Identification of Potential Biomarkers for Diagnosis of Patients with Methamphetamine Use Disorder
Abstract
1. Introduction
1.1. Contribution
- This study is a novel approach to identify non-invasive transcriptome biomarkers using human hair follicles for the accurate diagnosis of MUD.
- A two-step predictive model that can monitor and identify the progression of MUD treatment was developed and validated.
- The proposed biomarkers and predictive model have the potential to improve the accuracy of expert diagnoses and provide better treatment options for MUD patients.
1.2. Literature Review
2. Results
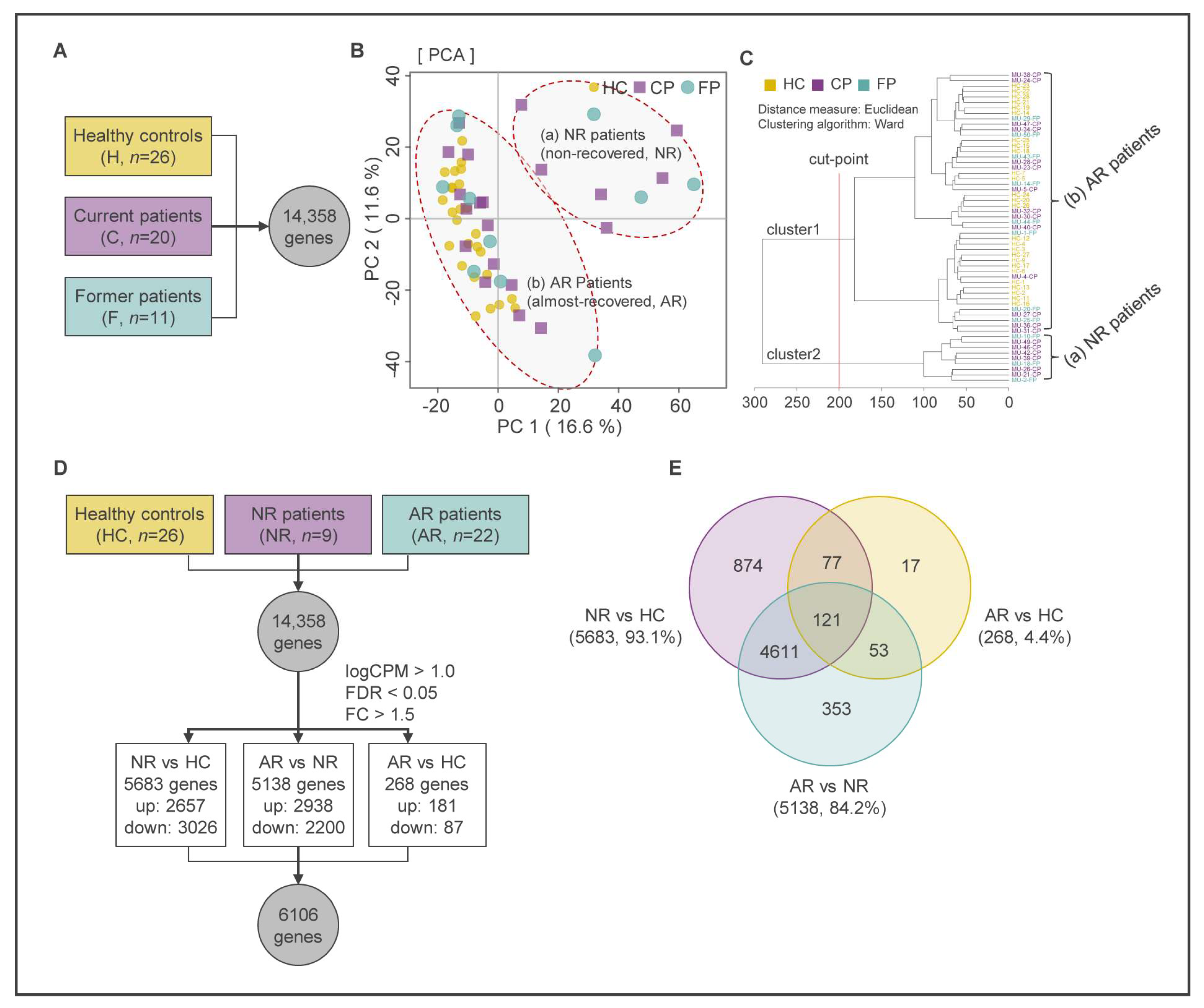
2.1. RNA-seq Data Processing
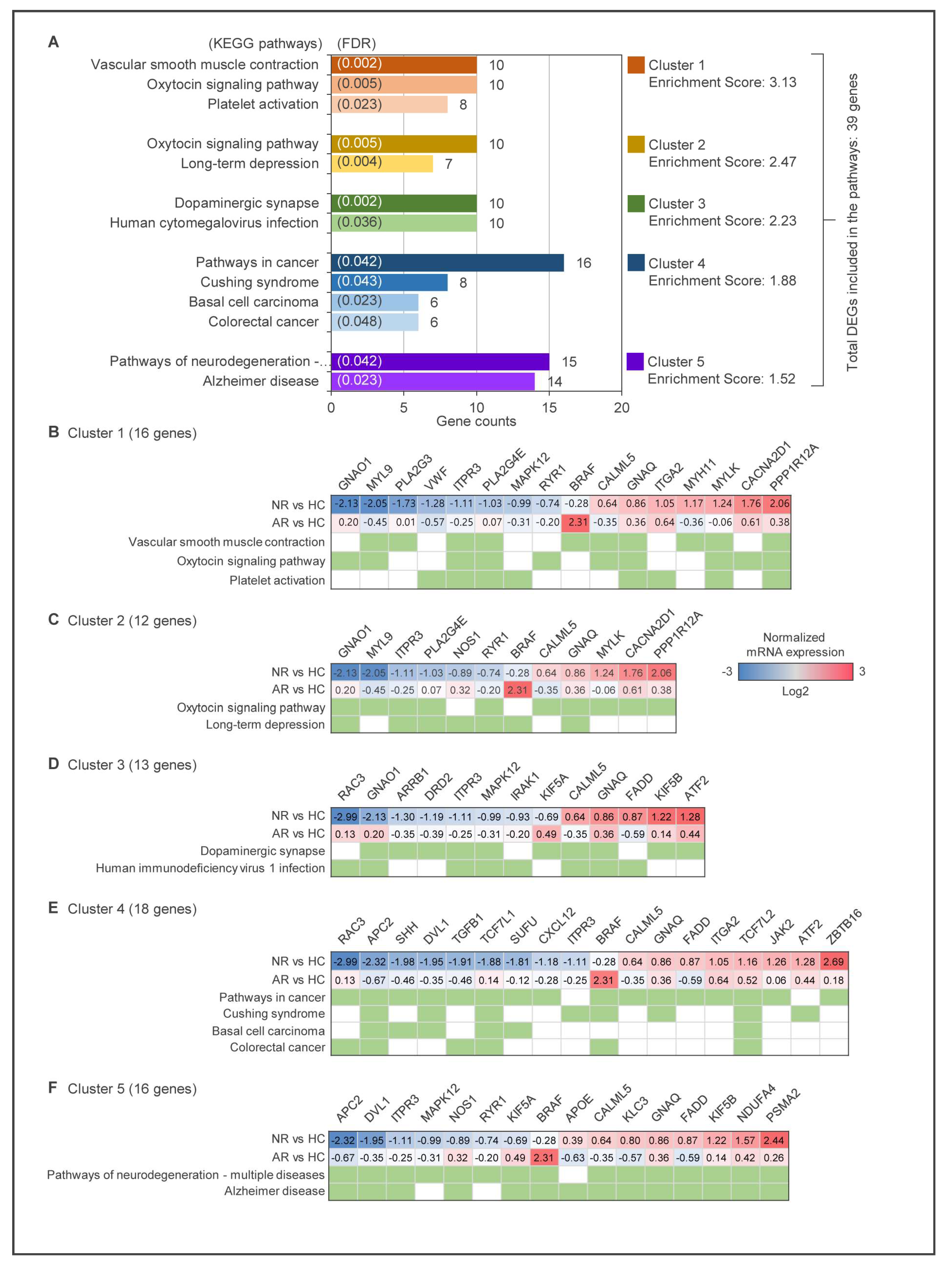
2.2. Identification of DEGs Using Multivariate Analysis and Keg Pathway Enrichment Analysis
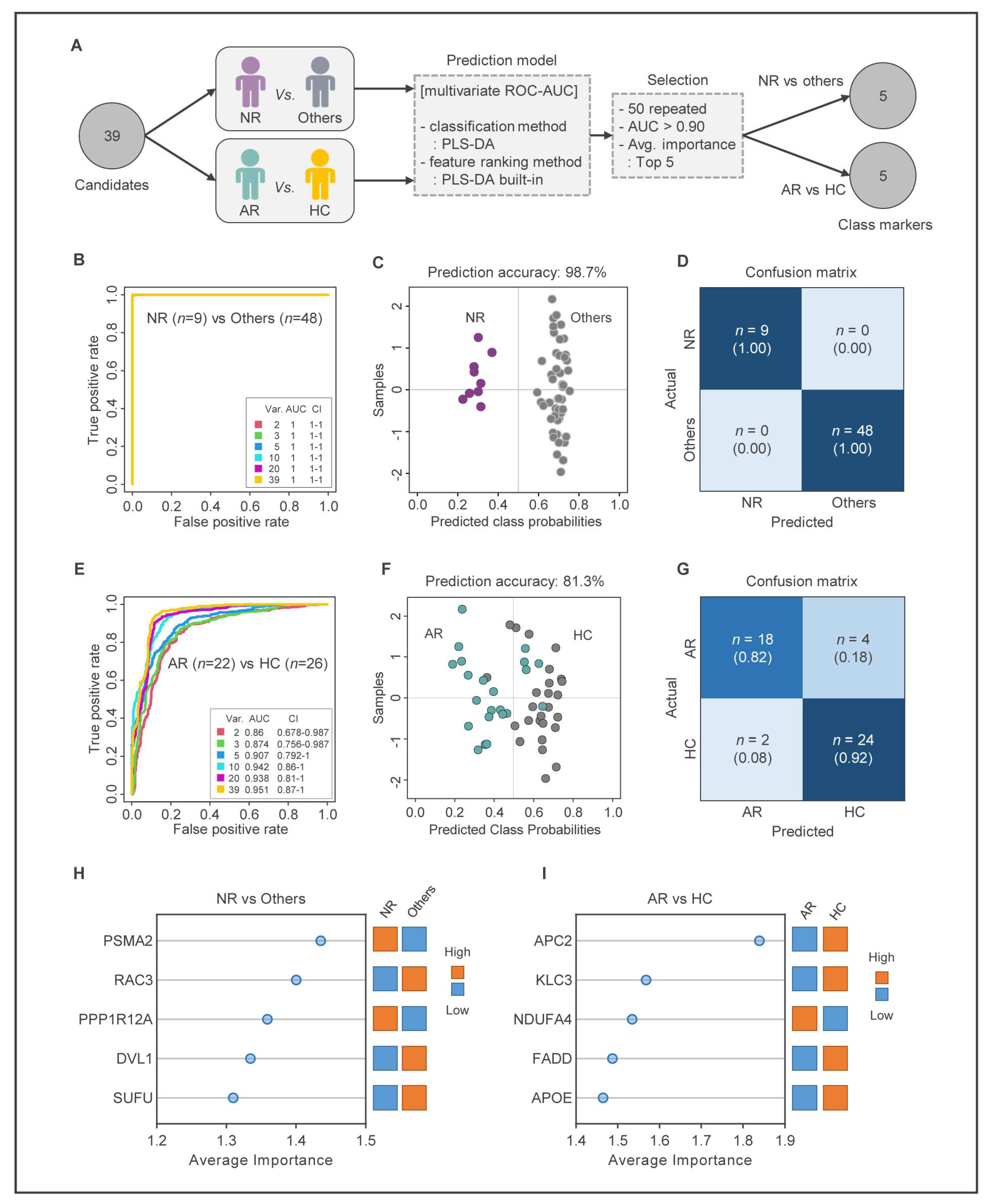
2.3. Partial Least Squares-Discriminant Analysis (PLS-DA)
2.4. Construction of Protein-Protein Interaction (PPI) Network and Network Centrality Analysis
2.5. Functional Annotation Analysis
2.6. Construction of MUD Prediction Models
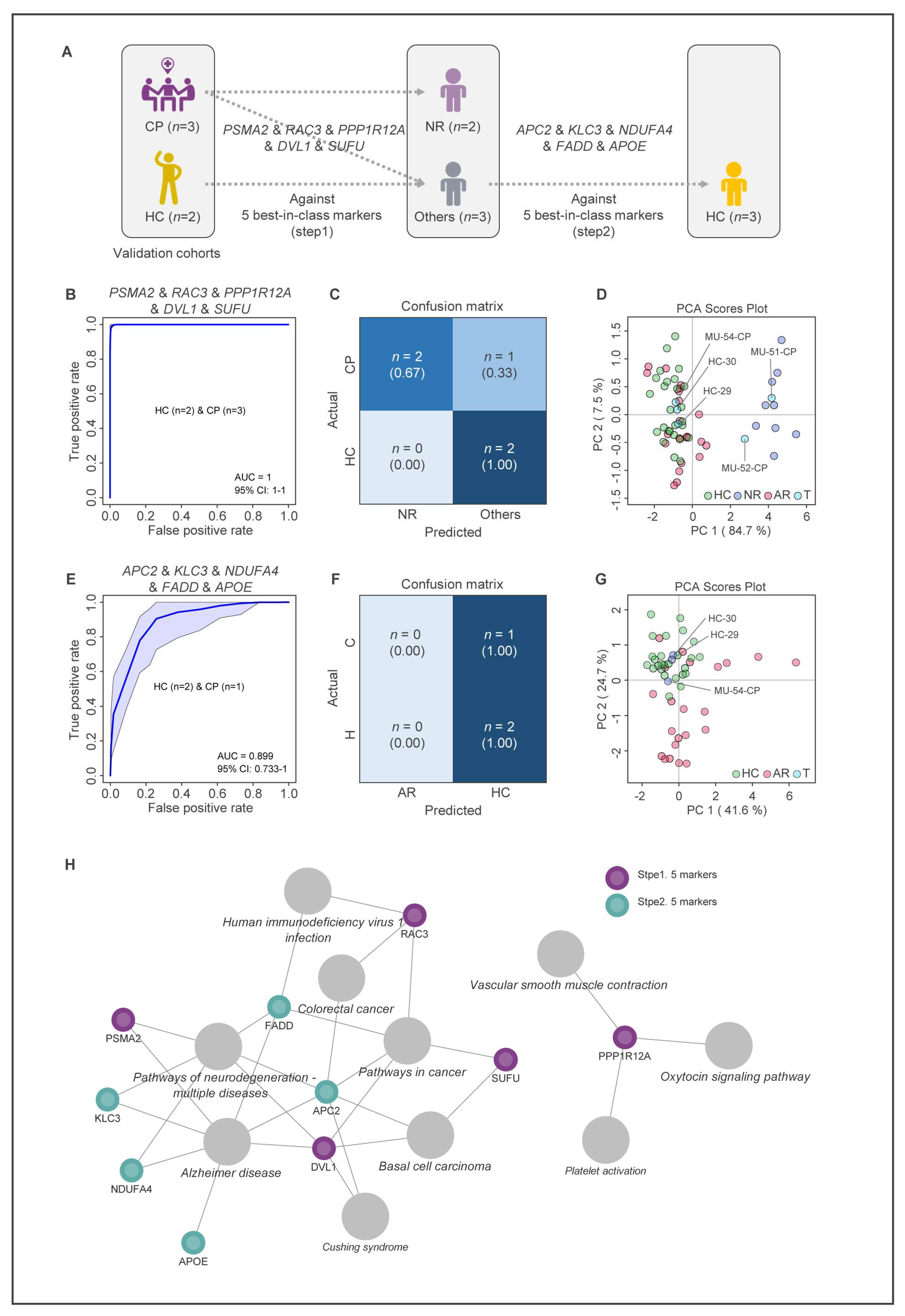
2.7. Validation of MUD Prediction Models
3. Discussion
4. Materials and Methods
4.1. Participants
4.2. Sample Collection and RNA Extraction
4.3. RNA Sequencing and Data Processing
4.4. Differential Gene Expression Analysis
4.5. Principal Component Analysis and Partial Least Square Discriminant Analysis
4.6. KEGG Pathway Enrichment Analysis
4.7. Protein–Protein Interaction Network Analysis
4.8. Construction and Validation of MUD Prediction Model
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Koob, G.F.; Le Moal, M. Drug addiction, dysregulation of reward, and allostasis. Neuropsychopharmacology 2001, 24, 97–129. [Google Scholar] [CrossRef]
- Kim, S.; Jang, W.J.; Yu, H.; Ryu, I.S.; Jeong, C.H.; Lee, S. Integrated Non-targeted and Targeted Metabolomics Uncovers Dynamic Metabolic Effects during Short-Term Abstinence in Methamphetamine Self-Administering Rats. J. Proteome Res. 2019, 18, 3913–3925. [Google Scholar] [CrossRef]
- Zaitsu, K.; Hayashi, Y.; Kusano, M.; Tsuchihashi, H.; Ishii, A. Application of metabolomics to toxicology of drugs of abuse: A mini review of metabolomics approach to acute and chronic toxicity studies. Drug Metab. Pharmacokinet. 2016, 31, 21–26. [Google Scholar] [CrossRef]
- Ballester, J.; Valentine, G.; Sofuoglu, M. Pharmacological treatments for methamphetamine addiction: Current status and future directions. Expert Rev. Clin. Pharmacol. 2017, 10, 305–314. [Google Scholar] [CrossRef] [PubMed]
- Morley, K.C.; Cornish, J.L.; Faingold, A.; Wood, K.; Haber, P.S. Pharmacotherapeutic agents in the treatment of methamphetamine dependence. Expert Opin. Investig. Drugs 2017, 26, 563–578. [Google Scholar] [CrossRef] [PubMed]
- Longo, M.; Wickes, W.; Smout, M.; Harrison, S.; Cahill, S.; White, J.M. Randomized controlled trial of dexamphetamine maintenance for the treatment of methamphetamine dependence. Addiction 2010, 105, 146–154. [Google Scholar] [CrossRef] [PubMed]
- Ling, W.; Chang, L.; Hillhouse, M.; Ang, A.; Striebel, J.; Jenkins, J.; Hernandez, J.; Olaer, M.; Mooney, L.; Reed, S.; et al. Sustained-release methylphenidate in a randomized trial of treatment of methamphetamine use disorder. Addiction 2014, 109, 1489–1500. [Google Scholar] [CrossRef] [PubMed]
- Kohno, M.; Dennis, L.E.; McCready, H.; Schwartz, D.L.; Hoffman, W.F.; Korthuis, P.T. A preliminary randomized clinical trial of naltrexone reduces striatal resting state functional connectivity in people with methamphetamine use disorder. Drug Alcohol Depend. 2018, 192, 186–192. [Google Scholar] [CrossRef]
- Harada, T.; Tsutomi, H.; Mori, R.; Wilson, D.B. Cognitive-behavioural treatment for amphetamine-type stimulants (ATS)-use disorders. Cochrane Database Syst. Rev. 2018, 12, CD011315. [Google Scholar] [CrossRef]
- Polcin, D.L.; Bond, J.; Korcha, R.; Nayak, M.B.; Galloway, G.P.; Evans, K. Randomized trial of intensive motivational interviewing for methamphetamine dependence. J. Addict. Dis. 2014, 33, 253–265. [Google Scholar] [CrossRef]
- Collins, K.C.; Schlosburg, J.E.; Bremer, P.T.; Janda, K.D. Methamphetamine Vaccines: Improvement through Hapten Design. J. Med. Chem. 2016, 59, 3878–3885. [Google Scholar] [CrossRef] [PubMed]
- Howells, F.M.; Uhlmann, A.; Temmingh, H.; Sinclair, H.; Meintjes, E.; Wilson, D.; Stein, D.J. (1)H-magnetic resonance spectroscopy ((1)H-MRS) in methamphetamine dependence and methamphetamine induced psychosis. Schizophr. Res. 2014, 153, 122–128. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.E.; Kim, G.H.; Hwang, J.; Kim, J.Y.; Renshaw, P.F.; Yurgelun-Todd, D.A.; Kim, B.; Kang, I.; Jeon, S.; Ma, J.; et al. Metabolic alterations in the anterior cingulate cortex and related cognitive deficits in late adolescent methamphetamine users. Addict. Biol. 2018, 23, 327–336. [Google Scholar] [CrossRef]
- Yang, W.; Yang, R.; Luo, J.; He, L.; Liu, J.; Zhang, J. Increased Absolute Glutamate Concentrations and Glutamate-to-Creatine Ratios in Patients With Methamphetamine Use Disorders. Front. Psychiatry 2018, 9, 368. [Google Scholar] [CrossRef]
- Lee, S.; Han, E.; Park, Y.; Choi, H.; Chung, H. Distribution of methamphetamine and amphetamine in drug abusers’ head hair. Forensic Sci. Int. 2009, 190, 16–18. [Google Scholar] [CrossRef] [PubMed]
- Koob, G.F.; Volkow, N.D. Neurocircuitry of addiction. Neuropsychopharmacology 2010, 35, 217–238. [Google Scholar] [CrossRef] [PubMed]
- Paulus, M.P. Pragmatism Instead of Mechanism: A Call for Impactful Biological Psychiatry. JAMA Psychiatry 2015, 72, 631–632. [Google Scholar] [CrossRef]
- Robinson, T.E.; Berridge, K.C. The neural basis of drug craving: An incentive-sensitization theory of addiction. Brain Res. Rev. 1993, 18, 247–291. [Google Scholar] [CrossRef] [PubMed]
- Bough, K.J.; Pollock, J.D. Defining Substance Use Disorders: The Need for Peripheral Biomarkers. Trends Mol. Med. 2018, 24, 109–120. [Google Scholar] [CrossRef]
- Ghanbari, R.; Sumner, S. Using Metabolomics to Investigate Biomarkers of Drug Addiction. Trends Mol. Med. 2018, 24, 197–205. [Google Scholar] [CrossRef]
- Song, S.H.; Jang, W.J.; Hwang, J.; Park, B.; Jang, J.H.; Seo, Y.H.; Yang, C.H.; Lee, S.; Jeong, C.H. Transcriptome profiling of whisker follicles in methamphetamine self-administered rats. Sci. Rep. 2018, 8, 11420. [Google Scholar] [CrossRef] [PubMed]
- Jang, W.J.; Son, T.; Song, S.H.; Ryu, I.S.; Lee, S.; Jeong, C.H. Transcriptional Profiling of Whisker Follicles and of the Striatum in Methamphetamine Self-Administered Rats. Int. J. Mol. Sci. 2020, 21, 8856. [Google Scholar] [CrossRef]
- Song, S.-H.; Jang, W.-J.; Jang, E.Y.; Kim, O.-H.; Kim, H.; Son, T.; Choi, D.-Y.; Lee, S.; Jeong, C.-H. Striatal miR-183-5p inhibits methamphetamine-induced locomotion by regulating glucocorticoid receptor signaling. Front. Pharmacol. 2022, 13, 997701. [Google Scholar] [CrossRef] [PubMed]
- Fathi, S.; Soltanzadeh, H.; Tanomand, A.; Asadi, Z.; Rezai Moradali, S. Investigation of miR-222 as a potential biomarker in diagnosis of patients with methamphetamine abuse disorder. Egypt. J. Med. Hum. Genet. 2022, 23, 1–6. [Google Scholar] [CrossRef]
- Chand, S.; Gowen, A.; Savine, M.; Moore, D.; Clark, A.; Huynh, W.; Wu, N.; Odegaard, K.; Weyrich, L.; Bevins, R.A. A comprehensive study to delineate the role of an extracellular vesicle-associated microRNA-29a in chronic methamphetamine use disorder. J. Extracell. Vesicles 2021, 10, e12177. [Google Scholar] [CrossRef] [PubMed]
- Shabbir, S.; Asif, M.S.; Alam, T.M.; Ramzan, Z. Early prediction of malignant mesothelioma: An approach towards non-invasive method. Curr. Bioinform. 2021, 16, 1257–1277. [Google Scholar] [CrossRef]
- Alam, T.M.; Shaukat, K.; Mahboob, H.; Sarwar, M.U.; Iqbal, F.; Nasir, A.; Hameed, I.A.; Luo, S. A machine learning approach for identification of malignant mesothelioma etiological factors in an imbalanced dataset. Comput. J. 2022, 65, 1740–1751. [Google Scholar] [CrossRef]
- Seo, M.J.; Song, S.H.; Kim, S.; Jang, W.J.; Jeong, C.H.; Lee, S. Characteristics of Korean patients with methamphetamine use disorder based on the quantitative analysis of methamphetamine and amphetamine in hair. Arch. Pharm. Res. 2020, 43, 798–807. [Google Scholar] [CrossRef]
- Seo, M.J.; Song, S.H.; Kim, S.; Jang, W.J.; Jeong, C.H.; Lee, S. Mass spectrometry-based metabolomics in hair from current and former patients with methamphetamine use disorder. Arch. Pharm. Res. 2021, 44, 890–901. [Google Scholar] [CrossRef]
- Lukk, M.; Kapushesky, M.; Nikkila, J.; Parkinson, H.; Goncalves, A.; Huber, W.; Ukkonen, E.; Brazma, A. A global map of human gene expression. Nat. Biotechnol. 2010, 28, 322–324. [Google Scholar] [CrossRef]
- Kahraman, M.; Roske, A.; Laufer, T.; Fehlmann, T.; Backes, C.; Kern, F.; Kohlhaas, J.; Schrors, H.; Saiz, A.; Zabler, C.; et al. MicroRNA in diagnosis and therapy monitoring of early-stage triple-negative breast cancer. Sci. Rep. 2018, 8, 11584. [Google Scholar] [CrossRef] [PubMed]
- Everitt, B.; Hothorn, T.; Everitt, B.; Hothorn, T. Multivariate Data and Multivariate Analysis. In An Introduction to Applied Multivariate Analysis with R; Springer: Berlin/Heidelberg, Germany, 2011; pp. 1–24. [Google Scholar]
- Jolliffe, I.T.; Cadima, J. Principal component analysis: A review and recent developments. Philos. Trans. R. Soc. A Math. Phys. Eng. Sci. 2016, 374, 20150202. [Google Scholar] [CrossRef]
- Barker, M.; Rayens, W. Partial least squares for discrimination. J. Chemom. A J. Chemom. Soc. 2003, 17, 166–173. [Google Scholar] [CrossRef]
- Zhao, L.; Yang, S.; Cheng, Y.; Hou, C.; You, X.; Zhao, J.; Zhang, Y.; He, W. Identification of transcriptional biomarkers by RNA-sequencing for improved detection of β2-agonists abuse in goat skeletal muscle. PLoS ONE 2017, 12, e0181695. [Google Scholar] [CrossRef] [PubMed]
- Tomkins, J.E.; Manzoni, C. Advances in protein-protein interaction network analysis for Parkinson’s disease. Neurobiol. Dis. 2021, 155, 105395. [Google Scholar] [CrossRef]
- Xia, J.; Broadhurst, D.I.; Wilson, M.; Wishart, D.S. Translational biomarker discovery in clinical metabolomics: An introductory tutorial. Metabolomics 2013, 9, 280–299. [Google Scholar] [CrossRef]
- Kim, Y.G.; Hwang, J.; Choi, H.; Lee, S. Development of a Column-Switching HPLC-MS/MS Method and Clinical Application for Determination of Ethyl Glucuronide in Hair in Conjunction with AUDIT for Detecting High-Risk Alcohol Consumption. Pharmaceutics 2018, 10, 84. [Google Scholar] [CrossRef]
- Lendoiro, E.; de Castro, A.; Jimenez-Morigosa, C.; Gomez-Fraguela, X.A.; Lopez-Rivadulla, M.; Cruz, A. Usefulness of hair analysis and psychological tests for identification of alcohol and drugs of abuse consumption in driving license regranting. Forensic Sci. Int. 2018, 286, 239–244. [Google Scholar] [CrossRef]
- Frampton, J.P.; Guo, C.; Pierchala, B.A. Expression of axonal protein degradation machinery in sympathetic neurons is regulated by nerve growth factor. J. Neurosci. Res. 2012, 90, 1533–1546. [Google Scholar] [CrossRef] [PubMed]
- Vingill, S.; Brockelt, D.; Lancelin, C.; Tatenhorst, L.; Dontcheva, G.; Preisinger, C.; Schwedhelm-Domeyer, N.; Joseph, S.; Mitkovski, M.; Goebbels, S. Loss of FBXO 7 (PARK 15) results in reduced proteasome activity and models a parkinsonism-like phenotype in mice. EMBO J. 2016, 35, 2008–2025. [Google Scholar] [CrossRef] [PubMed]
- Grünblatt, E.; Zehetmayer, S.; Jacob, C.P.; Müller, T.; Jost, W.H.; Riederer, P. Pilot study: Peripheral biomarkers for diagnosing sporadic Parkinson’s disease. J. Neural Transm. 2010, 117, 1387–1393. [Google Scholar] [CrossRef] [PubMed]
- Tivodar, S.; Kalemaki, K.; Kounoupa, Z.; Vidaki, M.; Theodorakis, K.; Denaxa, M.; Kessaris, N.; De Curtis, I.; Pachnis, V.; Karagogeos, D. Rac-GTPases regulate microtubule stability and axon growth of cortical GABAergic interneurons. Cereb. Cortex 2015, 25, 2370–2382. [Google Scholar] [CrossRef] [PubMed]
- Corbetta, S.; Gualdoni, S.; Ciceri, G.; Monari, M.; Zuccaro, E.; Tybulewicz, V.L.; De Curtis, I. Essential role of Rac1 and Rac3 GTPases in neuronal development. FASEB J. 2009, 23, 1347–1357. [Google Scholar] [CrossRef] [PubMed]
- Pennucci, R.; Talpo, F.; Astro, V.; Montinaro, V.; Morè, L.; Cursi, M.; Castoldi, V.; Chiaretti, S.; Bianchi, V.; Marenna, S. Loss of either Rac1 or Rac3 GTPase differentially affects the behavior of mutant mice and the development of functional GABAergic networks. Cereb. Cortex 2016, 26, 873–890. [Google Scholar] [CrossRef]
- De Curtis, I. Roles of Rac1 and Rac3 GTPases during the development of cortical and hippocampal GABAergic interneurons. Front. Cell. Neurosci. 2014, 8, 307. [Google Scholar] [CrossRef]
- Maekawa, M.; Yamada, K.; Toyoshima, M.; Ohnishi, T.; Iwayama, Y.; Shimamoto, C.; Toyota, T.; Nozaki, Y.; Balan, S.; Matsuzaki, H.; et al. Utility of Scalp Hair Follicles as a Novel Source of Biomarker Genes for Psychiatric Illnesses. Biol. Psychiatry 2015, 78, 116–125. [Google Scholar] [CrossRef]
- Dobin, A.; Davis, C.A.; Schlesinger, F.; Drenkow, J.; Zaleski, C.; Jha, S.; Batut, P.; Chaisson, M.; Gingeras, T.R. STAR: Ultrafast universal RNA-seq aligner. Bioinformatics 2013, 29, 15–21. [Google Scholar] [CrossRef]
- Liao, Y.; Smyth, G.K.; Shi, W. featureCounts: An efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 2014, 30, 923–930. [Google Scholar] [CrossRef]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef]
- Jones, C.M.; Athanasiou, T. Summary receiver operating characteristic curve analysis techniques in the evaluation of diagnostic tests. Ann. Thorac. Surg. 2005, 79, 16–20. [Google Scholar] [CrossRef]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Jang, W.-J.; Song, S.-H.; Son, T.; Bae, J.W.; Lee, S.; Jeong, C.-H. Identification of Potential Biomarkers for Diagnosis of Patients with Methamphetamine Use Disorder. Int. J. Mol. Sci. 2023, 24, 8672. https://doi.org/10.3390/ijms24108672
Jang W-J, Song S-H, Son T, Bae JW, Lee S, Jeong C-H. Identification of Potential Biomarkers for Diagnosis of Patients with Methamphetamine Use Disorder. International Journal of Molecular Sciences. 2023; 24(10):8672. https://doi.org/10.3390/ijms24108672
Chicago/Turabian StyleJang, Won-Jun, Sang-Hoon Song, Taekwon Son, Jung Woo Bae, Sooyeun Lee, and Chul-Ho Jeong. 2023. "Identification of Potential Biomarkers for Diagnosis of Patients with Methamphetamine Use Disorder" International Journal of Molecular Sciences 24, no. 10: 8672. https://doi.org/10.3390/ijms24108672
APA StyleJang, W.-J., Song, S.-H., Son, T., Bae, J. W., Lee, S., & Jeong, C.-H. (2023). Identification of Potential Biomarkers for Diagnosis of Patients with Methamphetamine Use Disorder. International Journal of Molecular Sciences, 24(10), 8672. https://doi.org/10.3390/ijms24108672




