef]
- Agarwal, M.; Hao, Y.; Kapoor, A.; Dong, C.-H.; Fujii, H.; Zheng, X.; Zhu, J.-K. A R2R3 type MYB transcription factor is involved in the cold regulation of CBF genes and in acquired freezing tolerance. J. Biol. Chem. 2006, 281, 37636–37645. [Google Scholar] [CrossRef]
- Chen, Y.; Chen, Z.; Kang, J.; Kang, D.; Gu, H.; Qin, G. AtMYB14 regulates cold tolerance in Arabidopsis. Plant Mol. Biol. Rep. 2013, 31, 87–97. [Google Scholar] [CrossRef]
- Huang, H.; Ullah, F.; Zhou, D.-X.; Yi, M.; Zhao, Y. Mechanisms of ROS regulation of plant development and stress responses. Front. Plant Sci. 2019, 10, 800. [Google Scholar] [CrossRef]
- Mittler, R. Oxidative stress, antioxidants and stress tolerance [Review]. Trends Plant Sci. 2002, 7, 405–410. [Google Scholar] [CrossRef]
- Agati, G. Multiple functional roles of flavonoids in photoprotection. New Phytol. 2010, 186, 786–793. [Google Scholar] [CrossRef]
- Gautam, H.; Sharma, A.; Trivedi, P.K. The role of flavonols in insect resistance and stress response. Curr. Opin. Plant Biol. 2023, 73, 102353. [Google Scholar] [CrossRef]
- Pourcel, L.; Irani, N.G.; Koo, A.; Bohorquez-Restrepo, A.; Howe, G.A.; Grotewold, E. A chemical complementation approach reveals genes and interactions of flavonoids with other pathways. Plant J. 2013, 74, 383–397. [Google Scholar] [CrossRef] [PubMed]
- Yu, J.; Yang, X.D.; Wang, Q.; Gao, L.W.; Yang, Y.; Xiao, D.; Liu, T.K.; Li, Y.; Hou, X.L.; Zhang, C.W. Efficient virus-induced gene silencing in Brassica rapa using a turnip yellow mosaic virus vector. Biol. Plant. 2018, 62, 826–834. [Google Scholar] [CrossRef]
- Clough, S.J.; Bent, A.F. Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 1998, 16, 735–743. [Google Scholar] [CrossRef] [PubMed]




Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chen, X.; Wu, Y.; Yu, Z.; Gao, Z.; Ding, Q.; Shah, S.H.A.; Lin, W.; Li, Y.; Hou, X. BcMYB111 Responds to BcCBF2 and Induces Flavonol Biosynthesis to Enhance Tolerance under Cold Stress in Non-Heading Chinese Cabbage. Int. J. Mol. Sci. 2023, 24, 8670. https://doi.org/10.3390/ijms24108670
Chen X, Wu Y, Yu Z, Gao Z, Ding Q, Shah SHA, Lin W, Li Y, Hou X. BcMYB111 Responds to BcCBF2 and Induces Flavonol Biosynthesis to Enhance Tolerance under Cold Stress in Non-Heading Chinese Cabbage. International Journal of Molecular Sciences. 2023; 24(10):8670. https://doi.org/10.3390/ijms24108670
Chicago/Turabian StyleChen, Xiaoshan, Ying Wu, Zhanghong Yu, Zhanyuan Gao, Qiang Ding, Sayyed Hamad Ahmad Shah, Wenyuan Lin, Ying Li, and Xilin Hou. 2023. "BcMYB111 Responds to BcCBF2 and Induces Flavonol Biosynthesis to Enhance Tolerance under Cold Stress in Non-Heading Chinese Cabbage" International Journal of Molecular Sciences 24, no. 10: 8670. https://doi.org/10.3390/ijms24108670
APA StyleChen, X., Wu, Y., Yu, Z., Gao, Z., Ding, Q., Shah, S. H. A., Lin, W., Li, Y., & Hou, X. (2023). BcMYB111 Responds to BcCBF2 and Induces Flavonol Biosynthesis to Enhance Tolerance under Cold Stress in Non-Heading Chinese Cabbage. International Journal of Molecular Sciences, 24(10), 8670. https://doi.org/10.3390/ijms24108670







