DT-PICS: An Efficient and Cost-Effective SNP Selection Method for the Germplasm Identification of Arabidopsis
Abstract
1. Introduction
2. Results
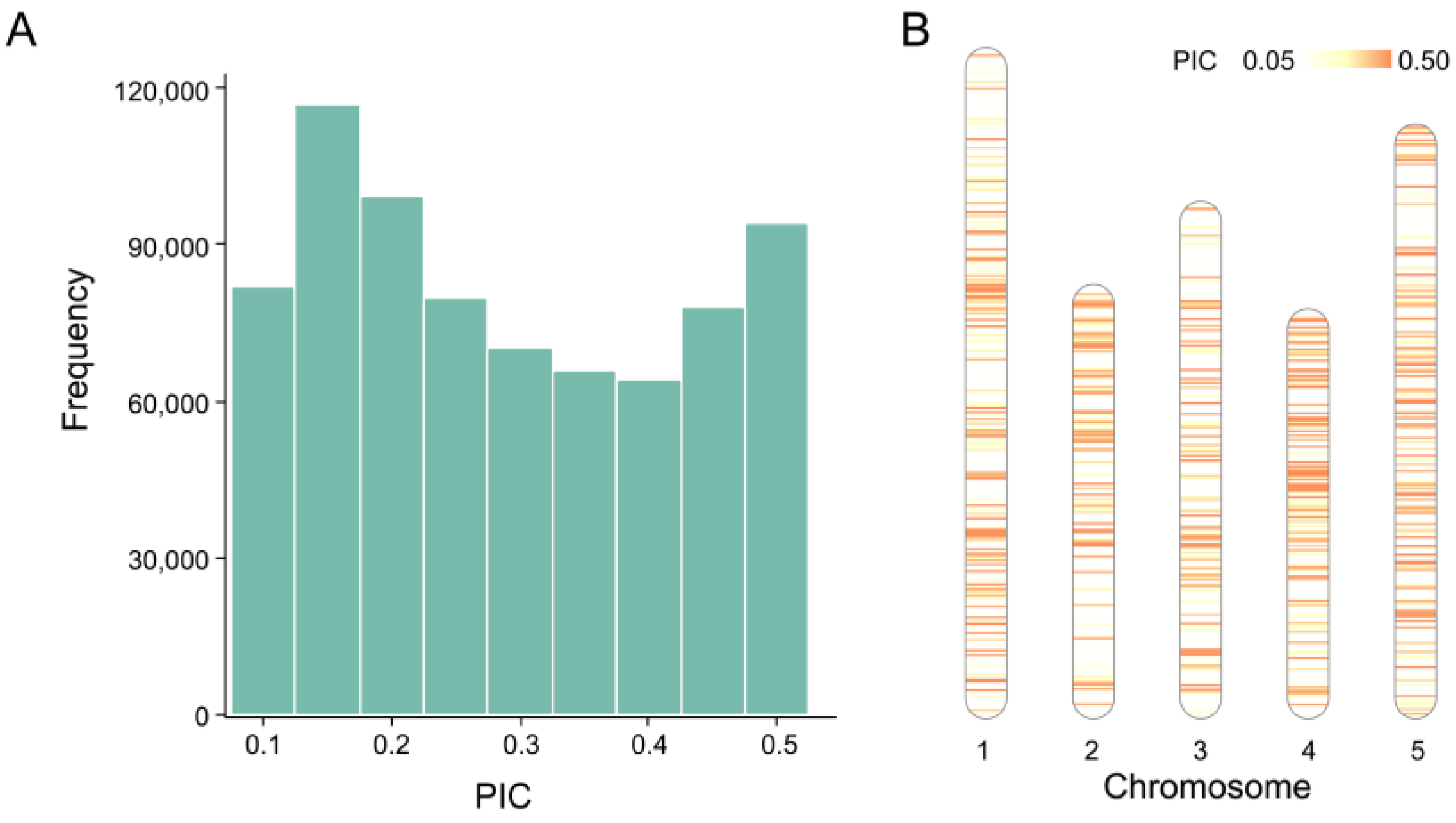
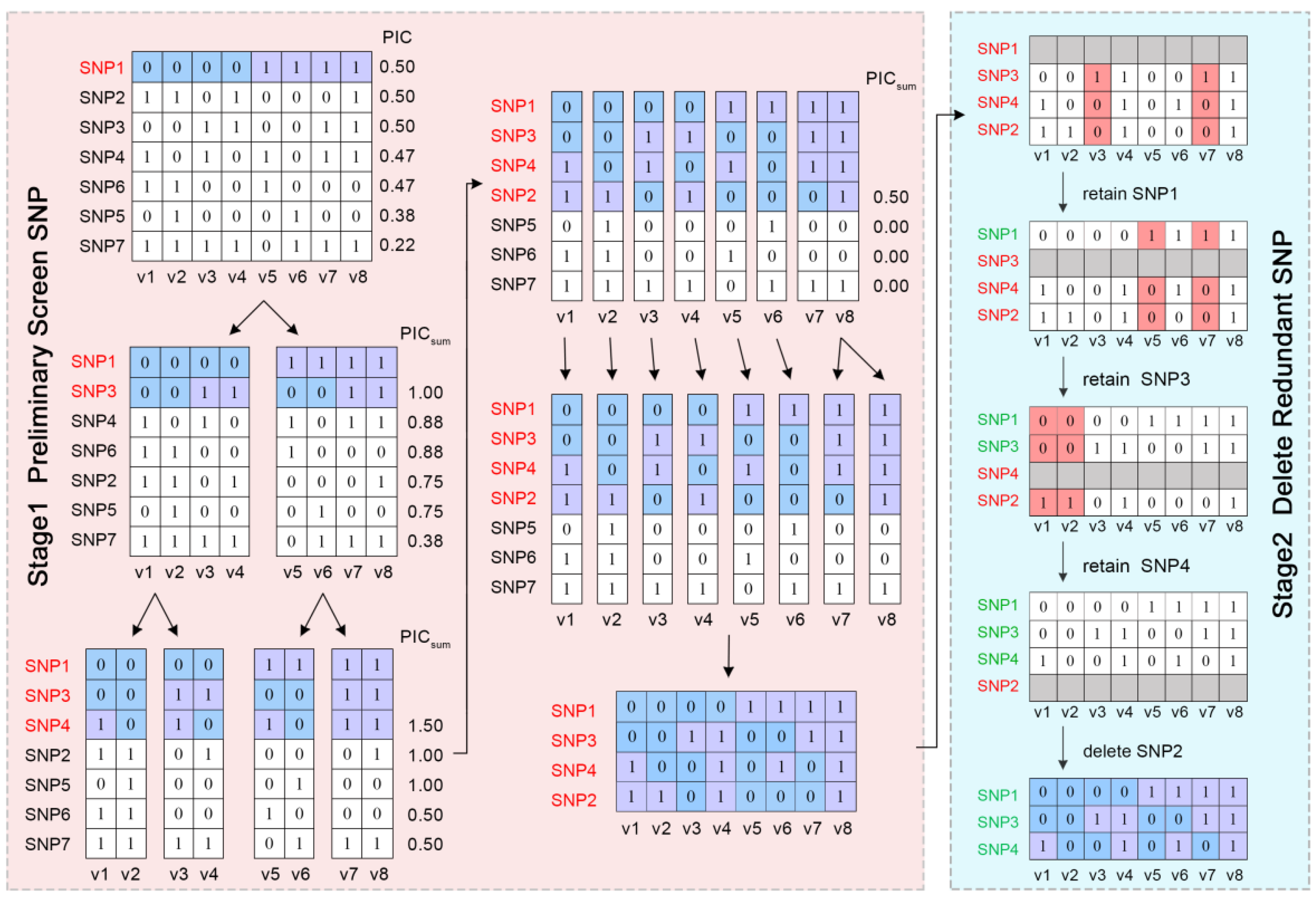
2.1. Characteristics of SNPs in Training Data
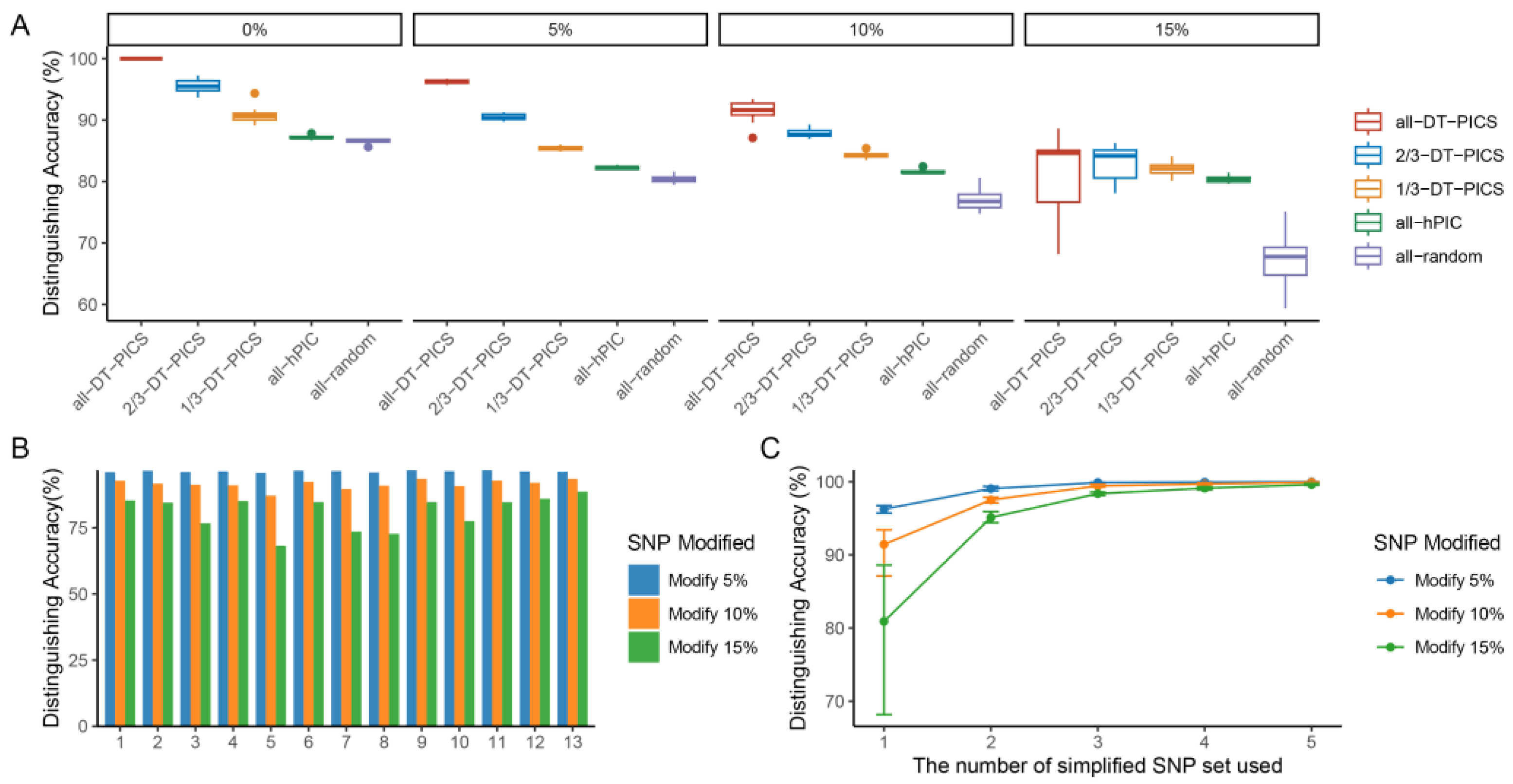
2.2. Fingerprint of 1135 Arabidopsis Varieties Established by DT-PICS
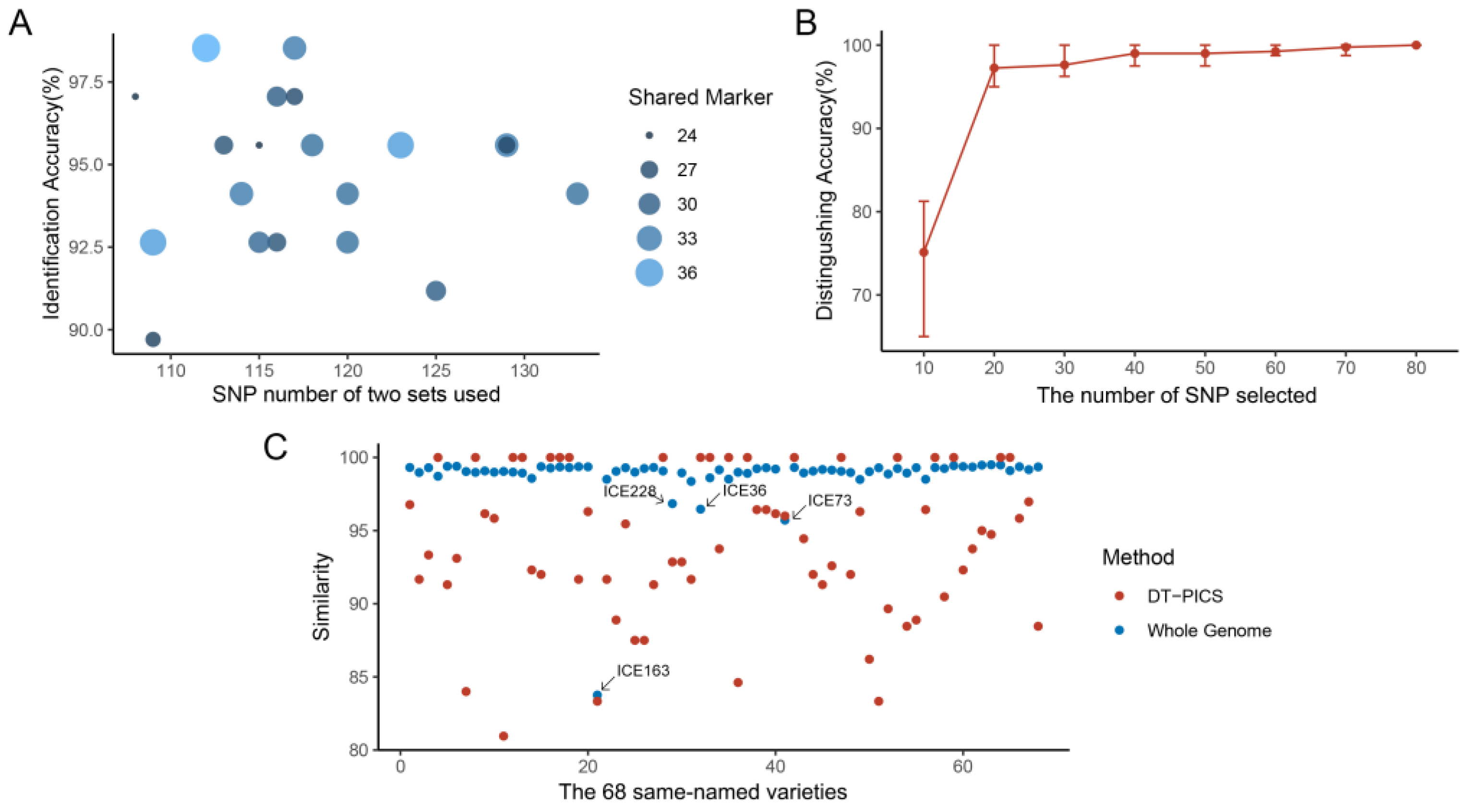
2.3. Fingerprint of the Identification Power of Same-Named and Different-Named Varieties in Test Datasets
2.4. Generation of QR Codes
3. Discussion
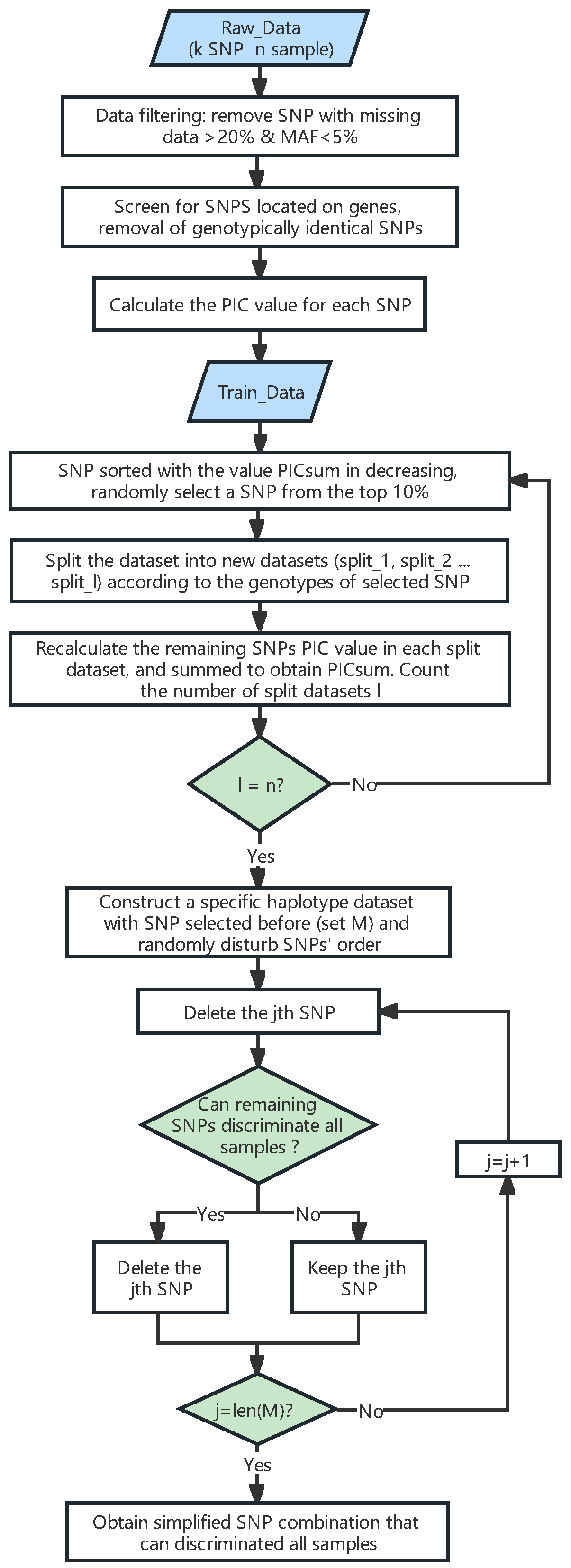
4. Materials and Methods
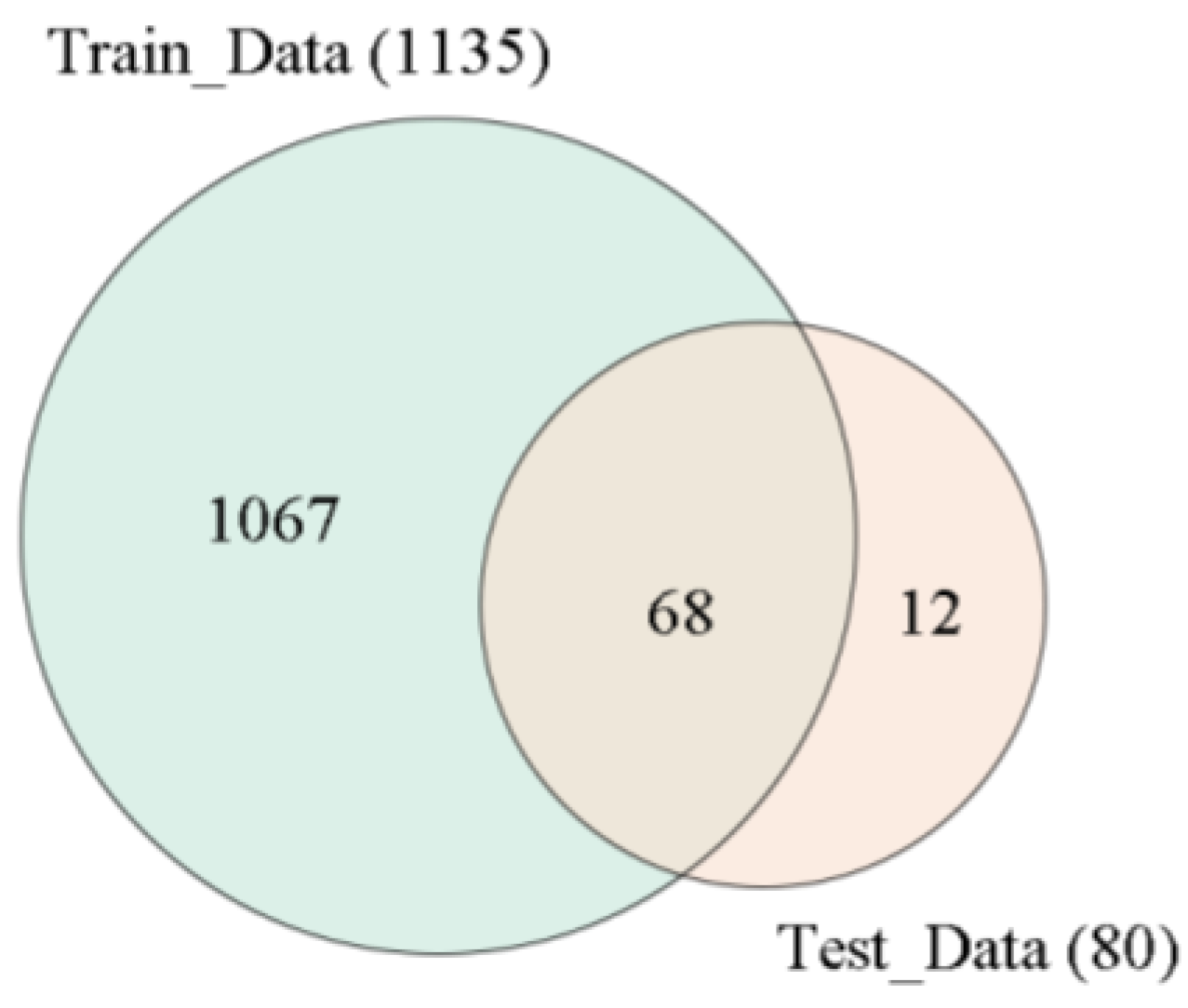
4.1. Genotype Dataset of Materials
4.2. Marker Polymorphism Analysis
4.3. Selecting SNPs from the Training Dataset to Construct a Fingerprint Map
4.4. Variety Identification in the Training Set Using the DT-PICS Method
4.5. Independent Testing of Variety Identification
4.6. Generation of QR Codes
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Parry, G.; Provart, N.J.; Brady, S.M.; Uzilday, B.; Adams, K.; Araújo, W.; The Multinational Arabidopsis Steering Committee. Current status of the multinational Arabidopsis community. Plant Direct 2020, 4, e00248. [Google Scholar] [CrossRef]
- Pisupati, R.; Reichardt, I.; Seren, Ü.; Korte, P.; Nizhynska, V.; Kerdaffrec, E.; Uzunova, K.; Rabanal, F.A.; Filiault, D.L.; Nordborg, M.; et al. Verification of Arabidopsis stock collectionpsis stock collections using SNPmatch, a tool for genotyping high-plexed samples. Sci. Data 2017, 4, 170184. [Google Scholar] [CrossRef] [PubMed]
- Simon, M.; Simon, A.; Martins, F.; Botran, L.; Tisné, S.; Granier, F.; Loudet, O.; Camilleri, C. DNA fingerprinting and new tools for fine-scale discrimination of Arabidopsis thaliana accessions. Plant J. Cell Mol. Biol. 2012, 69, 1094–1101. [Google Scholar] [CrossRef] [PubMed]
- El Bakkali, A.; Essalouh, L.; Tollon, C.; Rivallan, R.; Mournet, P.; Moukhli, A.; Zaher, H.; Mekkaoui, A.; Hadidou, A.; Sikaoui, L.; et al. Characterization of worldwide olive germplasm banks of Marrakech (Morocco) and Córdoba (Spain): Towards management and use of olive germplasm in breeding programs. PLoS ONE 2019, 14, e0223716. [Google Scholar] [CrossRef] [PubMed]
- Dar, A.A.; Mahajan, R.; Sharma, S. Molecular markers for characterization and conservation of plant genetic resources. Indian J. Agric. Sci. 2019, 89, 1755–1763. [Google Scholar] [CrossRef]
- Wu, C.C.; Chang, S.H.; Tung, C.W.; Ho, C.K.; Gogorcena, Y.; Chu, F.H. Identification of hybridization and introgression between Cinnamomum kanehirae Hayata and C. camphora (L.) Presl using genotyping-by-sequencing. Sci. Rep. 2020, 10, 15995. [Google Scholar] [CrossRef]
- Morales, K.Y.; Singh, N.; Perez, F.A.; Ignacio, J.C.; Thapa, R.; Arbelaez, J.D.; Tabien, R.E.; Famoso, A.; Wang, D.R.; Septiningsih, E.M.; et al. An improved 7K SNP array, the C7AIR, provides a wealth of validated SNP markers for rice breeding and genetics studies. PLoS ONE 2020, 15, e0232479. [Google Scholar] [CrossRef]
- Zhang, J.; Yang, J.; Zhang, L.; Luo, J.; Zhao, H.; Zhang, J.; Wen, C. A new SNP genotyping technology Target SNP-seq and its application in genetic analysis of cucumber varieties. Sci. Rep. 2020, 10, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Song, Q.; Hyten, D.L.; Jia, G.; Quigley, C.V.; Fickus, E.W.; Nelson, R.L.; Cregan, P.B. Fingerprinting soybean germplasm and its utility in genomic research. G3 Genes Genomes Genet. 2015, 5, 1999–2006. [Google Scholar] [CrossRef]
- Melo, A.T.O.; Bartaula, R.; Hale, I. GBS-SNP-CROP: A reference-optional pipeline for SNP discovery and plant germplasm characterization using variable length, paired-end genotyping-by-sequencing data. BMC Bioinformatics 2016, 17, 29. [Google Scholar] [CrossRef]
- Singh, R.; Iquebal, M.A.; Mishra, C.N.; Jaiswal, S.; Kumar, D.; Raghav, N.; Paul, S.; Sheoran, S.; Sharma, P.; Gupta, A.; et al. Development of model web-server for crop variety identifcation using throughput SNP genotyping data. Sci. Rep. 2019, 9, 5122. [Google Scholar] [CrossRef]
- Carvalho, J.; Yadav, S.; Garrido-Maestu, A.; Azinheiro, S.; Trujillo, I.; Barros-Velázquez, J.; Prado, M. Evaluation of simple sequence repeats (SSR) and single nucleotide polymorphism (SNP)-based methods in olive varieties from the Northwest of Spain and potential for miniaturization. Food Chem. Mol. Sci. 2021, 3, 2666–5662. [Google Scholar] [CrossRef]
- Valliyodan, B.; Brown, A.V.; Wang, J.; Patil, G.; Liu, Y.; Otyama, P.I.; Nelson, R.T.; Vuong, T.; Song, Q.; Musket, T.A.; et al. Genetic variation among 481 diverse soybean accessions, inferred from genomic re-sequencing. Sci. Data 2021, 8, 1–9. [Google Scholar] [CrossRef]
- Ellis, D.; Chavez, O.; Coombs, J.; Soto, J.; Gomez, R.; Douches, D.; Panta, A.; Silvestre, R.; Noelle, L. Genetic identity in genebanks: Application of the SolCAP 12K SNP array in fingerprinting and diversity analysis in the global in trust potato collection. Genome 2018, 61, 523–537. [Google Scholar] [CrossRef]
- Zhang, Z.; Xie, W.; Zhang, J.; Wang, N.; Zhao, Y.; Wang, Y.; Bai, S. Construction of the first high-density genetic linkage map and identification of seed yield-related QTLs and candidate genes in Elymus sibiricus, an important forage grass in Qinghai-Tibet Plateau. BMC Genomics 2019, 20, 861. [Google Scholar] [CrossRef]
- Serrote, C.M.L.; Reiniger, L.R.S.; Silva, K.B.; Rabaiolli, S.M.D.S.; Stefanel, C.M. Determining the Polymorphism Information Content of a molecular marker. Gene 2020, 726, 144–175. [Google Scholar] [CrossRef] [PubMed]
- Priyanka; Kumar, D. Decision tree classifier: A detailed survey. Int. J. Inf. Decis. Sci. 2020, 12, 246–269. [Google Scholar] [CrossRef]
- Song, Y.Y.; Lu, Y. Decision tree methods: Applications for classification and prediction. Shanghai Arch. Psychiatry 2015, 27, 130–135. [Google Scholar] [PubMed]
- Vasseur, F.; Sartori, K.; Baron, E.; Fort, F.; Kazakou, E.; Segrestin, J.; Garnier, E.; Vile, D.; Violle, C. Climate as a driver of adaptive variations in ecological strategies in Arabidopsis thaliana. Ann. Bot. 2018, 122, 935–945. [Google Scholar] [CrossRef] [PubMed]
- ZhiYuan, L.; HaiLong, Y.; ZhiYuan, F.; LiMei, Y.; YuMei, L.; Mu, Z.; Zhang, Y. Development of SNP markers in cabbage and construction of DNA fingerprinting of main varieties. Sci. Agric. Sin. 2018, 51, 2771–2787. [Google Scholar] [CrossRef]
- Zhao, R.X.; Li, S.Y.; Guo, R.X.; Zeng, X.H.; Wen, J.; Ma, C.Z.; Shen, J.X.; Tu, J.X.; Fu, T.D.; Yi, B. Construction of DNA fingerprinting for Brassica napus varieties based on SNP chip. Zuowu Xuebao (Acta Agron. Sin.) 2018, 44, 956–965. [Google Scholar] [CrossRef]
- GuoZhong, Z.; Fang, Z.; Jie, F.; LeChen, L.; ErLi, N.; WangZhen, G. Genome-wide screening and evaluation of SNP core loci for identification of upland cotton varieties. Acta Agron. Sin. 2018, 44, 1631–1639. [Google Scholar] [CrossRef]
- Kawakatsu, T.; Huang, S.-S.C.; Jupe, F.; Sasaki, E.; Schmitz, R.; Urich, M.; Castanon, R.; Nery, J.; Barragan, C.; He, Y.; et al. Epigenomic Diversity in a Global Collection of Arabidopsis thaliana Accessions. Cell 2016, 166, 492–505. [Google Scholar] [CrossRef] [PubMed]
- Alonso-Blanco, C.; Andrade, J.; Becker, C.; Bemm, F.; Bergelson, J.; Borgwardt, K.M.M.; Cao, J.; Chae, E.; Dezwaan, T.M.; Ding, W.; et al. 1135 Genomes Reveal the Global Pattern of Polymorphism in Arabidopsis thaliana. Cell 2016, 166, 481. [Google Scholar] [CrossRef]
- Chang, C.C.; Chow, C.C.; Tellier, L.C.A.M.; Vattikuti, S.; Purcell, S.M.; Lee, J.J. Second-generation PLINK: Rising to the challenge of larger and richer datasets. GigaScience 2015, 4, 7. [Google Scholar] [CrossRef] [PubMed]
- Browning, B.L.; Zhou, Y.; Browning, S.R. A One-Penny Imputed Genome from Next-Generation Reference Panels. American J. Hum. Genet. 2018, 103, 338–348. [Google Scholar] [CrossRef]
- Cao, J.; Schneeberger, K.; Ossowski, S.; Günther, T.; Bender, S.; Fitz, J.; Koenig, D.; Lanz, C.; Stegle, O.; Lippert, C.; et al. Whole-genome sequencing of multiple Arabidopsis thaliana populations. Nat. Genet. 2011, 43, 956–963. [Google Scholar] [CrossRef]
- Chesnokov, Y.V.; Artemyeva, A.M. Evaluation of the measure of polymorphism information of genetic diversity. Sel’skokhozyaistvennaya Biol. 2015, 50, 571–578. [Google Scholar] [CrossRef]

| SNP Combination | Distinguishing Accuracy (%) | Mean Accuracy (%) | |||
|---|---|---|---|---|---|
| Raw SNP | Modify 5% SNP | Modify 10% SNP | Modify 15% SNP | ||
| 59 DT-PICS SNPs | 100 | 96.74 | 92.78 | 84.64 | 93.54 |
| 39 DT-PICS + 20 hPIC | 94.19 | 91.28 | 88.65 | 85.21 | 89.83 |
| 20 DT-PICS + 39 hPIC | 91.28 | 85.94 | 84.68 | 82.67 | 86.14 |
| 59 hPICS SNPs | 87.14 | 82.32 | 81.71 | 80.51 | 82.92 |
| 59 random SNPs | 86.70 | 80.42 | 77.90 | 68.26 | 78.32 |
| SNP Combination | Distinguishing Accuracy (%) | Mean Accuracy (%) | |||
|---|---|---|---|---|---|
| Raw SNP | Modify 5% SNP | Modify 10% SNP | Modify 15% SNP | ||
| 109 DT-PIC SNPs | 100 | 99.44 | 97.94 | 95.64 | 98.26 |
| 73 DT-PIC + 36 hPIC | 99.56 | 97.28 | 94.82 | 92.56 | 96.06 |
| 36 DT-PIC + 73 hPIC | 95.51 | 92.02 | 90.19 | 88.58 | 91.58 |
| 109 hPIC SNPs | 87.93 | 83.86 | 83.57 | 83.14 | 84.63 |
| 109 random SNPs | 87.58 | 83.10 | 82.57 | 81.59 | 83.71 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Xiong, L.; Li, Z.; Li, W.; Li, L. DT-PICS: An Efficient and Cost-Effective SNP Selection Method for the Germplasm Identification of Arabidopsis. Int. J. Mol. Sci. 2023, 24, 8742. https://doi.org/10.3390/ijms24108742
Xiong L, Li Z, Li W, Li L. DT-PICS: An Efficient and Cost-Effective SNP Selection Method for the Germplasm Identification of Arabidopsis. International Journal of Molecular Sciences. 2023; 24(10):8742. https://doi.org/10.3390/ijms24108742
Chicago/Turabian StyleXiong, Liwen, Zirong Li, Weihua Li, and Lanzhi Li. 2023. "DT-PICS: An Efficient and Cost-Effective SNP Selection Method for the Germplasm Identification of Arabidopsis" International Journal of Molecular Sciences 24, no. 10: 8742. https://doi.org/10.3390/ijms24108742
APA StyleXiong, L., Li, Z., Li, W., & Li, L. (2023). DT-PICS: An Efficient and Cost-Effective SNP Selection Method for the Germplasm Identification of Arabidopsis. International Journal of Molecular Sciences, 24(10), 8742. https://doi.org/10.3390/ijms24108742




