Extent of N-Terminus Folding of Semenogelin 1 Cleavage Product Determines Tendency to Amyloid Formation
Abstract
1. Introduction
2. Results
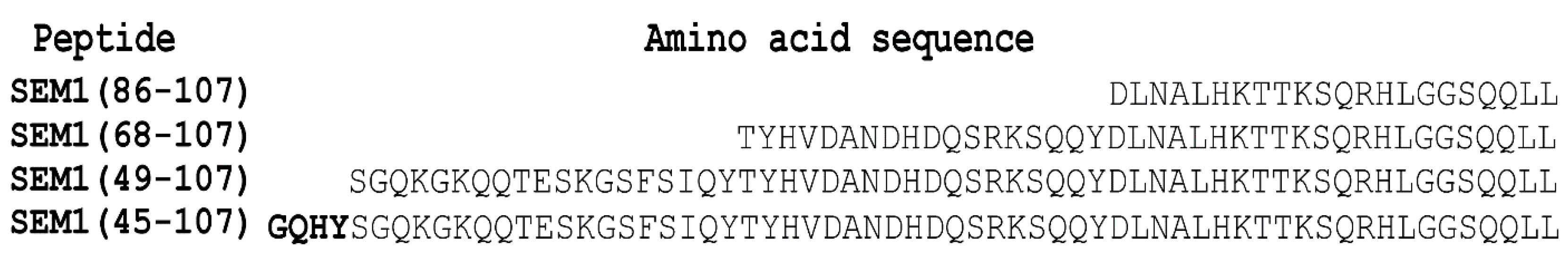
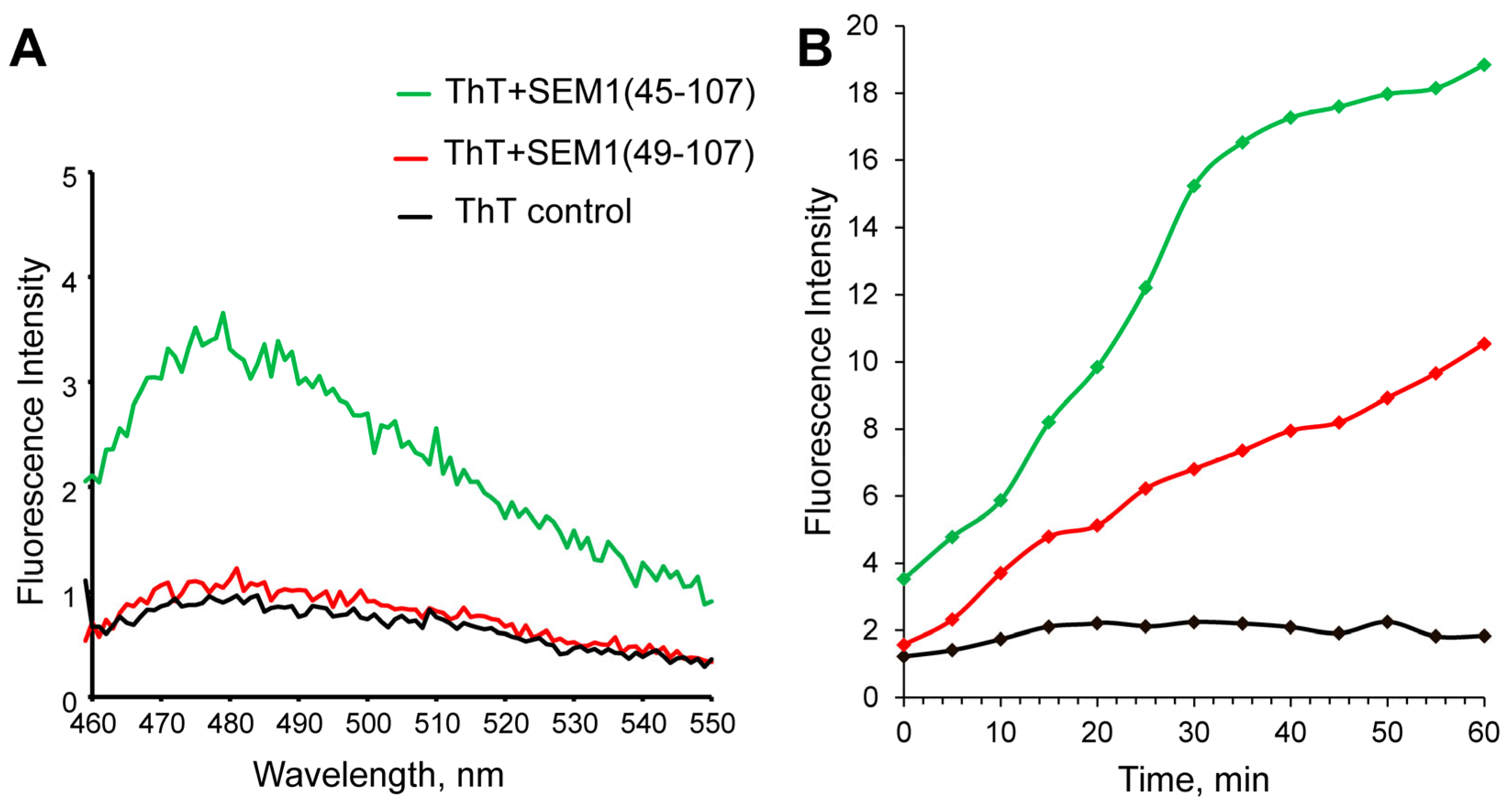
2.1. Thioflavin T Fluorescence Measurements of SEM1(49–107) and SEM1(45–107)
2.2. Nuclear Magnetic Resonance Spectroscopy
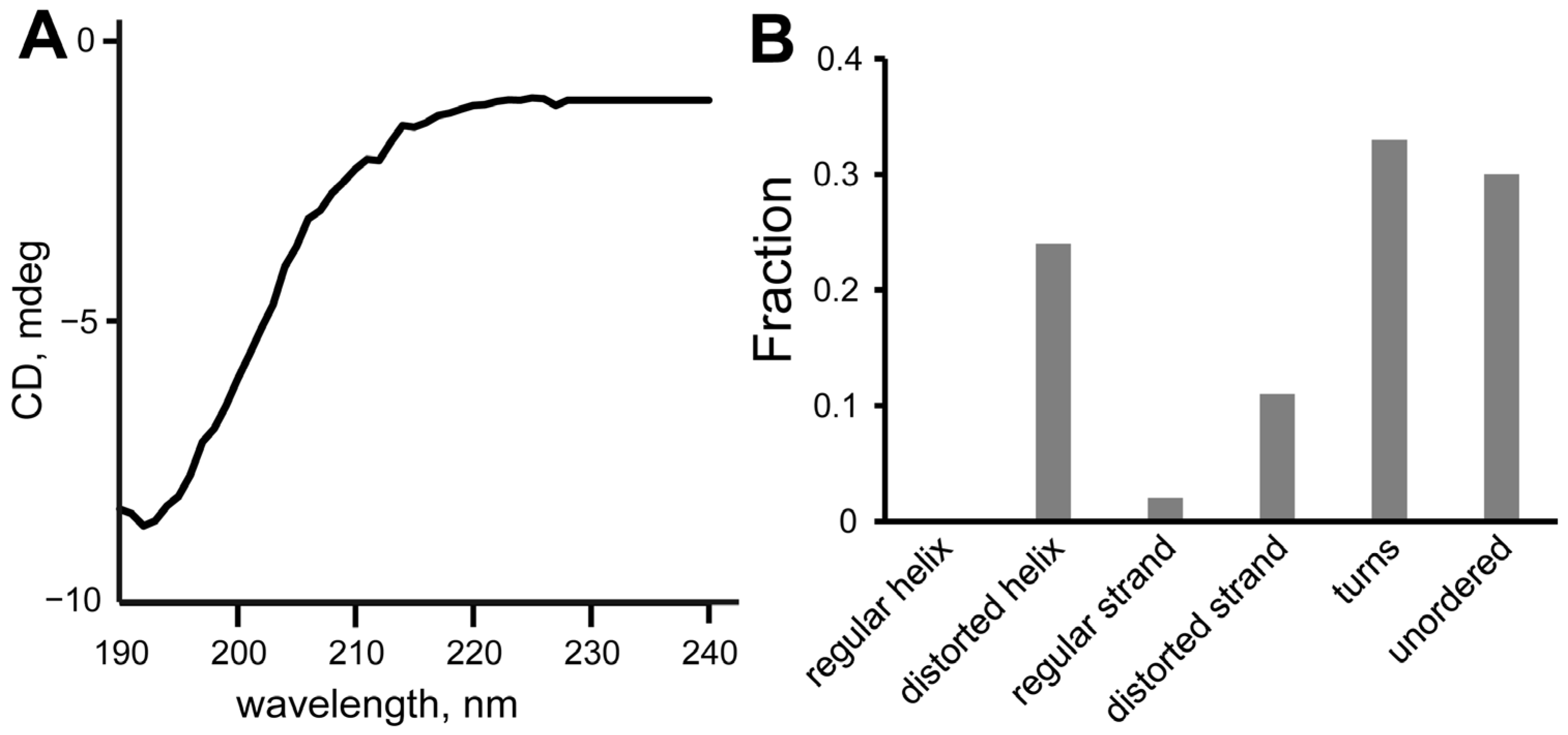
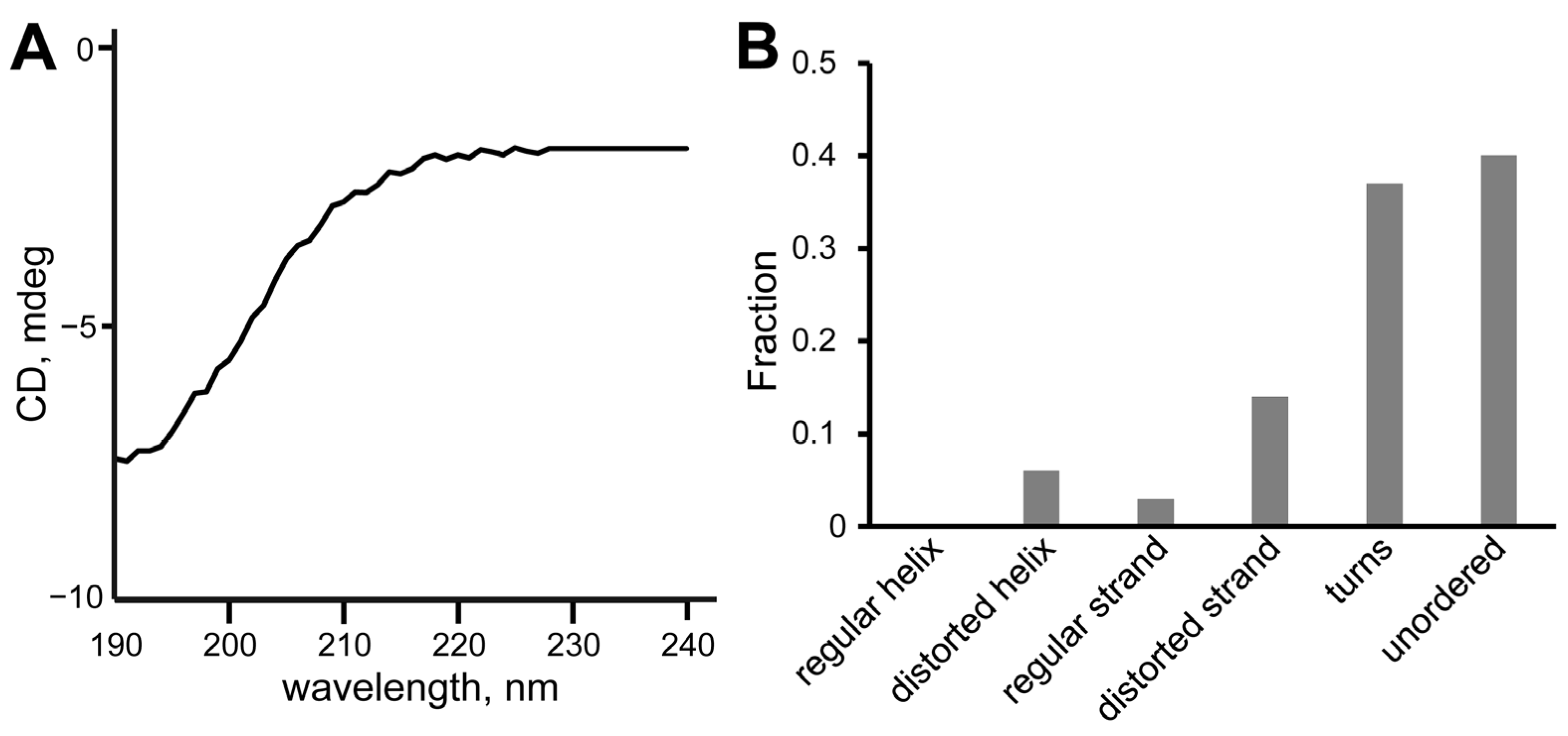
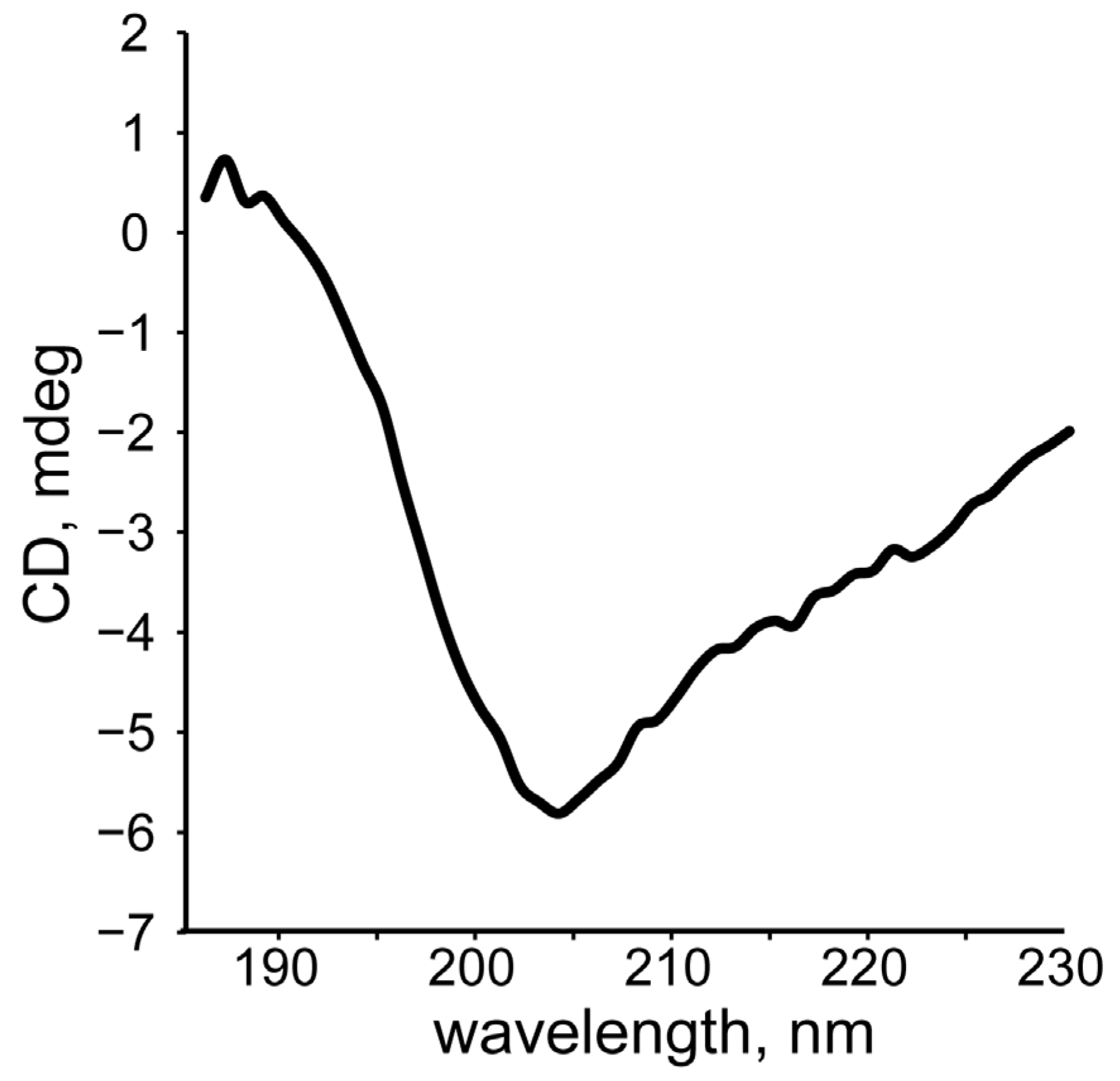
2.3. Circular Dichroism Spectroscopy
2.4. PFG-NMR Spectroscopy of SEM1(45–67) and SEM1(49–67)
2.5. DLS Spectroscopy of SEM1(45–67) and SEM1(49–67)
2.6. MD Simulation of SEM1(45–107) and SEM1(49–107)
3. Discussion
4. Materials and Methods
4.1. Protein Expression and Purification
4.2. ThT Fluorescence
4.3. PFG-NMR Spectroscopy
4.4. NMR Spectroscopy
4.5. CD Spectroscopy
4.6. Calculation and Visualization
4.7. DLS Spectroscopy
4.8. MD Simulation
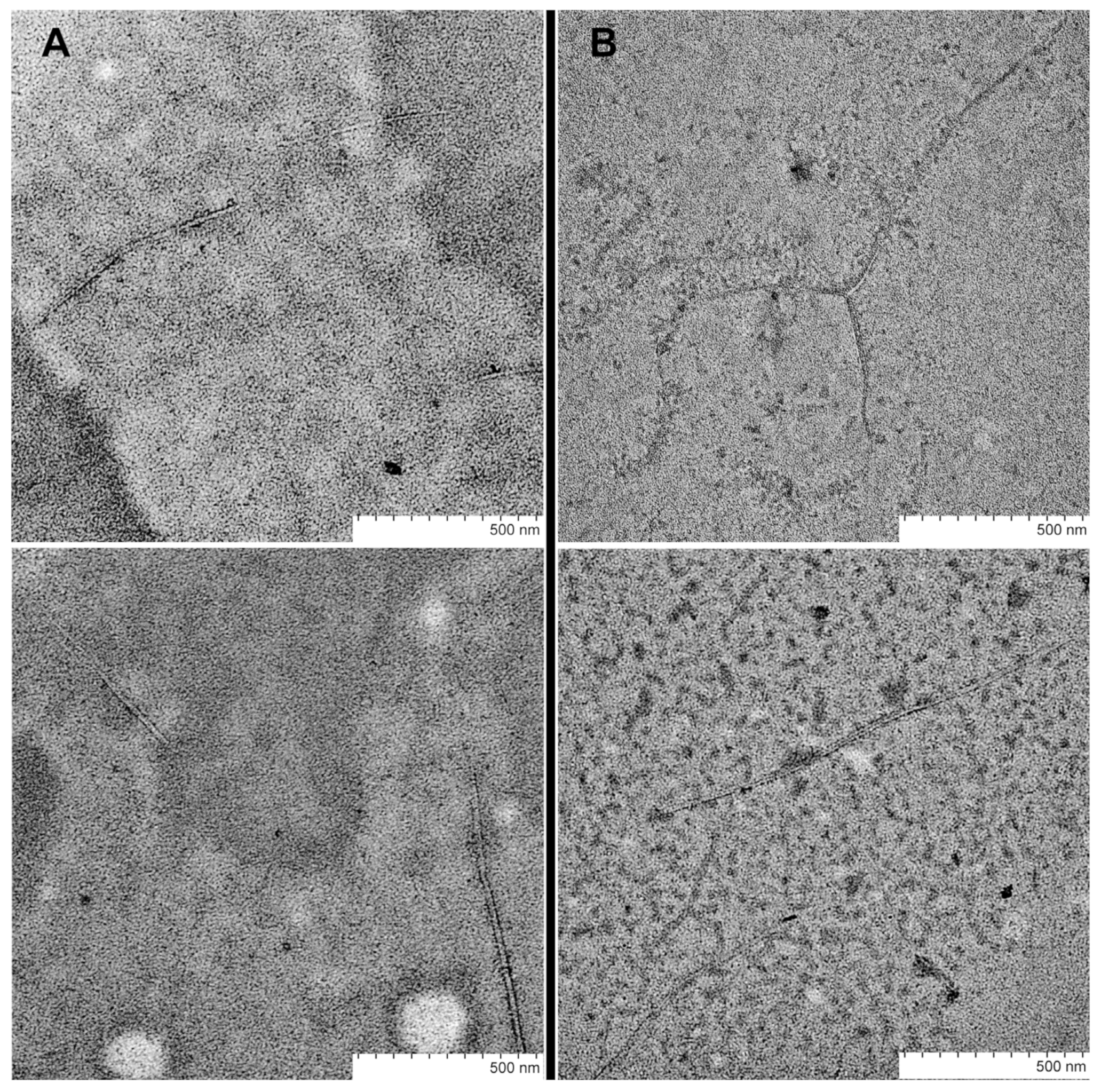
4.9. Transmission Electron Microscopy
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Simon, V.; Ho, D.D.; Karim, Q.A. HIV/AIDS epidemiology, pathogenesis, prevention, and treatment. Lancet 2006, 368, 489–504. [Google Scholar] [CrossRef]
- Dimitrov, D.S.; Willey, R.L.; Sato, H.; Chang, L.J.; Blumenthal, R.; Martin, M.A. Quantitation of human immunodeficiency virus type 1 infection kinetics. J. Virol. 1993, 67, 2182–2190. [Google Scholar] [CrossRef] [PubMed]
- Pomerantz, R.J. HIV: Cross-talk and viral reservoirs. Nature 2003, 424, 136–137. [Google Scholar] [CrossRef] [PubMed]
- Lilja, H.; Abrahamsson, P.A.; Lundwall, A. Semenogelin, the predominant protein in human semen. Primary structure and identification of closely related proteins in the male accessory sex glands and on the spermatozoa. J. Biol. Chem. 1989, 264, 1894–1900. [Google Scholar] [CrossRef]
- Malm, J.; Hellman, J.; Magnusson, H.; Laurell, C.B.; Lilja, H. Isolation and characterization of the major gel proteins in human semen, semenogelin I and semenogelin II. Eur. J. Biochem. 1996, 238, 48–53. [Google Scholar] [CrossRef]
- de Lamirande, E. Semenogelin, the main protein of the human semen coagulum, regulates sperm function. Semin. Thromb. Hemost. 2007, 33, 60–68. [Google Scholar] [CrossRef]
- Robert, M.; Gagnon, C. Semenogelin I: A coagulum forming, multifunctional seminal vesicle protein. Cell. Mol. Life Sci. 1999, 55, 944–960. [Google Scholar] [CrossRef] [PubMed]
- Robert, M.; Gagnon, C. Purification and characterization of the active precursor of a human sperm motility inhibitor secreted by the seminal vesicles: Identity with semenogelin. Biol. Reprod. 1996, 55, 813–821. [Google Scholar] [CrossRef]
- Vickram, A.S.; Rohini, K.; Anbarasu, K.; Dey, N.; Jeyanthi, P.; Thanigaivel, S.; Issac, P.K.; Arockiaraj, J. Semenogelin, a coagulum macromolecule monitoring factor involved in the first step of fertilization: A prospective review. Int. J. Biol. Macromol. 2022, 209, 951–962. [Google Scholar] [CrossRef]
- Lilja, H. A kallikrein-like serine protease in prostatic fluid cleaves the predominant seminal vesicle protein. J. Clin. Investig. 1985, 76, 1899–1903. [Google Scholar] [CrossRef]
- Kim, K.A.; Yolamanova, M.; Zirafi, O.; Roan, N.R.; Staendker, L.; Forssmann, W.G.; Burgener, A.; Dejucq-Rainsford, N.; Hahn, B.H.; Shaw, G.M.; et al. Semen-mediated enhancement of HIV infection is donor-dependent and correlates with the levels of SEVI. Retrovirology 2010, 7, 55. [Google Scholar] [CrossRef] [PubMed]
- Roan, N.R.; Müller, J.A.; Liu, H.; Chu, S.; Arnold, F.; Stürzel, C.M.; Walther, P.; Dong, M.; Witkowska, H.E.; Kirchhoff, F.; et al. Peptides released by physiological cleavage of semen coagulum proteins form amyloids that enhance HIV infection. Cell Host Microbe 2011, 10, 541–550. [Google Scholar] [CrossRef] [PubMed]
- Roan, N.R.; Liu, H.; Usmani, S.M.; Neidleman, J.; Müller, J.A.; Avila-Herrera, A.; Gawanbacht, A.; Zirafi, O.; Chu, S.; Dong, M.; et al. Liquefaction of Semen Generates and Later Degrades a Conserved Semenogelin Peptide That Enhances HIV Infection. J. Virol. 2014, 88, 7221–7234. [Google Scholar] [CrossRef] [PubMed]
- Roan, N.R.; Munch, J.; Arhel, N.; Mothes, W.; Neidleman, J.; Kobayashi, A.; Smith-McCune, K.; Kirchhoff, F.; Greene, W.C. The cationic properties of SEVI underlie its ability to enhance human immunodeficiency virus infection. J. Virol. 2009, 83, 73–80. [Google Scholar] [CrossRef]
- Roan, N.R.; Sowinski, S.; Munch, J.; Kirchhoff, F.; Greene, W.C. Aminoquinoline surfen inhibits the action of SEVI (semen-derived enhancer of viral infection). J. Biol. Chem. 2010, 285, 1861–1869. [Google Scholar] [CrossRef]
- Roan, N.R.; Sandi-Monroy, N.; Kohgadai, N.; Usmani, S.M.; Hamil, K.G.; Neidleman, J.; Montano, M.; Ständker, L.; Röcker, A.; Cavrois, M.; et al. Semen amyloids participate in spermatozoa selection and clearance. eLife 2017, 6, e24888. [Google Scholar] [CrossRef]
- Castellano, L.M.; Shorter, J. The Surprising Role of Amyloid Fibrils in HIV Infection. Biology 2012, 1, 58–80. [Google Scholar] [CrossRef]
- Jones, R. Interaction of zona pellucida glycoproteins, sulphated carbohydrates and synthetic polymers with proacrosin, the putative egg-binding protein from mammalian spermatozoa. Development 1991, 111, 1155–1163. [Google Scholar] [CrossRef]
- Cohen, F.S.; Melikyan, G.B. The Energetics of Membrane Fusion from Binding, through Hemifusion, Pore Formation, and Pore Enlargement. J. Membr. Biol. 2004, 199, 1–14. [Google Scholar] [CrossRef]
- Evans, J.P.; Florman, H.M. The state of the union: The cell biology of fertilization. Nat. Cell Biol. 2002, 4, s57–s63. [Google Scholar] [CrossRef]
- Sanchugova, D.; Kusova, A.; Bikmullin, A.; Yulmetov, A.; Mukhametzyanov, T.; Klochkov, V.; Blokhin, D. Conformational ensemble of amyloid-forming semenogelin 1 peptide SEM1(68–107) by NMR spectroscopy and MD simulations. J. Struct. Biol. 2022, 214, 107900. [Google Scholar] [CrossRef] [PubMed]
- Blokhin, D.S.; Fayzullina, A.R.; Filippov, A.V.; Karataeva, F.K.; Klochkov, V.V. Spatial structure of fibrinopeptide B in water solution with DPC micelles by NMR spectroscopy. J. Mol. Struct. 2015, 1102, 91–94. [Google Scholar] [CrossRef]
- Kononova, O.; Litvinov, R.; Blokhin, D.; Klochkov, V.; Weisel, J.; Bennett, J.; Barsegov, V. Mechanistic Basis for the Binding of RGD- and AGDV-Peptides to the Platelet Integrin αIIbβ3. Biochemistry 2017, 56, 1932–1942. [Google Scholar] [CrossRef] [PubMed]
- Kazimierczuk, K.; Kasprzak, P.; Georgoulia, P.S.; Matečko-Burmann, I.; Burmann, B.M.; Isaksson, L.; Gustavsson, E.; Westenhoff, S.; Orekhov, V.Y. Resolution enhancement in NMR spectra by deconvolution with compressed sensing reconstruction. Chem. Commun. 2020, 56, 14585–14588. [Google Scholar] [CrossRef] [PubMed]
- Lawrence, C.; Grishaev, A. Chemical shifts-based similarity restraints improve accuracy of RNA structures determined via NMR. RNA 2020, 26, 2051–2061. [Google Scholar] [CrossRef]
- Puthenveetil, R.; Vinogradova, O. Solution NMR: A powerful tool for structural and functional studies of membrane proteins in reconstituted environments. J. Biol. Chem. 2019, 294, 15914–15931. [Google Scholar] [CrossRef]
- Yao, S.; Babon, J.J.; Norton, R.S. Protein effective rotational correlation times from translational self-diffusion coefficients measured by PFG-NMR. Biophys. Chem. 2008, 136, 145–151. [Google Scholar] [CrossRef]
- Lucas, L.H.; Larive, C.K. Measuring ligand-protein binding using NMR diffusion experiments. Concepts Magn. Reson. 2004, 20A, 24–41. [Google Scholar] [CrossRef]
- Kusova, A.M.; Sitnitsky, A.E.; Uversky, V.N.; Zuev, Y.F. Effect of Protein–Protein Interactions on Translational Diffusion of Spheroidal Proteins. Int. J. Mol. Sci. 2022, 23, 9240. [Google Scholar] [CrossRef]
- Kusova, A.M.; Sitnitsky, A.E.; Zuev, Y.F. The Role of pH and Ionic Strength in the Attraction–Repulsion Balance of Fibrinogen Interactions. Langmuir 2021, 37, 10394–10401. [Google Scholar] [CrossRef]
- Lorber, B.; Fischer, F.; Bailly, M.; Roy, H.; Kern, D. Protein analysis by dynamic light scattering: Methods and techniques for students. Biochem. Mol. Biol. Educ. 2012, 40, 372–382. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.; Lv, H.; Zhao, Y.; Liu, D.; Wang, Y.; Zhang, Y. DLS: A Link Prediction Method Based on Network Local Structure for Predicting Drug-Protein Interactions. Front. Bioeng. Biotechnol. 2020, 8, 330. [Google Scholar] [CrossRef] [PubMed]
- Whitmore, L.; Wallace, B.A. Protein secondary structure analyses from circular dichroism spectroscopy: Methods and reference databases. Biopolymers 2008, 89, 392–400. [Google Scholar] [CrossRef]
- Liu, G.; Prabhakar, A.; Aucoin, D.; Simon, M.; Sparks, S.; Robbins, K.J.; Sheen, A.; Petty, S.A.; Lazo, N.D. Mechanistic studies of peptide self-assembly: Transient α-helices to stable β-sheets. J. Am. Chem. Soc. 2010, 132, 18223–18232. [Google Scholar] [CrossRef] [PubMed]
- Karplus, M. Contact Electron-Spin Coupling of Nuclear Magnetic Moments. J. Phys. Chem. 1959, 30, 11–15. [Google Scholar] [CrossRef]
- Lovell, S.C.; Davis, I.W.; Arendall III, W.B.; de Bakker, P.I.W.; Word, J.M.; Prisant, M.G.; Richardson, J.S.; Richardson, D.C. Structure validation by Calpha geometry: Phi, psi and Cbeta deviation. Proteins 2003, 50, 437–450. [Google Scholar] [CrossRef] [PubMed]
- Kabsch, W.; Sander, C. Dictionary of protein secondary structure: Pattern recognition of hydrogen-bonded and geometrical features. Biopolymers 1983, 22, 2577–2637. [Google Scholar] [CrossRef]
- Wüthrich, K. NMR of Proteins and Nucleic Acids; Wiley-Interscience: New York, NY, USA, 1986. [Google Scholar]
- Leite, J.P.; Gimeno, A.; Taboada, P.; Jiménez-Barbero, J.J.; Gales, L. Dissection of the key steps of amyloid-β peptide 1-40 fibrillogenesis. Int. J. Biol. Macromol. 2020, 164, 2240–2246. [Google Scholar] [CrossRef]
- Kusova, A.M.; Iskhakova, A.K.; Zuev, Y.F. NMR and dynamic light scattering give different diffusion information for short-living protein oligomers. Human serum albumin in water solutions of metal ions. Eur. Biophys. J. 2022, 51, 375–383. [Google Scholar] [CrossRef]
- Pronk, S.; Páll, S.; Schulz, R.; Larsson, P.; Bjelkmar, P.; Apostolov, R.; Shirts, M.R.; Smith, J.C.; Kasson, P.M.; van der Spoel, D.; et al. GROMACS 4.5: A high-throughput and highly parallel open source molecular simulation toolkit. J. Bioinform. 2013, 29, 845–854. [Google Scholar] [CrossRef]
- Hess, B.; Kutzner, C.; van der Spoel, D.; Lindahl, E. GROMACS 4: Algorithms for Highly Efficient, Load-Balanced, and Scalable Molecular Simulation. J. Chem. Theory Comput. 2008, 4, 435–447. [Google Scholar] [CrossRef] [PubMed]
- Blokhin, D.S.; Filippov, A.V.; Antzutkin, O.N.; Karataeva, F.K.; Klochkov, V.V. Spatial structure of oligopeptide PAP (248–261), the N-terminal fragment of the HIV enhancer prostatic acid phosphatase peptide PAP (248–286), in aqueous and SDS micelle solutions. J. Mol. Struct. 2014, 1070, 38–42. [Google Scholar] [CrossRef]
- Blokhin, D.S.; Filippov, A.V.; Antzutkin, O.N.; Afonin, S.; Klochkov, V.V. Spatial Structures of PAP (262–270) and PAP (274–284), Two Selected Fragments of PAP (248–286), an Enhancer of HIV Infectivity. Appl. Magn. Reson. 2015, 46, 757–769. [Google Scholar] [CrossRef]
- Nanga, R.P.R.; Brender, J.R.; Vivekanandan, S.; Popovych, N.; Ramamoorthy, A. NMR Structure in a Membrane Environment Reveals Putative Amyloidogenic Regions of the SEVI Precursor Peptide PAP248-286. J. Am. Chem. Soc. 2009, 131, 17972–17979. [Google Scholar] [CrossRef]
- Sanchugova, D.; Kusova, A.; Bikmullin, A.; Klochkov, V.; Blokhin, D. The Structure of Fibril-Forming SEM1 (86–107) Peptide Increasing the HIV Infectivity. BioNanoScience 2021, 11, 182–188. [Google Scholar] [CrossRef]
- Ziehm, T.; Buell, A.K.; Willbold, D. Role of Hydrophobicity and Charge of Amyloid-Beta Oligomer Eliminating d-Peptides in the Interaction with Amyloid-Beta Monomers. ACS Chem. Neurosci. 2018, 9, 2679–2688. [Google Scholar] [CrossRef]
- Itoh, S.G.; Yagi-Utsumi, M.; Kato, K.; Okumura, H. Effects of a Hydrophilic/Hydrophobic Interface on Amyloid-β Peptides Studied by Molecular Dynamics Simulations and NMR Experiments. J. Phys. Chem. B 2019, 123, 160–169. [Google Scholar] [CrossRef]
- Ye, Z.; French, K.C.; Popova, L.A.; Lednev, I.K.; Lopez, M.M.; Makhatadze, G.I. Mechanism of fibril formation by a 39-residue peptide (PAPf39) from human prostatic acidic phosphatase. Biochemistry 2009, 48, 11582–11591. [Google Scholar] [CrossRef]
- Kallberg, Y.; Gustafsson, M.; Persson, B.; Thyberg, J.; Johansson, J. Prediction of amyloid fibril-forming proteins. J. Biol. Chem. 2001, 276, 12945–12950. [Google Scholar] [CrossRef]
- Bogomolovas, J.; Simon, B.; Sattler, M.; Stier, G. Screening of fusion partners for high yield expression and purification of bioactive viscotoxins. Protein Expr. Purif. 2009, 64, 16–23. [Google Scholar] [CrossRef]
- Bikmullin, A.; Klochkova, E.; Krasnovid, F.; Blokhin, D. The data of heterologous expression protocol for synthesis of 15N, 13C-labeled SEM1 (68–107) peptide fragment of homo sapiens semenogelin 1. MethodsX 2021, 8, 101512. [Google Scholar] [CrossRef] [PubMed]
- Tropea, J.E.; Cherry, S.; Waugh, D.S. Expression and purification of soluble His(6)-tagged TEV protease. High Throughput Protein Expr. Purif. 2009, 498, 297–307. [Google Scholar] [CrossRef]
- Raran-Kurussi, S.; Cherry, S.; Zhang, D.; Waugh, D.S. Removal of affinity tags with TEV protease. Methods Mol. Biol. 2017, 1586, 221–230. [Google Scholar] [CrossRef] [PubMed]
- Laemmli, U.K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 1970, 227, 680–685. [Google Scholar] [CrossRef]
- Abelein, A.; Jarvet, J.; Barth, A.; Gräslund, A.; Danielsson, J. Ionic strength modulation of the free energy landscape of Aβ40 peptide fibril formation. J. Am. Chem. Soc. 2016, 138, 6893–6902. [Google Scholar] [CrossRef]
- Sklenar, V.; Piotto, M.; Leppik, R.; Saudek, V. Gradient-Tailored Water Suppression for 1H- 15N HSQC Experiments Optimized to Retain Full Sensitivity. J. Magn. Reson. Ser. A. 1993, 102, 241–245. [Google Scholar] [CrossRef]
- Avni, A.; Swasthi, H.M.; Majumdar, A.; Mukhopadhyay, S. Intrinsically disordered proteins in the formation of functional amyloids from bacteria to humans. Prog. Mol. Biol. Transl. Sci. 2019, 166, 109–143. [Google Scholar] [CrossRef]
- Berger, S.; Braun, S. 200 and More NMR Experiments; Wiley-VCH: Weinheim, Germany, 2004. [Google Scholar]
- Rule, G.S.; Hitchens, T.K. Fundamentals of Protein NMR Spectroscopy; Springer: Dordrecht, The Netherlands, 2006. [Google Scholar]
- Schleucher, J.; Schwendinger, M.; Sattler, M.; Schmidt, P.; Schedletzky, O.; Glaser, S.J.; Sørensen, O.W.; Griesinger, C. A general enhancement scheme in heteronuclear multidimensional NMR employing pulsed field gradients. J. Biomol. NMR 1994, 4, 301–306. [Google Scholar] [CrossRef]
- Nolis, P.; Parella, T. Spin-edited 2D HSQC-TOCSY experiments for the measurement of homonuclear and heteronuclear coupling constants: Application to carbohydrates and peptides. J. Magn. Reson. 2005, 176, 15–26. [Google Scholar] [CrossRef]
- Huang, Y.; Zhang, Z.; Chen, H.; Feng, J.; Cai, S.; Chen, Z. A high-resolution 2D J-resolved NMR detection technique for metabolite analyses of biological samples. Sci. Rep. 2015, 5, 8390. [Google Scholar] [CrossRef]
- Vranken, W.F.; Boucher, W.; Stevens, T.J.; Fogh, R.H.; Pajon, A.; Llinas, M.; Ulrich, E.L.; Markley, J.L.; Ionides, J.; Laue, E.D. The CCPN data model for NMR spectroscopy: Development of a software pipeline. Proteins 2005, 59, 687–696. [Google Scholar] [CrossRef] [PubMed]
- Sreerama, N.; Woody, R.W. Estimation of protein secondary structure from circular dichroism spectra: Comparison of CONTIN, SELCON, and CDSSTR methods with an expanded reference set. Anal. Biochem. 2000, 287, 252–260. [Google Scholar] [CrossRef] [PubMed]
- Schwieters, C.D.; Kuszewski, J.J.; Tjandra, N.; Clore, G.M. The Xplor-NIH NMR molecular structure determination package. J. Magn. Reson. 2003, 160, 65–73. [Google Scholar] [CrossRef] [PubMed]
- Shen, Y.; Delaglio, F.; Cornilescu, G.; Bax, A. TALOS+: A hybrid method for predicting protein backbone torsion angles from NMR chemical shifts. J. Biomol. NMR 2009, 44, 213–223. [Google Scholar] [CrossRef] [PubMed]
- Schwieters, C.D.; Kuszewski, J.J.; Clore, G.M. Using Xplor–NIH for NMR molecular structure determination. Prog. Nucl. Magn. Reson. Spectrosc. 2006, 48, 47–62. [Google Scholar] [CrossRef]
- Engh, R.A.; Huber, R. Accurate bond and angle parameters for X-ray protein structure refinement. Acta Crystallogr. A 1991, 47, 392–400. [Google Scholar] [CrossRef]
- Nederveen, A.J.; Doreleijers, J.F.; Vranken, W.; Miller, Z.; Spronk, C.A.; Nabuurs, S.B.; Güntert, P.; Livny, M.; Markley, J.L.; Nilges, M.; et al. RECOORD: A REcalculated COORdinates Database of 500+ proteins from the PDB using restraints from the BioMagResBank. Proteins 2005, 59, 662–672. [Google Scholar] [CrossRef]
- Pettersen, E.F.; Goddard, T.D.; Huang, C.C.; Couch, G.S.; Greenblatt, D.M.; Meng, E.C.; Ferrin, T.E. UCSF Chimera—A visualization system for exploratory research and analysis. J. Comput. Chem. 2004, 25, 1605–1612. [Google Scholar] [CrossRef]
- Koradi, R.; Billeter, M.; Wüthrich, K. MOLMOL: A program for display and analysis of macromolecular structures. J. Mol. Graph. 1996, 14, 51–55. [Google Scholar] [CrossRef]
- Scotti, A.E.; Liu, W.; Hyatt, J.S.; Herman, E.S.; Choi, H.S.; Kim, J.W.; Lyon, L.A.; Gasser, U.; Fernandez-Nieves, A. The CONTIN algorithm and its application to determine the size distribution of microgel suspensions. J. Chem. Phys. 2015, 142, 234905. [Google Scholar] [CrossRef]
- Sinelnikova, A.; Spoel, D.V.D. NMR refinement and peptide folding using the GROMACS software. J. Biomol. NMR 2021, 75, 143–149. [Google Scholar] [CrossRef] [PubMed]
- Huang, J.; MacKerell, A.D., Jr. CHARMM36 All-Atom Additive Protein Force Field: Validation Based on Comparison to NMR Data. J. Comput. Chem. 2013, 34, 2135–2145. [Google Scholar] [CrossRef] [PubMed]
- Jorgensen, W.L.; Chandrasekhar, J.; Madura, J.D. Comparison of simple potential functions for simulating liquid water. J. Chem. Phys. 1983, 79, 926–935. [Google Scholar] [CrossRef]
- Berendsen, H.J.; Postma, J.V.; van Gunsteren, W.F.; DiNola, A.; Haak, J. Molecular dynamics with coupling to an external bath. J. Chem. Phys. 1984, 81, 3684–3690. [Google Scholar] [CrossRef]
- Parrinello, M.; Rahman, A. Polymorphic transitions in single crystals: A new molecular dynamics method. J. Appl. Phys. 1981, 52, 7182–7190. [Google Scholar] [CrossRef]
- Daura, X.; Gademann, K.; Jaun, B.; Seebach, D.; Gunsteren, W.F.V.; Mark, A.E. Peptide folding: When simulation meets experiment. Angew. Chem. Int. Ed. Engl. 1999, 38, 236–240. [Google Scholar] [CrossRef]






| Distance Constraints | |
|---|---|
| total | 143 |
| intra-residual | 88 |
| inter-residual | 55 |
| sequential (i–j = 1) | 24 |
| medium (i–j = 2–6) | 16 |
| long-range (i–j > 6) | 15 |
| 3JHα-HN alpha coupling constants (jna) | |
| Restraints | 22 |
| Structural Statistics | |
| number of NOE violations | 0 |
| number of jna restraint violations | 0 |
| RMSD for bond deviations (Å) | 0.005 |
| RMSD for angle deviations (deg) | 0.598 |
| RMSD of all backbone atoms (Å) | |
| E58-K60 | 0.13 ± 0.04 |
| S49-Y67 | 0.39 ± 0.11 |
| Y48-K52 | 0.14 ± 0.06 |
| Ramachandran Plot | |
| residues in the most favored region (%) | 80.1 |
| residues in the additionally allowed region (%) | 19.9 |
| residues in the disallowed region (%) | 0.0 |
| Distance Constraints | |
|---|---|
| total | 67 |
| intra-residual | 54 |
| inter-residual | 13 |
| sequential (i–j = 1) | 8 |
| medium (i–j = 3) | 2 |
| long-range (i–j > 13, 14) | 3 |
| Dihedral Angle Constraints | |
| Phi restraints | 17 |
| Psi restraints | 17 |
| 3JHα-HN alpha coupling constants (jna) | |
| Restraints | 16 |
| Structural Statistics | |
| number of NOE violations | 0 |
| number of dihedral angle restraint violations | 0 |
| number of jna restraint violations | 0 |
| RMSD for bond deviations (Å) | 0.004 |
| RMSD for angle deviations (deg) | 0.794 |
| RMSD of all backbone atoms (Å) | |
| E58-K60 | 0.24 ± 0.10 |
| S49-Y67 | 1.12 ± 0.30 |
| Y49-K52 | 0.69 ± 0.26 |
| T57-S59 | 0.17 ± 0.08 |
| G50-T57 | 0.70 ± 0.25 |
| Ramachandran Plot | |
| residues in the most favored region (%) | 94.1 |
| residues in the additionally allowed region (%) | 5.9 |
| residues in the disallowed region (%) | 0.0 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Osetrina, D.A.; Kusova, A.M.; Bikmullin, A.G.; Klochkova, E.A.; Yulmetov, A.R.; Semenova, E.A.; Mukhametzyanov, T.A.; Usachev, K.S.; Klochkov, V.V.; Blokhin, D.S. Extent of N-Terminus Folding of Semenogelin 1 Cleavage Product Determines Tendency to Amyloid Formation. Int. J. Mol. Sci. 2023, 24, 8949. https://doi.org/10.3390/ijms24108949
Osetrina DA, Kusova AM, Bikmullin AG, Klochkova EA, Yulmetov AR, Semenova EA, Mukhametzyanov TA, Usachev KS, Klochkov VV, Blokhin DS. Extent of N-Terminus Folding of Semenogelin 1 Cleavage Product Determines Tendency to Amyloid Formation. International Journal of Molecular Sciences. 2023; 24(10):8949. https://doi.org/10.3390/ijms24108949
Chicago/Turabian StyleOsetrina, Daria A., Aleksandra M. Kusova, Aydar G. Bikmullin, Evelina A. Klochkova, Aydar R. Yulmetov, Evgenia A. Semenova, Timur A. Mukhametzyanov, Konstantin S. Usachev, Vladimir V. Klochkov, and Dmitriy S. Blokhin. 2023. "Extent of N-Terminus Folding of Semenogelin 1 Cleavage Product Determines Tendency to Amyloid Formation" International Journal of Molecular Sciences 24, no. 10: 8949. https://doi.org/10.3390/ijms24108949
APA StyleOsetrina, D. A., Kusova, A. M., Bikmullin, A. G., Klochkova, E. A., Yulmetov, A. R., Semenova, E. A., Mukhametzyanov, T. A., Usachev, K. S., Klochkov, V. V., & Blokhin, D. S. (2023). Extent of N-Terminus Folding of Semenogelin 1 Cleavage Product Determines Tendency to Amyloid Formation. International Journal of Molecular Sciences, 24(10), 8949. https://doi.org/10.3390/ijms24108949









