Epigenetic Alterations in DCIS Progression: What Can lncRNAs Teach Us?
Abstract
1. Introduction
2. LncRNAs and Breast Cancer
3. LncRNAs and DCIS
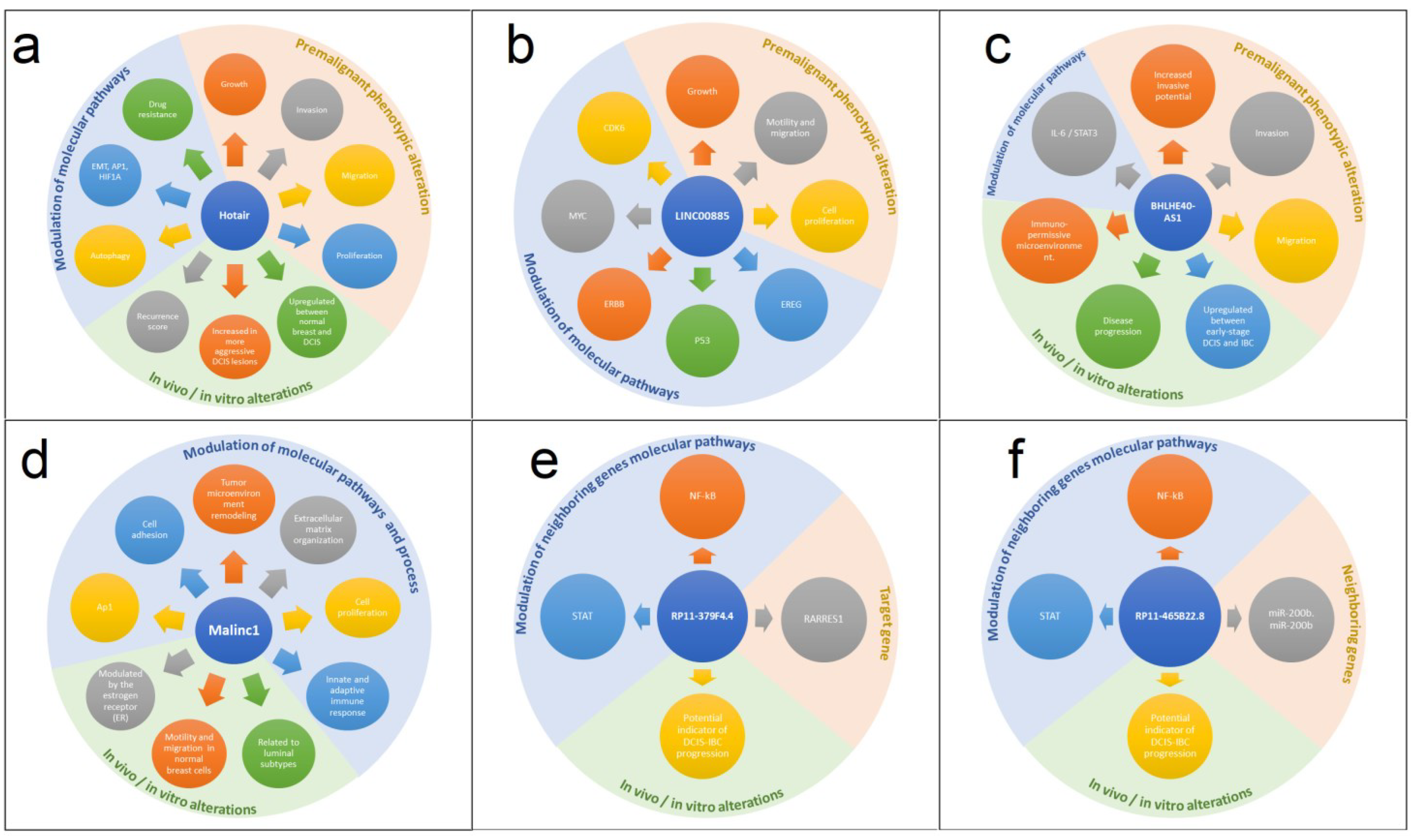
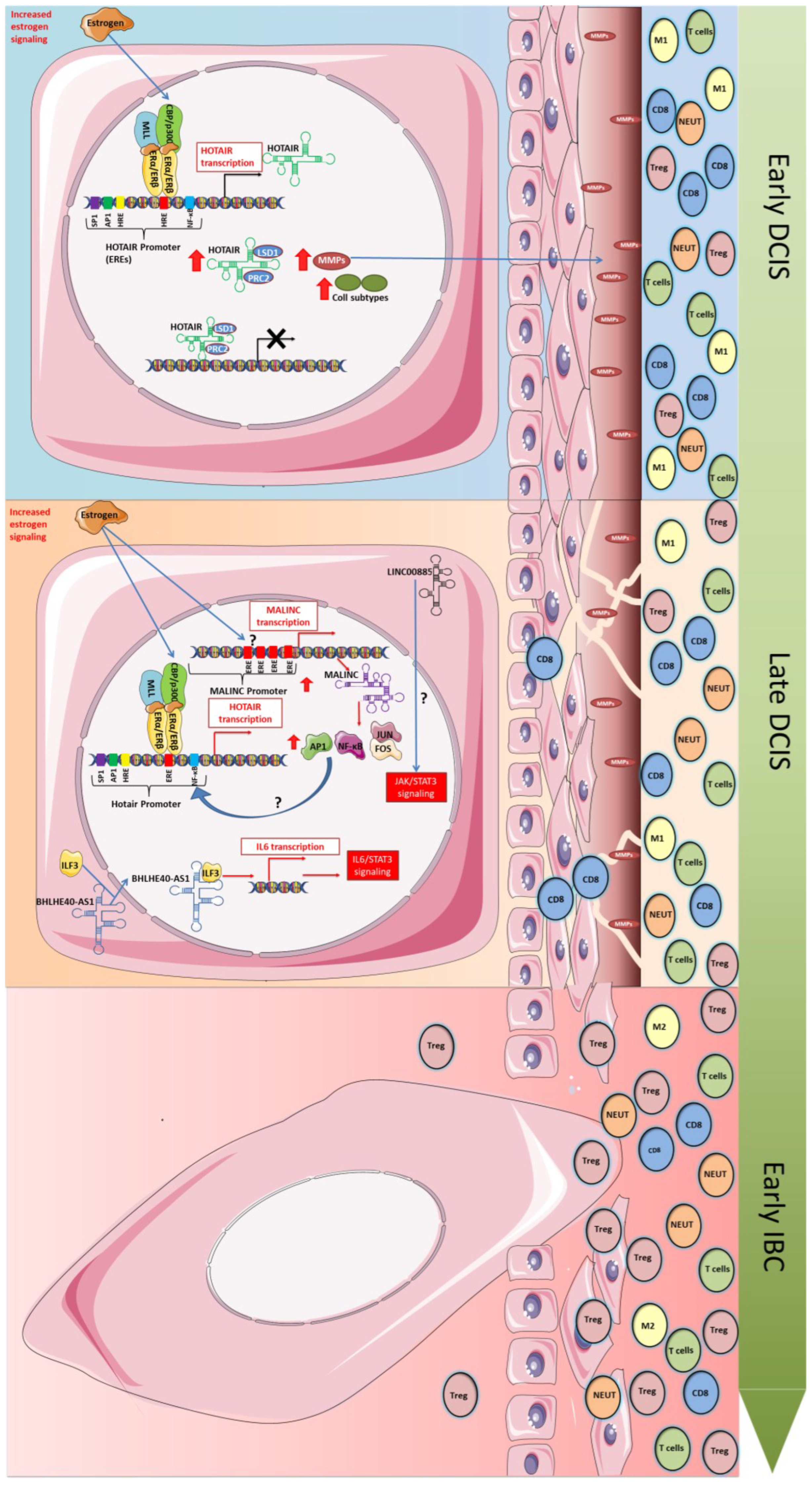
3.1. HOTAIR
3.2. LINC00885
3.3. BHLHE40-AS1
3.4. MALINC1
3.5. SE-lncRNAs
| lncRNA | Function | Expression | Effect in DCIS | Invasive BC | References |
|---|---|---|---|---|---|
| HOTAIR | Modulate the chromatin state by epigenetically repressing the transcription of its target genes | Upregulated | Role in initiating malignant of breast transformation | EMT and Metastatic Potential | [48,52,53,54,55] |
| LINC00885 | A potential novel BC driver lncRNA | Upregulated | Induction of premalignant phenotypic changes and BC progression | Remaining up-modulated in primary invasive BC and could be associated on resistance to hormone therapy | [59] |
| BHLHE40-AS1 | Has not been functionally characterized yet | Upregulated | Role in proliferation, motility, inflammation, and invasive potential | Expression profile and function unknown | [66] |
| MALINC1 | Associated with cell cycle progression | Upregulated | Role in immune response, extracellular matrix remodeling, cell adhesion, and activation of the AP1 signaling pathway | Possible role in cell cycle progression of ER+ subtypes | [72,73] |
| RP11-379F4.4 | Cis-acting SE-lncRNA to its target RARRES1 | Upregulated | DCIS-invasive BC progression | Effect not yet identified | [88] |
| RP11-465B22.8 | Regulation of miR-200b | Upregulated | DCIS-invasive BC progression | Effect not yet identified | [88] |
4. Open Questions
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Dey, B.K.; Mueller, A.C.; Dutta, A. Long Non-Coding RNAs as Emerging Regulators of Differentiation, Development, and Disease. Transcription 2014, 5, e944014. [Google Scholar] [CrossRef] [PubMed]
- Wapinski, O.; Chang, H.Y. Long Noncoding RNAs and Human Disease. Trends Cell Biol. 2011, 21, 354–361. [Google Scholar] [CrossRef]
- Bhan, A.; Mandal, S.S. Long Noncoding RNAs: Emerging Stars in Gene Regulation, Epigenetics and Human Disease. ChemMedChem 2014, 9, 1932–1956. [Google Scholar] [CrossRef]
- Gagan, J.; Dey, B.K.; Dutta, A. MicroRNAs Regulate and Provide Robustness to the Myogenic Transcriptional Network. Curr. Opin. Pharmacol. 2012, 12, 383–388. [Google Scholar] [CrossRef]
- Dey, B.K.; Mueller, A.C.; Dutta, A. Non-Micro-Short RNAs: The New Kids on the Block. Mol. Biol. Cell 2012, 23, 4664–4667. [Google Scholar] [CrossRef] [PubMed]
- Ameres, S.L.; Zamore, P.D. Diversifying MicroRNA Sequence and Function. Nat. Rev. Mol. Cell Biol. 2013, 14, 475–488. [Google Scholar] [CrossRef] [PubMed]
- Dey, B.K.; Gagan, J.; Dutta, A. MiR-206 and -486 Induce Myoblast Differentiation by Downregulating Pax7. Mol. Cell. Biol. 2011, 31, 203–214. [Google Scholar] [CrossRef]
- Panni, S.; Lovering, R.C.; Porras, P.; Orchard, S. Non-Coding RNA Regulatory Networks. Biochim. Et Biophys. Acta (BBA) -Gene Regul. Mech. 2020, 1863, 194417. [Google Scholar] [CrossRef]
- Guttman, M.; Rinn, J.L. Modular Regulatory Principles of Large Non-Coding RNAs. Nature 2012, 482, 339–346. [Google Scholar] [CrossRef]
- Vieira, L.; Grativol, C.; Thiebaut, F.; Carvalho, T.; Hardoim, P.; Hemerly, A.; Lifschitz, S.; Ferreira, P.; Walter, M. PlantRNA_Sniffer: A SVM-Based Workflow to Predict Long Intergenic Non-Coding RNAs in Plants. Non-Coding RNA 2017, 3, 11. [Google Scholar] [CrossRef]
- Dahariya, S.; Paddibhatla, I.; Kumar, S.; Raghuwanshi, S.; Pallepati, A.; Gutti, R.K. Long Non-Coding RNA: Classification, Biogenesis and Functions in Blood Cells. Mol. Immunol. 2019, 112, 82–92. [Google Scholar] [CrossRef]
- Statello, L.; Guo, C.-J.; Chen, L.-L.; Huarte, M. Gene Regulation by Long Non-Coding RNAs and Its Biological Functions. Nat. Rev. Mol. Cell Biol. 2021, 22, 96–118. [Google Scholar] [CrossRef] [PubMed]
- Haase, A.D. An Introduction to PIWI-Interacting RNAs (PiRNAs) in the Context of Metazoan Small RNA Silencing Pathways. RNA Biol. 2022, 19, 1094–1102. [Google Scholar] [CrossRef] [PubMed]
- Khandelwal, A.; Bacolla, A.; Vasquez, K.M.; Jain, A. Long Non-Coding RNA: A New Paradigm for Lung Cancer. Mol. Carcinog. 2015, 54, 1235–1251. [Google Scholar] [CrossRef]
- Guo, X.; Gao, L.; Wang, Y.; Chiu, D.K.Y.; Wang, T.; Deng, Y. Advances in Long Noncoding RNAs: Identification, Structure Prediction and Function Annotation. Brief. Funct. Genom. 2015, 15, 38–46. [Google Scholar] [CrossRef]
- Quinn, J.J.; Chang, H.Y. Unique Features of Long Non-Coding RNA Biogenesis and Function. Nat. Rev. Genet. 2015, 17, 47–62. [Google Scholar] [CrossRef]
- Sun, M.; Kraus, W.L. From Discovery to Function: The Expanding Roles of Long NonCoding RNAs in Physiology and Disease. Endocr. Rev. 2015, 36, 25–64. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.-L. Linking Long Noncoding RNA Localization and Function. Trends Biochem. Sci. 2016, 41, 761–772. [Google Scholar] [CrossRef] [PubMed]
- Fang, Y.; Fullwood, M.J. Roles, Functions, and Mechanisms of Long Non-Coding RNAs in Cancer. Genom. Proteom. Bioinform. 2016, 14, 42–54. [Google Scholar] [CrossRef]
- Kallen, A.N.; Zhou, X.-B.; Xu, J.; Qiao, C.; Ma, J.; Yan, L.; Lu, L.; Liu, C.; Yi, J.-S.; Zhang, H.; et al. The Imprinted H19 LncRNA Antagonizes Let-7 MicroRNAs. Mol. Cell 2013, 52, 101–112. [Google Scholar] [CrossRef]
- Lam, M.T.Y.; Li, W.; Rosenfeld, M.G.; Glass, C.K. Enhancer RNAs and Regulated Transcriptional Programs. Trends Biochem. Sci. 2014, 39, 170–182. [Google Scholar] [CrossRef]
- Ounzain, S.; Pedrazzini, T. The Promise of Enhancer-Associated Long Noncoding RNAs in Cardiac Regeneration. Trends Cardiovasc. Med. 2015, 25, 592–602. [Google Scholar] [CrossRef]
- Li, W.; Notani, D.; Ma, Q.; Tanasa, B.; Nunez, E.; Chen, A.Y.; Merkurjev, D.; Zhang, J.; Ohgi, K.; Song, X.; et al. Functional Roles of Enhancer RNAs for Oestrogen-Dependent Transcriptional Activation. Nature 2013, 498, 516–520. [Google Scholar] [CrossRef]
- Ørom, U.; Shiekhattar, R. Long Noncoding RNAs Usher in a New Era in the Biology of Enhancers. Cell 2013, 154, 1190–1193. [Google Scholar] [CrossRef]
- Sanyal, A.; Lajoie, B.R.; Jain, G.; Dekker, J. The Long-Range Interaction Landscape of Gene Promoters. Nature 2012, 489, 109–113. [Google Scholar] [CrossRef]
- Li, G.; Ruan, X.; Auerbach, R.K.; Sandhu, K.; Zheng, M.; Wang, P.; Poh, H.; Goh, Y.; Lim, J.; Zhang, J.; et al. Extensive Promoter-Centered Chromatin Interactions Provide a Topological Basis for Transcription Regulation. Cell 2012, 148, 84–98. [Google Scholar] [CrossRef] [PubMed]
- Panis, C.; Cecílio-da-Silva, A.P.; Takakura, E.T.; Jumes, J.J.; Willhelm-dos-Santos, J.; Herrera, A.C.; Victorino, V.J. Breast Cancer in Brazil: Epidemiology and Treatment Challenges. Breast Cancer Targets Ther. 2015, 7, 43. [Google Scholar] [CrossRef]
- Ciriello, G.; Sinha, R.; Hoadley, K.A.; Jacobsen, A.S.; Reva, B.; Perou, C.M.; Sander, C.; Schultz, N. The Molecular Diversity of Luminal a Breast Tumors. Breast Cancer Res. Treat. 2013, 141, 409–420. [Google Scholar] [CrossRef]
- Koboldt, D.C.; Fulton, R.S.; McLellan, M.D.; Schmidt, H.; Kalicki-Veizer, J.; McMichael, J.F.; Fulton, L.L.; Dooling, D.J.; Ding, L.; Mardis, E.R.; et al. Comprehensive Molecular Portraits of Human Breast Tumours. Nature 2012, 490, 61–70. [Google Scholar] [CrossRef]
- Goldhirsch, A.; Winer, E.P.; Coates, A.S.; Gelber, R.D.; Piccart-Gebhart, M.; Thürlimann, B.; Senn, H.-J.; Panel members. Personalizing the Treatment of Women with Early Breast Cancer: Highlights of the St Gallen International Expert Consensus on the Primary Therapy of Early Breast Cancer 2013. Ann. Oncol. Off. J. Eur. Soc. Med. Oncol. 2013, 24, 2206–2223. [Google Scholar] [CrossRef] [PubMed]
- Petrone, I.; Rodrigues, F.R.; Fernandes, P.V.; Abdelhay, E. Immunohistochemical Biomarkers in Ductal Carcinoma in Situ. Open J. Pathol. 2020, 10, 129–146. [Google Scholar] [CrossRef]
- Zhang, T.; Hu, H.; Yan, G.; Wu, T.; Liu, S.; Chen, W.; Ning, Y.; Lu, Z. Long Non-Coding RNA and Breast Cancer. Technol. Cancer Res. Treat. 2019, 18, 153303381984388. [Google Scholar] [CrossRef] [PubMed]
- Nagini, S. Breast Cancer: Current Molecular Therapeutic Targets and New Players. Anti-Cancer Agents Med. Chem. 2017, 17, 152–163. [Google Scholar] [CrossRef] [PubMed]
- Collette, J.; Le Bourhis, X.; Adriaenssens, E. Regulation of Human Breast Cancer by the Long Non-Coding RNA H19. Int. J. Mol. Sci. 2017, 18, 2319. [Google Scholar] [CrossRef]
- Vrba, L.; Futscher, B.W. Epigenetic Silencing of MORT Is an Early Event in Cancer and Is Associated with Luminal, Receptor Positive Breast Tumor Subtypes. J. Breast Cancer 2017, 20, 198. [Google Scholar] [CrossRef]
- Yang, F.; Lv, S.; Lv, L.; Liu, Y.; Dong, S.; Yao, Z.; Dai, X.; Zhang, X.; Wang, O. Identification of LncRNA FAM83H-AS1 as a Novel Prognostic Marker in Luminal Subtype Breast Cancer. OncoTargets Ther. 2016, 9, 7039–7045. [Google Scholar] [CrossRef]
- Marini, A.; Lena, A.M.; Panatta, E.; Ivan, C.; Han, L.; Liang, H.; Annicchiarico-Petruzzelli, M.; Di Daniele, N.; Calin, G.A.; Candi, E.; et al. Ultraconserved Long Non-Coding RNA Uc.63 in Breast Cancer. Oncotarget 2016, 8, 35669–35680. [Google Scholar] [CrossRef] [PubMed]
- Bhan, A.; Hussain, I.; Ansari, K.I.; Kasiri, S.; Bashyal, A.; Mandal, S.S. Antisense-Transcript Long Noncoding RNA (LncRNA) HOTAIR Is Transcriptionally Induced by Estradiol. J. Mol. Biol. 2013, 425, 3707–3722. [Google Scholar] [CrossRef]
- Xue, X.; Yang, Y.A.; Zhang, A.; Fong, K.; Kim, J.; Song, B.; Li, S.; Zhao, J.C.; Yu, J. LncRNA HOTAIR Enhances ER Signaling and Confers Tamoxifen Resistance in Breast Cancer. Oncogene 2016, 35, 2746–2755. [Google Scholar] [CrossRef]
- Guffanti, A.; Iacono, M.; Pelucchi, P.; Kim, N.; Soldà, G.; Croft, L.J.; Taft, R.J.; Rizzi, E.; Askarian-Amiri, M.; Bonnal, R.J.; et al. A Transcriptional Sketch of a Primary Human Breast Cancer by 454 Deep Sequencing. BMC Genom. 2009, 10, 163. [Google Scholar] [CrossRef]
- Zhang, Y.; Wang, D.-L.; Yan, H.-Y.; Liao, J.-Y.; He, J.-H.; Hu, K.-S.; Deng, W.-X.; Wang, Y.-J.; Xing, H.-T.; Koeffler, H.P.; et al. Genome-Wide Study of ER-Regulated LncRNAs Shows AP000439.3 May Function as a Key Regulator of Cell Cycle in Breast Cancer. Oncol. Rep. 2017, 38, 3227–3237. [Google Scholar] [CrossRef] [PubMed]
- Dong, H.; Wang, W.; Mo, S.; Liu, Q.; Chen, X.; Chen, R.; Zhang, Y.; Zou, K.; Ye, M.; He, X.; et al. Long Non-Coding RNA SNHG14 Induces Trastuzumab Resistance of Breast Cancer via Regulating PABPC1 Expression through H3K27 Acetylation. J. Cell. Mol. Med. 2018, 22, 4935–4947. [Google Scholar] [CrossRef]
- Augoff, K.; McCue, B.; Plow, E.F.; Sossey-Alaoui, K. MiR-31 and Its Host Gene LncRNA LOC554202 Are Regulated by Promoter Hypermethylation in Triple-Negative Breast Cancer. Mol. Cancer 2012, 11, 1–13. [Google Scholar] [CrossRef]
- Tian, T.; Gong, Z.; Wang, M.; Hao, R.; Lin, S.; Liu, K.; Guan, F.; Xu, P.; Deng, Y.; Song, D.; et al. Identification of Long Non-Coding RNA Signatures in Triple-Negative Breast Cancer. Cancer Cell Int. 2018, 18, 1–10. [Google Scholar] [CrossRef]
- Nekhlyudov, L.; Habel, L.A.; Achacoso, N.; Jung, I.; Haque, R.; Collins, L.C.; Schnitt, S.J.; Quesenberry, C.P.; Fletcher, S.W. Ten-Year Risk of Diagnostic Mammograms and Invasive Breast Procedures after Breast-Conserving Surgery for DCIS. J. Natl. Cancer Inst. 2012, 104, 614–621. [Google Scholar] [CrossRef]
- Sanders, M.E.; Schuyler, P.A.; Simpson, J.F.; Page, D.L.; Dupont, W.D. Continued Observation of the Natural History of Low-Grade Ductal Carcinoma in Situ Reaffirms Proclivity for Local Recurrence Even after More than 30 Years of Follow-Up. Mod. Pathol. 2014, 28, 662–669. [Google Scholar] [CrossRef]
- Allred, D.C. Ductal Carcinoma in Situ: Terminology, Classification, and Natural History. J. Natl. Cancer Institute. Monogr. 2010, 2010, 134–138. [Google Scholar] [CrossRef] [PubMed]
- Abba, M.C.; Gong, T.; Lu, Y.; Lee, J.; Zhong, Y.; Lacunza, E.; Butti, M.; Takata, Y.; Gaddis, S.; Shen, J.; et al. A Molecular Portrait of High-Grade Ductal Carcinoma in Situ. Cancer Res. 2015, 75, 3980–3990. [Google Scholar] [CrossRef] [PubMed]
- Bhan, A.; Mandal, S.S. LncRNA HOTAIR: A Master Regulator of Chromatin Dynamics and Cancer. Biochim. Et Biophys. Acta (BBA)-Rev. Cancer 2015, 1856, 151–164. [Google Scholar] [CrossRef]
- Yoon, J.-H.; Abdelmohsen, K.; Kim, J.; Yang, X.; Martindale, J.L.; Tominaga-Yamanaka, K.; White, E.J.; Orjalo, A.V.; Rinn, J.L.; Kreft, S.G.; et al. Scaffold Function of Long Non-Coding RNA HOTAIR in Protein Ubiquitination. Nat. Commun. 2013, 4, 2939. [Google Scholar] [CrossRef]
- Cantile, M.; Di Bonito, M.; Tracey De Bellis, M.; Botti, G. Functional Interaction among LncRNA HOTAIR and MicroRNAs in Cancer and Other Human Diseases. Cancers 2021, 13, 570. [Google Scholar] [CrossRef] [PubMed]
- Rinn, J.L.; Kertesz, M.; Wang, J.K.; Squazzo, S.L.; Xu, X.; Brugmann, S.A.; Goodnough, L.H.; Helms, J.A.; Farnham, P.J.; Segal, E.; et al. Functional Demarcation of Active and Silent Chromatin Domains in Human HOX Loci by Noncoding RNAs. Cell 2007, 129, 1311–1323. [Google Scholar] [CrossRef]
- Zhang, Z.; Weaver, D.L.; Olsen, D.; deKay, J.; Peng, Z.; Ashikaga, T.; Evans, M.F. Long Non-Coding RNA Chromogenic in Situ Hybridisation Signal Pattern Correlation with Breast Tumour Pathology. J. Clin. Pathol. 2016, 69, 76–81. [Google Scholar] [CrossRef]
- Cantile, M.; Di Bonito, M.; Cerrone, M.; Collina, F.; De Laurentiis, M.; Botti, G. Long Non-Coding RNA HOTAIR in Breast Cancer Therapy. Cancers 2020, 12, 1197. [Google Scholar] [CrossRef] [PubMed]
- Abba, M.C.; Fabre, M.L.; Lee, J.; Tatineni, P.; Kil, H.; Aldaz, C.M. HOTAIR Modulated Pathways in Early-Stage Breast Cancer Progression. Front. Oncol. 2021, 11, 783211. [Google Scholar] [CrossRef]
- Gupta, R.A.; Shah, N.; Wang, K.C.; Kim, J.; Horlings, H.M.; Wong, D.J.; Tsai, M.-C.; Hung, T.; Argani, P.; Rinn, J.L.; et al. Long Non-Coding RNA HOTAIR Reprograms Chromatin State to Promote Cancer Metastasis. Nature 2010, 464, 1071–1076. [Google Scholar] [CrossRef]
- Gökmen-Polar, Y.; Vladislav, I.T.; Neelamraju, Y.; Janga, S.C.; Badve, S. Prognostic Impact of HOTAIR Expression Is Restricted to ER-Negative Breast Cancers. Sci. Rep. 2015, 5, 8765. [Google Scholar] [CrossRef]
- Pádua Alves, C.; Fonseca, A.S.; Muys, B.R.; de Barros, E.; Lima Bueno, R.; Bürger, M.C.; de Souza, J.E.S.; Valente, V.; Zago, M.A.; Silva, W.A. Brief Report: The LincRNA HOTAIR Is Required for Epithelial-To-Mesenchymal Transition and Stemness Maintenance of Cancer Cell Lines. Stem Cells (Dayton, Ohio) 2013, 31, 2827–2832. [Google Scholar] [CrossRef]
- Abba, M.C.; Canzoneri, R.; Gurruchaga, A.; Lee, J.; Tatineni, P.; Kil, H.; Lacunza, E.; Aldaz, C.M. LINC00885 a Novel Oncogenic Long Non-Coding RNA Associated with Early Stage Breast Cancer Progression. Int. J. Mol. Sci. 2020, 21, 7407. [Google Scholar] [CrossRef] [PubMed]
- Chen, H.; Chi, Y.; Chen, M.; Zhao, L. Long Intergenic Non-Coding RNA LINC00885 Promotes Tumorigenesis of Cervical Cancer by Upregulating MACC1 Expression through Serving as a Competitive Endogenous RNA for MicroRNA-432-5p. Cancer Manag. Res. 2021, 13, 1435–1447. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Tu, H.; Zhang, L.; Xiong, J.; Li, L. FOXP3-Induced LINC00885 Promotes the Proliferation and Invasion of Cervical Cancer Cells. Mol. Med. Rep. 2021, 23, 12097. [Google Scholar] [CrossRef] [PubMed]
- Li, M.; Liu, Y.; Zhang, X.; Liu, J.; Wang, P. Transcriptomic Analysis of High-Throughput Sequencing about CircRNA, LncRNA and MRNA in Bladder Cancer. Gene 2018, 677, 189–197. [Google Scholar] [CrossRef] [PubMed]
- Amsterdam, A. Epiregulin as a Marker for the Initial Steps of Ovarian Cancer Development. Int. J. Oncol. 2011, 39, 1165–1172. [Google Scholar] [CrossRef]
- Keshet, Y.; Seger, R. The MAP Kinase Signaling Cascades: A System of Hundreds of Components Regulates a Diverse Array of Physiological Functions. MAP Kinase Signal. Protoc. 2010, 661, 3–38. [Google Scholar] [CrossRef]
- Zhang, X.; Zhang, Q.; Liu, G. Genome-Wide Analysis of the FOXA1 Transcriptional Regulatory Network Identifies Super Enhancer Associated LncRNAs in Tamoxifen Resistance. Front. Genet. 2022, 13, 992444. [Google Scholar] [CrossRef]
- DeVaux, R.S.; Ropri, A.S.; Grimm, S.L.; Hall, P.A.; Herrera, E.O.; Chittur, S.V.; Smith, W.P.; Coarfa, C.; Behbod, F.; Herschkowitz, J.I. Long Noncoding RNA BHLHE40-AS1 Promotes Early Breast Cancer Progression through Modulating IL-6/STAT3 Signaling. J. Cell. Biochem. 2020, 121, 3465–3478. [Google Scholar] [CrossRef] [PubMed]
- Navarro Gonzalez, J.; Zweig, A.S.; Speir, M.L.; Schmelter, D.; Rosenbloom, K.; Raney, B.J.; Powell, C.C.; Nassar, L.R.; Maulding, N.; Lee, C.M.; et al. The UCSC Genome Browser Database: 2021 Update. Nucleic Acids Res. 2020, 50.D1, D1115–D1122. [Google Scholar] [CrossRef]
- Dethlefsen, C.; Højfeldt, G.; Hojman, P. The Role of Intratumoral and Systemic IL-6 in Breast Cancer. Breast Cancer Res. Treat. 2013, 138, 657–664. [Google Scholar] [CrossRef]
- Johnson, D.E.; O’Keefe, R.A.; Grandis, J.R. Targeting the IL-6/JAK/STAT3 Signalling Axis in Cancer. Nat. Rev. Clin. Oncol. 2018, 15, 234–248. [Google Scholar] [CrossRef]
- Segatto, I.; Baldassarre, G.; Belletti, B. STAT3 in Breast Cancer Onset and Progression: A Matter of Time and Context. Int. J. Mol. Sci. 2018, 19, 2818. [Google Scholar] [CrossRef]
- Yu, H.; Lee, H.; Herrmann, A.; Buettner, R.; Jove, R. Revisiting STAT3 Signalling in Cancer: New and Unexpected Biological Functions. Nat. Rev. Cancer 2014, 14, 736–746. [Google Scholar] [CrossRef] [PubMed]
- Fabre, M.L.; Canzoneri, R.; Gurruchaga, A.; Lee, J.; Tatineni, P.; Kil, H.; Lacunza, E.; Aldaz, C.M.; Abba, M.C. MALINC1 an Immune-Related Long Non-Coding RNA Associated with Early-Stage Breast Cancer Progression. Cancers 2022, 14, 2819. [Google Scholar] [CrossRef]
- Bida, O.; Gidoni, M.; Ideses, D.; Efroni, S.; Ginsberg, D. A Novel Mitosis-Associated LncRNA, MALINC 1, Is Required for Cell Cycle Progression and Sensitizes Cancer Cells to Paclitaxel. Oncotarget 2015, 6, 27880. [Google Scholar] [CrossRef]
- Eckert, R.L.; Adhikary, G.; Young, C.A.; Jans, R.; Crish, J.F.; Xu, W.; Rorke, E.A. AP1 Transcription Factors in Epidermal Differentiation and Skin Cancer. J. Ski. Cancer 2013, 2013, 1–9. [Google Scholar] [CrossRef]
- Vierbuchen, T.; Ling, E.; Cowley, C.J.; Couch, C.H.; Wang, X.; Harmin, D.A.; Roberts, C.W.M.; Greenberg, M.E. AP-1 Transcription Factors and the BAF Complex Mediate Signal-Dependent Enhancer Selection. Mol. Cell 2017, 68, 1067–1082.e12. [Google Scholar] [CrossRef]
- Baan, B.; Pardali, E.; ten Dijke, P.; van Dam, H. In Situ Proximity Ligation Detection of C-Jun/AP-1 Dimers Reveals Increased Levels of C-Jun/Fra1 Complexes in Aggressive Breast Cancer Cell Lines in Vitro and in Vivo. Mol. Cell. Proteom. MCP 2010, 9, 1982–1990. [Google Scholar] [CrossRef]
- Wang, Y.; Xu, H.; Zhu, B.; Qiu, Z.; Lin, Z. Systematic Identification of the Key Candidate Genes in Breast Cancer Stroma. Cell. Mol. Biol. Lett. 2018, 23, 1–5. [Google Scholar] [CrossRef]
- Binato, R.; Corrêa, S.; Panis, C.; Ferreira, G.; Petrone, I.; da Costa, I.R.; Abdelhay, E. NRIP1 Is Activated by C-JUN/C-FOS and Activates the Expression of PGR, ESR1 and CCND1 in Luminal a Breast Cancer. Sci. Rep. 2021, 11, 21159. [Google Scholar] [CrossRef] [PubMed]
- Abba, M.C.; Hu, Y.; Sun, H.; Drake, J.A.; Gaddis, S.; Baggerly, K.; Sahin, A.; Aldaz, C.M. Gene expression signature of estrogen receptor α status in breast cancer. BMC Genom. 2005, 6, 37. [Google Scholar] [CrossRef]
- Samavat, H.; Kurzer, M.S. Estrogen Metabolism and Breast Cancer. Cancer Lett. 2015, 356, 231–243. [Google Scholar] [CrossRef] [PubMed]
- Starek-Świechowicz, B.; Budziszewska, B.; Starek, A. Endogenous Estrogens—Breast Cancer and Chemoprevention. Pharmacol. Rep. 2021, 73, 1497–1512. [Google Scholar] [CrossRef] [PubMed]
- Clarke, R.B.; Anderson, E.; Howell, A. Steroid Receptors in Human Breast Cancer. Trends Endocrinol. Metab. TEM 2004, 15, 316–323. [Google Scholar] [CrossRef]
- Lesurf, R.; Aure, M.R.; Mørk, H.H.; Vitelli, V.; Oslo Breast Cancer Research Consortium (OSBREAC); Lundgren, S.; Børresen-Dale, A.-L.; Kristensen, V.; Wärnberg, F.; Hallett, M.; et al. Molecular Features of Subtype-Specific Progression from Ductal Carcinoma in Situ to Invasive Breast Cancer. Cell Rep. 2016, 16, 1166–1179. [Google Scholar] [CrossRef] [PubMed]
- Mansour, M.R.; Abraham, B.J.; Anders, L.; Berezovskaya, A.; Gutierrez, A.; Durbin, A.D.; Etchin, J.; Lawton, L.; Sallan, S.E.; Silverman, L.B.; et al. An Oncogenic Super-Enhancer Formed through Somatic Mutation of a Noncoding Intergenic Element. Science 2014, 346, 1373–1377. [Google Scholar] [CrossRef]
- Tang, F.; Yang, Z.; Tan, Y.; Li, Y. Super-Enhancer Function and Its Application in Cancer Targeted Therapy. NPJ Precis. Oncol. 2020, 4, 2. [Google Scholar] [CrossRef]
- Hnisz, D.; Abraham, B.J.; Lee, T.I.; Lau, A.; Saint-André, V.; Sigova, A.A.; Hoke, H.; Young, R.A. Transcriptional Super-Enhancers Connected to Cell Identity and Disease. Cell 2013, 155, 934–947. [Google Scholar] [CrossRef]
- Hou, Y.; Zhang, R.; Sun, X. Enhancer LncRNAs Influence Chromatin Interactions in Different Ways. Front. Genet. 2019, 10, 936. [Google Scholar] [CrossRef]
- Ropri, A.S.; DeVaux, R.S.; Eng, J.; Chittur, S.V.; Herschkowitz, J.I. Cis-Acting Super-Enhancer LncRNAs as Biomarkers to Early-Stage Breast Cancer. Breast Cancer Res. 2021, 23, 1–18. [Google Scholar] [CrossRef]
- Wang, X.; Saso, H.; Iwamoto, T.; Xia, W.; Gong, Y.; Pusztai, L.; Woodward, W.A.; Reuben, J.M.; Warner, S.L.; Bearss, D.J.; et al. TIG1 Promotes the Development and Progression of Inflammatory Breast Cancer through Activation of Axl Kinase. Cancer Res. 2013, 73, 6516–6525. [Google Scholar] [CrossRef] [PubMed]
- Peng, Z.; Shen, R.; Li, Y.-W.; Teng, K.-Y.; Shapiro, C.L.; Lin, H.-J.L. Epigenetic Repression of RARRES1 Is Mediated by Methylation of a Proximal Promoter and a Loss of CTCF Binding. PLoS ONE 2012, 7, e36891. [Google Scholar] [CrossRef]
- Pavlova, N.N.; Thompson, C.B. The Emerging Hallmarks of Cancer Metabolism. Cell Metab. 2016, 23, 27–47. [Google Scholar] [CrossRef]
- Wang, Z.; Humphries, B.; Xiao, H.; Jiang, Y.; Yang, C. MicroRNA-200b Suppresses Arsenic-Transformed Cell Migration by Targeting Protein Kinase Cα and Wnt5b-Protein Kinase Cα Positive Feedback Loop and Inhibiting Rac1 Activation. J. Biol. Chem. 2014, 289, 18373–18386. [Google Scholar] [CrossRef]
- Chang, S.-H.; Lu, Y.-C.; Li, X.; Hsieh, W.-Y.; Xiong, Y.; Ghosh, M.; Evans, T.; Elemento, O.; Hla, T. Antagonistic Function of the RNA-Binding Protein HuR and MiR-200b in Post-Transcriptional Regulation of Vascular Endothelial Growth Factor-A Expression and Angiogenesis. J. Biol. Chem. 2013, 288, 4908–4921. [Google Scholar] [CrossRef]
- Gregory, P.A.; Bert, A.G.; Paterson, E.L.; Barry, S.C.; Tsykin, A.; Farshid, G.; Vadas, M.A.; Khew-Goodall, Y.; Goodall, G.J. The MiR-200 Family and MiR-205 Regulate Epithelial to Mesenchymal Transition by Targeting ZEB1 and SIP1. Nat. Cell Biol. 2008, 10, 593–601. [Google Scholar] [CrossRef]
- Tsai, M.-C.; Manor, O.; Wan, Y.; Mosammaparast, N.; Wang, J.K.; Lan, F.; Shi, Y.; Segal, E.; Chang, H.Y. Long Noncoding RNA as Modular Scaffold of Histone Modification Complexes. Science 2010, 329, 689–693. [Google Scholar] [CrossRef]
- Wang, L.; Zeng, X.; Chen, S.; Ding, L.; Zhong, J.; Zhao, J.C.; Wang, L.; Sarver, A.; Koller, A.; Zhi, J.; et al. BRCA1 Is a Negative Modulator of the PRC2 Complex. EMBO J. 2013, 32, 1584–1597. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.-H.; Sun, M.; Nie, F.-Q.; Ge, Y.-B.; Zhang, E.-B.; Yin, D.-D.; Kong, R.; Xia, R.; Lu, K.-H.; Li, J.-H.; et al. Lnc RNA HOTAIR Functions as a Competing Endogenous RNA to Regulate HER2 Expression by Sponging MiR-331-3p in Gastric Cancer. Mol. Cancer 2014, 13, 92. [Google Scholar] [CrossRef]
- Shibata, H.; Spencer, T.E.; Oñate, S.A.; Jenster, G.; Tsai, S.Y.; Tsai, M.J.; O’Malley, B.W. Role of Co-Activators and Co-Repressors in the Mechanism of Steroid/Thyroid Receptor Action. Recent Prog. Horm. Res. 1997, 52, 141–164. [Google Scholar]
- Jarroux, J.; Foretek, D.; Bertrand, C.; Gabriel, M.; Szachnowski, U.; Saci, Z.; Guo, S.; Londoño-Vallejo, A.; Pinskaya, M.; Morillon, A. HOTAIR LncRNA Promotes Epithelial–Mesenchymal Transition by Redistributing LSD1 at Regulatory Chromatin Regions. EMBO Rep. 2021, 22, e50193. [Google Scholar] [CrossRef] [PubMed]
- Trinh, A.; Gil Del Alcazar, C.R.; Shukla, S.A.; Chin, K.; Chang, Y.H.; Thibault, G.; Eng, J.; Jovanović, B.; Aldaz, C.M.; Park, S.Y.; et al. Genomic Alterations during the in Situ to Invasive Ductal Breast Carcinoma Transition Shaped by the Immune System. Mol. Cancer Res. 2021, 19, 623–635. [Google Scholar] [CrossRef] [PubMed]
- Foletta, V.C.; Segal, D.H.; Cohen, D.R. Transcriptional Regulation in the Immune System: All Roads Lead to AP-1. J. Leukoc. Biol. 1998, 63, 139–152. [Google Scholar] [CrossRef] [PubMed]
- Jones, L.M.; Broz, M.L.; Ranger, J.J.; Ozcelik, J.; Ahn, R.; Zuo, D.; Ursini-Siegel, J.; Hallett, M.T.; Krummel, M.; Muller, W.J. STAT3 Establishes an Immunosuppressive Microenvironment during the Early Stages of Breast Carcinogenesis to Promote Tumor Growth and Metastasis. Cancer Res. 2015, 76, 1416–1428. [Google Scholar] [CrossRef] [PubMed]
- Chandra Gupta, S.; Nandan Tripathi, Y. Potential of Long Non-Coding RNAs in Cancer Patients: From Biomarkers to Therapeutic Targets. Int. J. Cancer 2016, 140, 1955–1967. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Petrone, I.; Santos, E.C.d.; Binato, R.; Abdelhay, E. Epigenetic Alterations in DCIS Progression: What Can lncRNAs Teach Us? Int. J. Mol. Sci. 2023, 24, 8733. https://doi.org/10.3390/ijms24108733
Petrone I, Santos ECd, Binato R, Abdelhay E. Epigenetic Alterations in DCIS Progression: What Can lncRNAs Teach Us? International Journal of Molecular Sciences. 2023; 24(10):8733. https://doi.org/10.3390/ijms24108733
Chicago/Turabian StylePetrone, Igor, Everton Cruz dos Santos, Renata Binato, and Eliana Abdelhay. 2023. "Epigenetic Alterations in DCIS Progression: What Can lncRNAs Teach Us?" International Journal of Molecular Sciences 24, no. 10: 8733. https://doi.org/10.3390/ijms24108733
APA StylePetrone, I., Santos, E. C. d., Binato, R., & Abdelhay, E. (2023). Epigenetic Alterations in DCIS Progression: What Can lncRNAs Teach Us? International Journal of Molecular Sciences, 24(10), 8733. https://doi.org/10.3390/ijms24108733






