Acute Myeloid Leukemia Causes Serious and Partially Irreversible Changes in Secretomes of Bone Marrow Multipotent Mesenchymal Stromal Cells
Abstract
1. Introduction
2. Results
2.1. Comparison of AML-MSC and D-MSC Secretomes
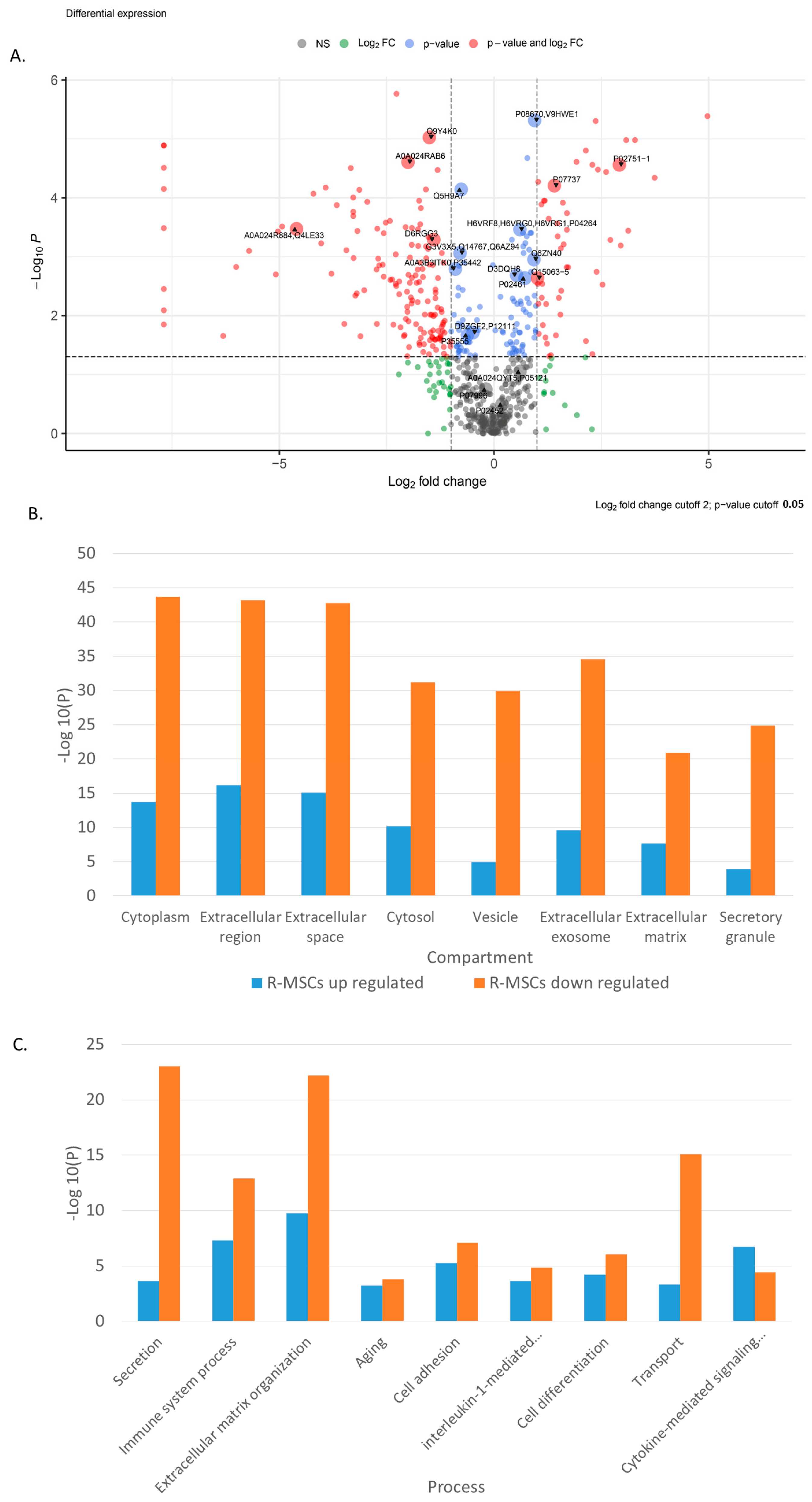
2.2. Comparison of R-MSCs and D-MSCs Secretome
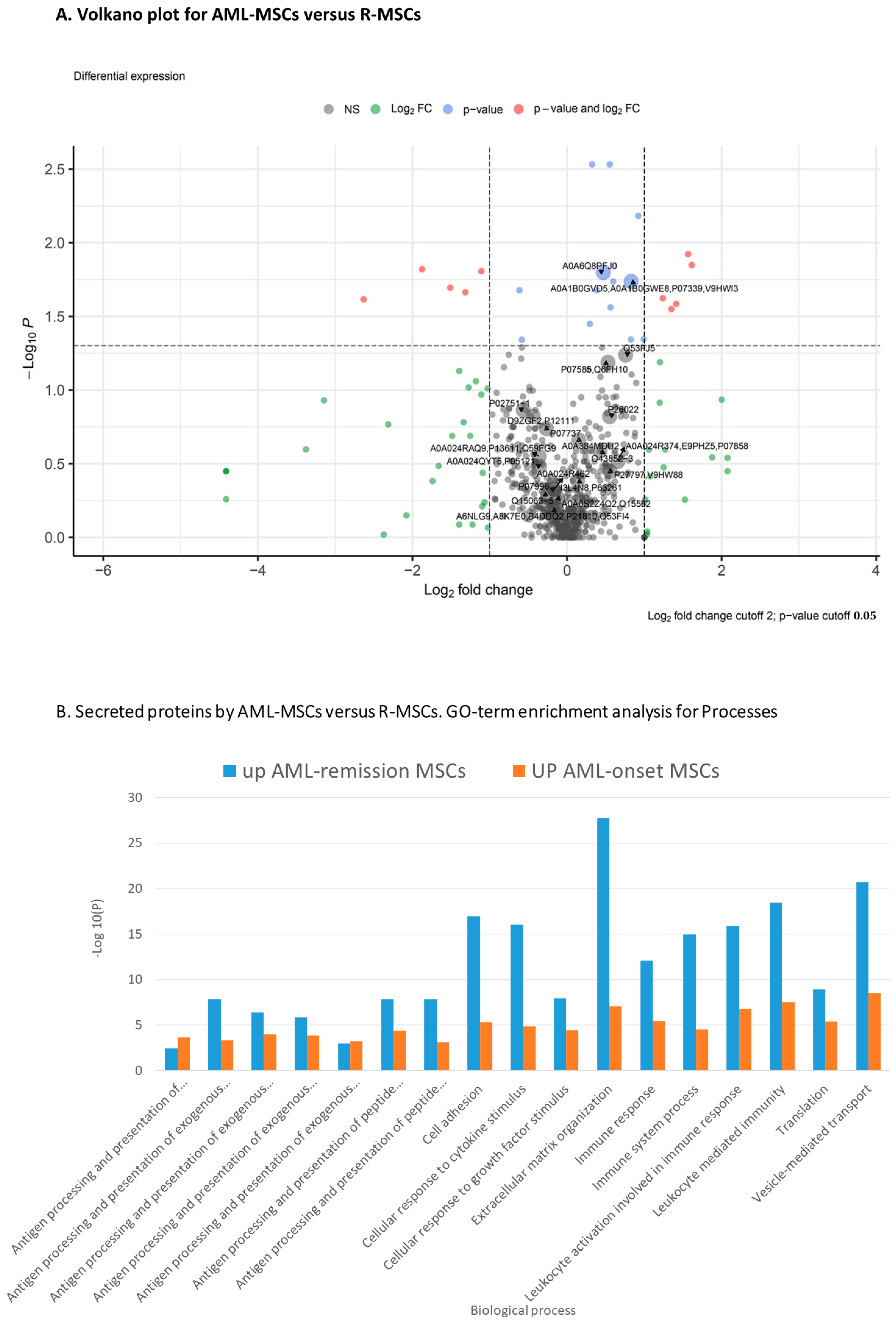
2.3. Comparison of AML-MSCs and R-MSCs Secretome
3. Discussion
4. Materials and Methods
4.1. Patients
4.2. MSCs
4.3. Preparation of MSC-Conditioned Medium
4.4. Proteomic Analysis of Secretomes
4.5. LC-MS/MS Analysis
4.6. Protein Identification and Bioinformatics Analysis
4.7. Statistics
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Goulard, M.; Dosquet, C.; Bonnet, D. Role of the microenvironment in myeloid malignancies. Cell. Mol. Life Sci. 2018, 75, 1377–1391. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Comazzetto, S.; Shen, B.; Morrison, S.J. Niches that regulate stem cells and hematopoiesis in adult bone marrow. Dev. Cell 2021, 56, 1848–1860. [Google Scholar] [CrossRef] [PubMed]
- Rasini, V.; Dominici, M.; Kluba, T.; Siegel, G.; Lusenti, G.; Northoff, H.; Horwitz, E.M.; Schäfer, R. Mesenchymal stromal/stem cells markers in the human bone marrow. Cytotherapy 2013, 15, 292–306. [Google Scholar] [CrossRef]
- Frenette, P.S.; Pinho, S.; Lucas, D.; Scheiermann, C. Mesenchymal stem cell: Keystone of the hematopoietic stem cell niche and a stepping-stone for regenerative medicine. Annu. Rev. Immunol. 2013, 31, 285–316. [Google Scholar] [CrossRef]
- Méndez-Ferrer, S.; Michurina, T.V.; Ferraro, F.; Mazloom, A.R.; Macarthur, B.D.; Lira, S.A.; Scadden, D.T.; Ma’ayan, A.; Enikolopov, G.N.; Frenette, P.S. Mesenchymal and haematopoietic stem cells form a unique bone marrow niche. Nature 2010, 466, 829–834. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Pittenger, M.F.; Discher, D.E.; Péault, B.M.; Phinney, D.G.; Hare, J.M.; Caplan, A.I. Mesenchymal stem cell perspective: Cell biology to clinical progress. NPJ Regen. Med. 2019, 4, 22. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Fontaine, M.J.; Shih, H.; Schäfer, R.; Pittenger, M.F. Unraveling the Mesenchymal Stromal Cells’ Paracrine Immunomodulatory Effects. Transfus. Med. Rev. 2016, 30, 37–43. [Google Scholar] [CrossRef]
- Jo, H.; Brito, S.; Kwak, B.M.; Park, S.; Lee, M.-G.; Bin, B.-H. Applications of Mesenchymal Stem Cells in Skin Regeneration and Rejuvenation. Int. J. Mol. Sci. 2021, 22, 2410. [Google Scholar] [CrossRef]
- Higuchi, H.; Yamakawa, N.; Imadome, K.-I.; Yahata, T.; Kotaki, R.; Ogata, J.; Kakizaki, M.; Fujita, K.; Lu, J.; Yokoyama, K.; et al. Role of exosomes as a proinflammatory mediator in the development of EBV-associated lymphoma. Blood 2018, 131, 2552–2567. [Google Scholar] [CrossRef][Green Version]
- Katagiri, W.; Sakaguchi, K.; Kawai, T.; Wakayama, Y.; Osugi, M.; Hibi, H. A defined mix of cytokines mimics conditioned medium from cultures of bone marrow-derived mesenchymal stem cells and elicits bone regeneration. Cell Prolif. 2017, 50, e12333. [Google Scholar] [CrossRef][Green Version]
- Wechsler, M.E.; Rao, V.V.; Borelli, A.N.; Anseth, K.S. Engineering the MSC Secretome: A Hydrogel Focused Approach. Adv. Healthc. Mater. 2021, 10, 2001948. [Google Scholar] [CrossRef] [PubMed]
- Viswanathan, S.; Shi, Y.; Galipeau, J.; Krampera, M.; Leblanc, K.; Martin, I.; Nolta, J.; Phinney, D.G.; Sensebe, L. Mesenchymal stem versus stromal cells: International Society for Cell & Gene Therapy (ISCT®) Mesenchymal Stromal Cell committee position statement on nomenclature. Cytotherapy 2019, 21, 1019–1024. [Google Scholar] [CrossRef] [PubMed]
- Arai, F.; Hirao, A.; Ohmura, M.; Sato, H.; Matsuoka, S.; Takubo, K.; Ito, K.; Koh, G.Y.; Suda, T. Tie2/angiopoietin-1 signaling regulates hematopoietic stem cell quiescence in the bone marrow niche. Cell 2004, 118, 149–161. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Calvi, L.M.; Adams, G.B.; Weibrecht, K.W.; Weber, J.M.; Olson, D.P.; Knight, M.C.; Martin, R.P.; Schipani, E.; Divieti, P.; Bringhurst, F.R.; et al. Osteoblastic cells regulate the haematopoietic stem cell niche. Nature 2003, 425, 841–846. [Google Scholar] [CrossRef][Green Version]
- Taichman, R.S. Blood and bone: Two tissues whose fates are intertwined to create the hematopoietic stem-cell niche. Blood 2005, 105, 2631–2639. [Google Scholar] [CrossRef][Green Version]
- Ding, L.; Saunders, T.L.; Enikolopov, G.; Morrison, S.J. Endothelial and perivascular cells maintain haematopoietic stem cells. Nature 2012, 481, 457. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Yamazaki, S.; Iwama, A.; Takayanagi, S.; Eto, K.; Ema, H.; Nakauchi, H. TGF-beta as a candidate bone marrow niche signal to induce hematopoietic stem cell hibernation. Blood 2009, 113, 1250–1256. [Google Scholar] [CrossRef][Green Version]
- Morrison, S.J.; Scadden, D.T. The bone marrow niche for haematopoietic stem cells. Nature 2014, 505, 327–334. [Google Scholar] [CrossRef][Green Version]
- Lane, S.W.; Scadden, D.T.; Gilliland, D.G. The leukemic stem cell niche: Current concepts and therapeutic opportunities. Blood 2009, 114, 1150–1157. [Google Scholar] [CrossRef][Green Version]
- Hira, V.V.V.; Van Noorden, C.J.F.; Carraway, H.E.; Maciejewski, J.P.; Molenaar, R.J. Novel therapeutic strategies to target leukemic cells that hijack compartmentalized continuous hematopoietic stem cell niches. Biochim. Biophys. Acta Rev. Cancer 2017, 1868, 183–198. [Google Scholar] [CrossRef]
- Maganti, H.; Visram, A.; Shorr, R.; Fulcher, J.; Sabloff, M.; Allan, D.S. Plerixafor in combination with chemotherapy and/or hematopoietic cell transplantation to treat acute leukemia: A systematic review and metanalysis of preclinical and clinical studies. Leuk. Res. 2020, 97, 106442. [Google Scholar] [CrossRef] [PubMed]
- Nervi, B.; Ramirez, P.; Rettig, M.P.; Uy, G.L.; Holt, M.S.; Ritchey, J.K.; Prior, J.L.; Piwnica-Worms, D.; Bridger, G.; Ley, T.J.; et al. Chemosensitization of acute myeloid leukemia (AML) following mobilization by the CXCR4 antagonist AMD3100. Blood 2009, 113, 6206–6214. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Boyd, A.L.; Reid, J.C.; Salci, K.R.; Aslostovar, L.; Benoit, Y.D.; Shapovalova, Z.; Nakanishi, M.; Porras, D.P.; Almakadi, M.; Campbell, C.J.V.; et al. Acute myeloid leukaemia disrupts endogenous myelo-erythropoiesis by compromising the adipocyte bone marrow niche. Nat. Cell Biol. 2017, 19, 1336–1347. [Google Scholar] [CrossRef] [PubMed]
- Kim, Y.; Jekarl, D.W.; Kim, J.; Kwon, A.; Choi, H.; Lee, S.; Kim, Y.-J.; Kim, H.-J.; Kim, Y.; Oh, I.-H.; et al. Genetic and epigenetic alterations of bone marrow stromal cells in myelodysplastic syndrome and acute myeloid leukemia patients. Stem Cell Res. 2015, 14, 177–184. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Huang, J.C.; Basu, S.K.; Zhao, X.; Chien, S.; Fang, M.; Oehler, V.G.; Appelbaum, F.R.; Becker, P.S. Mesenchymal stromal cells derived from acute myeloid leukemia bone marrow exhibit aberrant cytogenetics and cytokine elaboration. Blood Cancer J. 2015, 5, e302. [Google Scholar] [CrossRef]
- Geyh, S.; Rodríguez-Paredes, M.; Jäger, P.; Khandanpour, C.; Cadeddu, R.P.; Gutekunst, J.; Wilk, C.M.; Fenk, R.; Zilkens, C.; Hermsen, D.; et al. Functional inhibition of mesenchymal stromal cells in acute myeloid leukemia. Leukemia 2016, 30, 683–691. [Google Scholar] [CrossRef]
- Battula, V.L.; Le, P.M.; Sun, J.C.; Nguyen, K.; Yuan, B.; Zhou, X.; Sonnylal, S.; McQueen, T.; Ruvolo, V.; Michel, K.A.; et al. AML-induced osteogenic differentiation in mesenchymal stromal cells supports leukemia growth. JCI Insight 2017, 2, e90036. [Google Scholar] [CrossRef]
- Plaks, V.; Kong, N.; Werb, Z. The cancer stem cell niche: How essential is the niche in regulating stemness of tumor cells? Cell Stem Cell 2015, 16, 225–238. [Google Scholar] [CrossRef][Green Version]
- Plakhova, N.; Panagopoulos, V.; Vandyke, K.; Zannettino, A.C.W.; Mrozik, K.M. Mesenchymal stromal cell senescence in haematological malignancies. Cancer Metastasis Rev. 2023, 42, 277–296. [Google Scholar] [CrossRef]
- Zheng, K.; Chen, S.; Hu, X. Peroxisome Proliferator-activated Receptor Gamma Coactivator-1 Alpha: A Double-edged Sword in Prostate Cancer. Curr. Cancer Drug Targets 2022, 22, 541–559. [Google Scholar] [CrossRef]
- Sasca, D.; Szybinski, J.; Schüler, A.; Shah, V.; Heidelberger, J.; Haehnel, P.S.; Dolnik, A.; Kriege, O.; Fehr, E.-M.; Gebhardt, W.H.; et al. NCAM1 (CD56) promotes leukemogenesis and confers drug resistance in AML. Blood 2019, 133, 2305–2319. [Google Scholar] [CrossRef] [PubMed]
- Shi, Y.; Xia, Y.-Y.; Wang, L.; Liu, R.; Khoo, K.-S.; Feng, Z.-W. Neural cell adhesion molecule modulates mesenchymal stromal cell migration via activation of MAPK/ERK signaling. Exp. Cell Res. 2012, 318, 2257–2267. [Google Scholar] [CrossRef] [PubMed]
- Skog, M.S.; Nystedt, J.; Korhonen, M.; Anderson, H.; Lehti, T.A.; Pajunen, M.I.; Finne, J. Expression of neural cell adhesion molecule and polysialic acid in human bone marrow-derived mesenchymal stromal cells. Stem Cell Res. Ther. 2016, 7, 113. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Kim, Y.H.; Kim, Y.H.; Shin, Y.K.; Jo, Y.R.; Park, D.K.; Song, M.-Y.; Yoon, B.-A.; Nam, S.H.; Kim, J.H.; Choi, B.-O.; et al. p75 and neural cell adhesion molecule 1 can identify pathologic Schwann cells in peripheral neuropathies. Ann. Clin. Transl. Neurol. 2019, 6, 1292–1301. [Google Scholar] [CrossRef] [PubMed]
- Tsumagari, K.; Shirakabe, K.; Ogura, M.; Sato, F.; Ishihama, Y.; Sehara-Fujisawa, A. Secretome analysis to elucidate metalloprotease-dependent ectodomain shedding of glycoproteins during neuronal differentiation. Genes Cells 2017, 22, 237–244. [Google Scholar] [CrossRef][Green Version]
- Li, X.; Liang, Y.; Lian, C.; Peng, F.; Xiao, Y.; He, Y.; Ma, C.; Wang, Y.; Zhang, P.; Deng, Y.; et al. CST6 protein and peptides inhibit breast cancer bone metastasis by suppressing CTSB activity and osteoclastogenesis. Theranostics 2021, 11, 9821–9832. [Google Scholar] [CrossRef]
- Gai, D.; Chen, J.-R.; Stewart, J.P.; Nookaew, I.; Habelhah, H.; Ashby, C.; Sun, F.; Cheng, Y.; Li, C.; Xu, H.; et al. CST6 suppresses osteolytic bone disease in multiple myeloma by blocking osteoclast differentiation. J. Clin. Investig. 2022, 132, e159527. [Google Scholar] [CrossRef]
- Tan, H.-Y.; Wang, N.; Zhang, C.; Chan, Y.-T.; Yuen, M.-F.; Feng, Y. Lysyl Oxidase-Like 4 Fosters an Immunosuppressive Microenvironment During Hepatocarcinogenesis. Hepatology 2021, 73, 2326–2341. [Google Scholar] [CrossRef]
- Choi, S.K.; Kim, H.S.; Jin, T.; Moon, W.K. LOXL4 knockdown enhances tumor growth and lung metastasis through collagen-dependent extracellular matrix changes in triple-negative breast cancer. Oncotarget 2017, 8, 11977–11989. [Google Scholar] [CrossRef] [PubMed][Green Version]
- van der Meer, J.H.M.; van der Poll, T.; van ’t Veer, C. TAM receptors, Gas6, and protein S: Roles in inflammation and hemostasis. Blood 2014, 123, 2460–2469. [Google Scholar] [CrossRef]
- Lamandé, S.R.; Bateman, J.F. Collagen VI disorders: Insights on form and function in the extracellular matrix and beyond. Matrix Biol. 2018, 71–72, 348–367. [Google Scholar] [CrossRef]
- Seoane, J.; Gomis, R.R. TGF-β Family Signaling in Tumor Suppression and Cancer Progression. Cold Spring Harb. Perspect. Biol. 2017, 9, a022277. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Fredriksson, L.; Li, H.; Eriksson, U. The PDGF family: Four gene products form five dimeric isoforms. Cytokine Growth Factor Rev. 2004, 15, 197–204. [Google Scholar] [CrossRef] [PubMed]
- Andrae, J.; Gallini, R.; Betsholtz, C. Role of platelet-derived growth factors in physiology and medicine. Genes Dev. 2008, 22, 1276–1312. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Kong, D.-H.; Kim, Y.K.; Kim, M.R.; Jang, J.H.; Lee, S. Emerging Roles of Vascular Cell Adhesion Molecule-1 (VCAM-1) in Immunological Disorders and Cancer. Int. J. Mol. Sci. 2018, 19, 1057. [Google Scholar] [CrossRef][Green Version]
- Reis, E.S.; Falcão, D.A.; Isaac, L. Clinical aspects and molecular basis of primary deficiencies of complement component C3 and its regulatory proteins factor I and factor H. Scand. J. Immunol. 2006, 63, 155–168. [Google Scholar] [CrossRef]
- Pettigrew, H.D.; Teuber, S.S.; Gershwin, M.E. Clinical significance of complement deficiencies. Ann. N. Y. Acad. Sci. 2009, 1173, 108–123. [Google Scholar] [CrossRef] [PubMed]
- Albitar, M.; Vose, J.M.; Johnson, M.M.; Do, K.-A.; Day, A.; Jilani, I.; Kantarjian, H.; Keating, M.; O’Brien, S.M.; Verstovsek, S.; et al. Clinical relevance of soluble HLA-I and beta2-microglobulin levels in non-Hodgkin’s lymphoma and Hodgkin’s disease. Leuk. Res. 2007, 31, 139–145. [Google Scholar] [CrossRef]
- Puppo, F.; Scudeletti, M.; Indiveri, F.; Ferrone, S. Serum HLA class I antigens: Markers and modulators of an immune response? Immunol. Today 1995, 16, 124–127. [Google Scholar] [CrossRef]
- Tabayoyong, W.B.; Zavazava, N. Soluble HLA revisited. Leuk. Res. 2007, 31, 121–125. [Google Scholar] [CrossRef]
- Sadagopan, A.; Michelakos, T.; Boyiadzis, G.; Ferrone, C.; Ferrone, S. Human Leukocyte Antigen Class I Antigen-Processing Machinery Upregulation by Anticancer Therapies in the Era of Checkpoint Inhibitors: A Review. JAMA Oncol. 2022, 8, 462–473. [Google Scholar] [CrossRef] [PubMed]
- Luo, Y.; Wang, X.; Shen, J.; Yao, J. Macrophage migration inhibitory factor in the pathogenesis of leukemia (Review). Int. J. Oncol. 2021, 59, 62. [Google Scholar] [CrossRef] [PubMed]
- Ponta, H.; Sherman, L.; Herrlich, P.A. CD44: From adhesion molecules to signalling regulators. Nat. Rev. Mol. Cell Biol. 2003, 4, 33–45. [Google Scholar] [CrossRef] [PubMed]
- Yu, Y.; Tan, C.-M.; Jia, Y.-Y. Research status and the prospect of POSTN in various tumors. Neoplasma 2021, 68, 673–682. [Google Scholar] [CrossRef]
- González-González, L.; Alonso, J. Periostin: A Matricellular Protein With Multiple Functions in Cancer Development and Progression. Front. Oncol. 2018, 8, 225. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Waters, J.A.; Urbano, I.; Robinson, M.; House, C.D. Insulin-like growth factor binding protein 5: Diverse roles in cancer. Front. Oncol. 2022, 12, 1052457. [Google Scholar] [CrossRef]
- Dong, R.; Zhang, M.; Hu, Q.; Zheng, S.; Soh, A.; Zheng, Y.; Yuan, H. Galectin-3 as a novel biomarker for disease diagnosis and a target for therapy (Review). Int. J. Mol. Med. 2018, 41, 599–614. [Google Scholar] [CrossRef][Green Version]
- de Castro, L.L.; Lopes-Pacheco, M.; Weiss, D.J.; Cruz, F.F.; Rocco, P.R.M. Current understanding of the immunosuppressive properties of mesenchymal stromal cells. J. Mol. Med. 2019, 97, 605–618. [Google Scholar] [CrossRef]
- Georgiou, K.R.; Foster, B.K.; Xian, C.J. Damage and recovery of the bone marrow microenvironment induced by cancer chemotherapy—Potential regulatory role of chemokine CXCL12/receptor CXCR4 signalling. Curr. Mol. Med. 2010, 10, 440–453. [Google Scholar] [CrossRef]
- Geng, S.; Wang, J.; Zhang, X.; Zhang, J.J.; Wu, F.; Pang, Y.; Zhong, Y.; Wang, J.; Wang, W.; Lyu, X.; et al. Single-cell RNA sequencing reveals chemokine self-feeding of myeloma cells promotes extramedullary metastasis. FEBS Lett. 2020, 594, 452–465. [Google Scholar] [CrossRef][Green Version]
- Shi, Y.; Riese, D.J.; Shen, J. The Role of the CXCL12/CXCR4/CXCR7 Chemokine Axis in Cancer. Front. Pharmacol. 2020, 11, 574667. [Google Scholar] [CrossRef] [PubMed]
- Abdul-Aziz, A.M.; Sun, Y.; Hellmich, C.; Marlein, C.R.; Mistry, J.; Forde, E.; Piddock, R.E.; Shafat, M.S.; Morfakis, A.; Mehta, T.; et al. Acute myeloid leukemia induces protumoral p16INK4a-driven senescence in the bone marrow microenvironment. Blood 2019, 133, 446–456. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Ruiz-Aparicio, P.F.; Vernot, J.-P. Bone Marrow Aging and the Leukaemia-Induced Senescence of Mesenchymal Stem/Stromal Cells: Exploring Similarities. J. Pers. Med. 2022, 12, 716. [Google Scholar] [CrossRef]
- Llewellyn, J.; Mallikarjun, V.; Appleton, E.; Osipova, M.; Gilbert, H.T.J.; Richardson, S.M.; Hubbard, S.J.; Swift, J. Loss of regulation of protein synthesis and turnover underpins an attenuated stress response in senescent human mesenchymal stem cells. Proc. Natl. Acad. Sci. USA 2023, 120, e2210745120. [Google Scholar] [CrossRef]
- Ramasamy, R.; Lam, E.W.-F.; Soeiro, I.; Tisato, V.; Bonnet, D.; Dazzi, F. Mesenchymal stem cells inhibit proliferation and apoptosis of tumor cells: Impact on in vivo tumor growth. Leukemia 2007, 21, 304–310. [Google Scholar] [CrossRef]
- Skelding, K.A.; Barry, D.L.; Theron, D.Z.; Lincz, L.F. Bone Marrow Microenvironment as a Source of New Drug Targets for the Treatment of Acute Myeloid Leukaemia. Int. J. Mol. Sci. 2023, 24, 563. [Google Scholar] [CrossRef]
- Winer, E.S.; Stone, R.M. Novel therapy in Acute myeloid leukemia (AML): Moving toward targeted approaches. Ther. Adv. Hematol. 2019, 10, 2040620719860645. [Google Scholar] [CrossRef]
- Kuzmina, L.A.; Petinati, N.A.; Parovichnikova, E.N.; Lubimova, L.S.; Gribanova, E.O.; Gaponova, T.V.; Shipounova, I.N.; Zhironkina, O.A.; Bigildeev, A.E.; Svinareva, D.A.; et al. Multipotent Mesenchymal Stromal Cells for the Prophylaxis of Acute Graft-versus-Host Disease-A Phase II Study. Stem Cells Int. 2012, 2012, 968213. [Google Scholar] [CrossRef]
- Fastova, E.A.; Magomedova, A.U.; Petinati, N.A.; Sats, N.V.; Kapranov, N.M.; Davydova, Y.O.; Drize, N.I.; Kravchenko, S.K.; Savchenko, V.G. Bone Marrow Multipotent Mesenchymal Stromal Cells in Patients with Diffuse Large B-Cell Lymphoma. Bull. Exp. Biol. Med. 2019, 167, 150–153. [Google Scholar] [CrossRef] [PubMed]
- Kumar, P.; Kandoi, S.; Misra, R.; Vijayalakshmi, S.; Rajagopal, K.; Verma, R.S. The mesenchymal stem cell secretome: A new paradigm towards cell-free therapeutic mode in regenerative medicine. Cytokine Growth Factor Rev. 2019, 46, 1–9. [Google Scholar] [CrossRef]

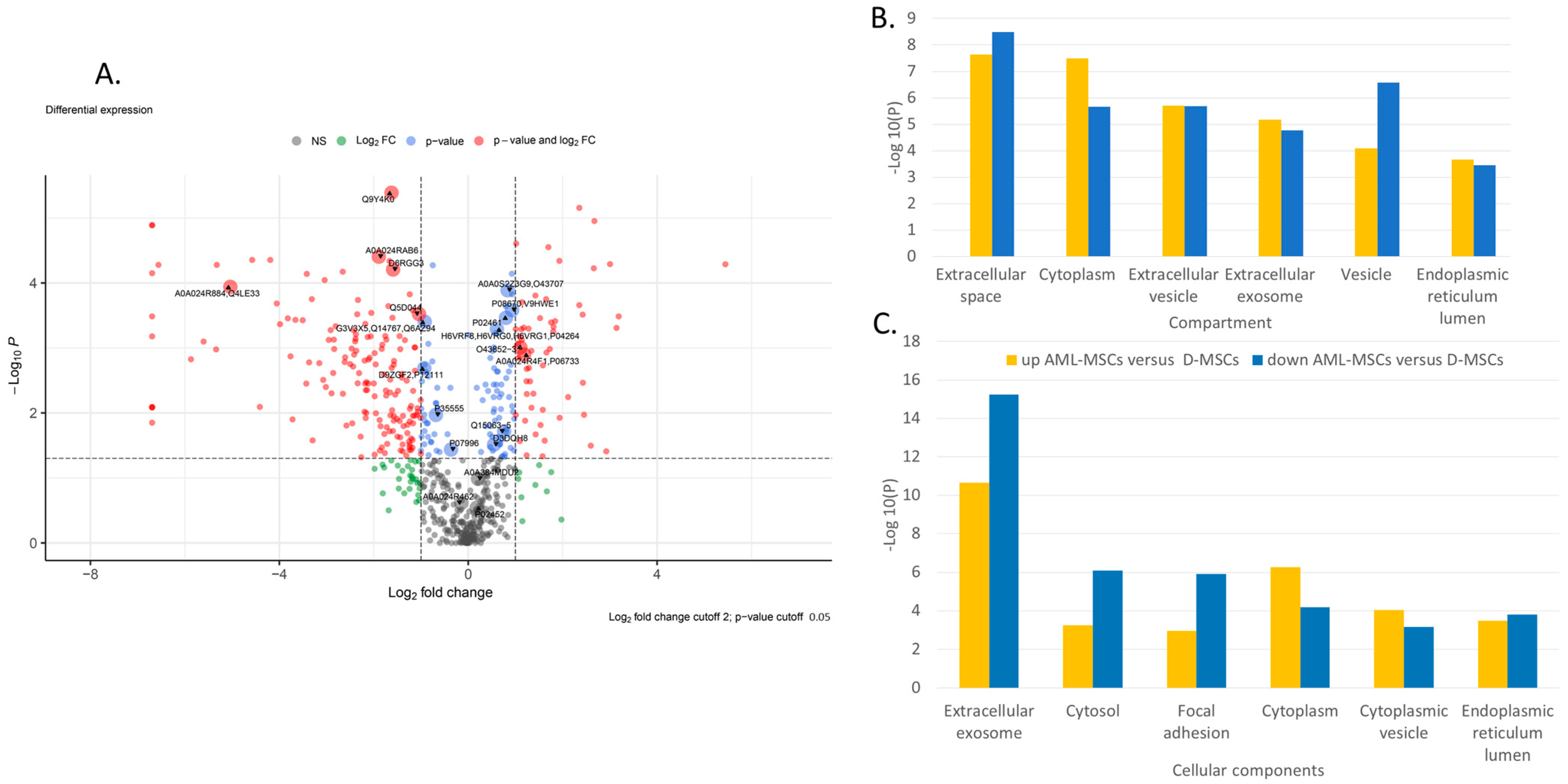
| Elements | Pathway Database | Term Description |
|---|---|---|
| Higher in AML-MSCs versus D-MSCs | GO COMPARTMENTS | Extracellular space, Cytoplasm, Extracellular region, Extracellular vesicle, Extracellular exosome, Proteasome core complex, Intracellular, Secretory granule, Integral component of endoplasmic reticulum membrane, Vesicle, Organelle, Membrane-bounded organelle, Intracellular organelle, Endomembrane system, ficolin-1-rich granule lumen, Intracellular organelle lumen, Intracellular membrane-bounded organelle, MHC class I peptide loading complex, Tertiary granule lumen, Endoplasmic reticulum lumen, Basement membrane, Proteasome core complex, beta-subunit complex, Cytoplasmic vesicle, Cellular anatomical entity, Collagen-containing extracellular matrix, Cytosol |
| GO PROCESS | Antigen processing and presentation of exogenous peptide antigen, Antigen processing and presentation of exogenous peptide antigen via MHC class I, tap-dependent, Cytokine-mediated signaling pathway, Regulated exocytosis, Proteasomal ubiquitin-independent protein catabolic process, Cellular response to cytokine stimulus, Vesicle-mediated transport, Immune system process, Secretion, interleukin-1-mediated signaling pathway, Cellular process | |
| GO COMPONENT | Extracellular region, Extracellular exosome, Extracellular space, Vesicle, Cytoplasm, Membrane-bounded organelle, Proteasome core complex, Secretory granule, Cytoplasmic vesicle, MHC class I peptide loading complex, ficolin-1-rich granule lumen, Tertiary granule lumen, Proteasome core complex, beta-subunit complex, Integral component of endoplasmic reticulum membrane, Endoplasmic reticulum lumen, Intracellular organelle lumen, Secretory granule lumen, Cytosol, Basement membrane, Endomembrane system, MHC protein complex, Collagen-containing extracellular matrix, Focal adhesion, Transport vesicle | |
| Lower in AML-MSCs versus D-MSCs | GO COMPARTMENTS | Extracellular space, Intermediate filament, Supramolecular fiber, Cytoskeleton, Vesicle, Polymeric cytoskeletal fiber, Keratin filament, Cytosol, Cytoplasm, Extracellular vesicle, Focal adhesion, Anchoring junction, Cornified envelope, Extracellular exosome, Intracellular non-membrane-bounded organelle, Pseudopodium, Blood microparticle, Intracellular organelle, Endoplasmic reticulum lumen, Cell surface |
| GO PROCESS | Cornification, Epithelial cell differentiation, Glycolytic process, Multicellular organism development, Peptide cross-linking, Multicellular organismal process, glyceraldehyde-3-phosphate biosynthetic process, Epithelium development, Supramolecular fiber organization, NAD metabolic process, interleukin-12-mediated signaling pathway, Monosaccharide biosynthetic process, Animal organ development, Cytoskeleton organization, Protein folding in the endoplasmic reticulum, Protein tetramerization, System development, Cell differentiation, Protein heterotetramerization, Oxidation-reduction process | |
| GO COMPONENT | Extracellular exosome, Cytosol, Intermediate filament, Focal adhesion, Supramolecular fiber, Melanosome, Anchoring junction, Cytoskeleton, Cornified envelope, Polymeric cytoskeletal fiber, Cytoplasm, Cell surface, Endoplasmic reticulum chaperone complex, Keratin filament, Pseudopodium, Endoplasmic reticulum lumen, Blood microparticle, Nucleus, Cytoplasmic vesicle | |
| GO FUNCTION | Structural molecule activity, Structural constituent of the cytoskeleton, Protein binding, Structural constituent of skin epidermis, Cell adhesion molecule binding, Cytoskeletal protein binding, Intramolecular oxidoreductase activity, Actin binding, Structural constituent of postsynapse, Peptide disulfide oxidoreductase activity, Protein disulfide isomerase activity, Identical protein binding, Cadherin binding, Protein dimerization activity |
| Elements | Pathway Database | Term Description |
|---|---|---|
| Higher in R-MSCs versus D-MSC | Biological process | Negative regulation of intrinsic apoptotic signaling pathway in response to DNA damage by p53 class mediator, Proteasomal ubiquitin-independent protein catabolic process, Endothelial cell development, Regulation of transcription from RNA polymerase II promoter in response to hypoxia, interleukin-1-mediated signaling pathway, Antigen processing and presentation of peptide antigen via MHC class I, Cellular response to interferon-gamma, Response to interleukin-1, Cellular response to hypoxia, T cell receptor signaling pathway, Extracellular matrix organization, Regulation of response to DNA damage stimulus, Positive regulation of growth, Cytokine-mediated signaling pathway, Immune response, Cellular response to tumor necrosis factor, Post-translational protein modification, Leukocyte mediated immunity, Response to cytokine, Cell adhesion, Secretion, Vesicle-mediated transport, Regulation of apoptotic process, Cell differentiation |
| Lower in R-MSCs versus D-MSCs | Biological process | Aging, Angiogenesis, Antigen processing and presentation, Blood coagulation, Blood vessel development, Bone development, Cell activation, Cell adhesion, Cell morphogenesis involved in differentiation, Chemotaxis, Chondrocyte development, Complement activation, Exocytosis, Extracellular matrix organization, Immune response, Innate immune response, Leukocyte activation, Myeloid leukocyte mediated immunity, Ossification, Plasminogen activation, Platelet-derived growth factor receptor signaling pathway, Posttranscriptional regulation of gene expression, Regeneration, Regulation of cell death, Regulation of cell differentiation, Regulation of cell growth, Signal transduction, Skeletal system development, Tissue homeostasis, Transforming growth factor beta receptor signaling pathway, Translation, Transport, Vesicle-mediated transport |
| Elements | Pathway Database | Term Description |
|---|---|---|
| Higher in R-MSCs versus AML-MSCs | GO Cellular component | Endomembrane system, Endoplasmic reticulum, Endoplasmic reticulum lumen, Extracellular exosome, Extracellular region, Extracellular space, Intracellular organelle lumen, Lysosome, Melanosome, Vesicle |
| Lower in R-MSCs versus AML-MSCs | Cellular component | Collagen-containing extracellular matrix, Extracellular matrix, Extracellular exosome, Extracellular space, Extracellular region |
| Elements | Pathway Database | Term Description |
|---|---|---|
| Equally expressed compared to D-MSCs | ||
| Higher in R-MSCs and AML-MSCs versus D-MSCs | GO Biological process | Extracellular matrix assembly, Platelet degranulation, Chondrocyte differentiation, interleukin-12-mediated signaling pathway, Platelet-derived growth factor receptor signaling pathway, Osteoclast differentiation, Regulation of epithelial to mesenchymal transition, Bone development, Cellular response to transforming growth factor beta stimulus, Cell adhesion, Angiogenesis, Blood coagulation, Mesenchymal cell differentiation, Secretion, Response to growth factor, Negative regulation of canonical WNT signaling pathway, Cell activation involved in immune response, Aging, Vesicle-mediated transport, Posttranscriptional regulation of gene expression, Regulation of translation, Immune system process, Regulation of cell death, Cellular protein modification process, Signaling |
| Lower in R-MSCs and AML-MSCs versus D-MSCs | Biological process | Response to interleukin-1, Extracellular matrix organization, Leukocyte migration, Response to hypoxia, Angiogenesis, Cytokine-mediated signaling pathway, Blood vessel morphogenesis, Cellular response to cytokine stimulus, Leukocyte mediated immunity, Cell adhesion, Secretion, Vesicle-mediated transport, Immune system process, Cell differentiation |
| Differently expressed compared to D-MSCs | ||
| Secreted by R-MSCs Lower than D-MSCs | Biological process | Regulation of complement activation, Cell-matrix adhesion, Leukocyte mediated immunity, Extracellular matrix organization, Angiogenesis, Secretion, Immune response, Vesicle-mediated transport, Cell adhesion |
| Secreted by R-MSCs Higher than D-MSCs | Cellular component | Extracellular exosome, Extracellular space, Extracellular region, Vesicle |
| Secreted by AML-MSCs Lower than D-MSCs | Biological process | interleukin-12-mediated signaling pathway, Extracellular matrix organization, Osteoblast differentiation, Ossification, Transmembrane receptor protein tyrosine kinase signaling pathway, Cell adhesion, Response to cytokine, Response to growth factor, Secretion, Vesicle-mediated transport, Immune system process |
| Secreted by AML-MSCs Higher than D-MSCs | Cellular component | Cytosolic ribosome, Secretory granule lumen, Collagen-containing extracellular matrix, Extracellular matrix, Secretory granule, Extracellular exosome, Extracellular space, Extracellular region, Vesicle |
| Acute Myeloid Leukemia | Donors | ||
|---|---|---|---|
| Onset of the Disease | Remission | ||
| Age, years (median) | 26–64 (38) | 26–64 (38) | 22–59 (36) |
| Gender male/female | 3/10 | 3/10 | 10/11 |
| Cumulative MSCs production for 3 passages, × 106 (M ± ME) | 12.5 ± 3.2 | 15.3 ± 2.2 | 8.7 ± 1.7 |
| MSCs-Time to P0, days (M ± ME) | 15.8 ± 1.2 | 14.1 ± 0.9 | 12.3 ± 0.4 |
| MSCs-Time to P3, days (M ± ME) | 35.2 ± 1.9 | 29.2 ± 1.5 | 24.4 ± 0.8 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sadovskaya, A.; Petinati, N.; Drize, N.; Smirnov, I.; Pobeguts, O.; Arapidi, G.; Lagarkova, M.; Belyavsky, A.; Vasilieva, A.; Aleshina, O.; et al. Acute Myeloid Leukemia Causes Serious and Partially Irreversible Changes in Secretomes of Bone Marrow Multipotent Mesenchymal Stromal Cells. Int. J. Mol. Sci. 2023, 24, 8953. https://doi.org/10.3390/ijms24108953
Sadovskaya A, Petinati N, Drize N, Smirnov I, Pobeguts O, Arapidi G, Lagarkova M, Belyavsky A, Vasilieva A, Aleshina O, et al. Acute Myeloid Leukemia Causes Serious and Partially Irreversible Changes in Secretomes of Bone Marrow Multipotent Mesenchymal Stromal Cells. International Journal of Molecular Sciences. 2023; 24(10):8953. https://doi.org/10.3390/ijms24108953
Chicago/Turabian StyleSadovskaya, Aleksandra, Nataliya Petinati, Nina Drize, Igor Smirnov, Olga Pobeguts, Georgiy Arapidi, Maria Lagarkova, Alexander Belyavsky, Anastasia Vasilieva, Olga Aleshina, and et al. 2023. "Acute Myeloid Leukemia Causes Serious and Partially Irreversible Changes in Secretomes of Bone Marrow Multipotent Mesenchymal Stromal Cells" International Journal of Molecular Sciences 24, no. 10: 8953. https://doi.org/10.3390/ijms24108953
APA StyleSadovskaya, A., Petinati, N., Drize, N., Smirnov, I., Pobeguts, O., Arapidi, G., Lagarkova, M., Belyavsky, A., Vasilieva, A., Aleshina, O., & Parovichnikova, E. (2023). Acute Myeloid Leukemia Causes Serious and Partially Irreversible Changes in Secretomes of Bone Marrow Multipotent Mesenchymal Stromal Cells. International Journal of Molecular Sciences, 24(10), 8953. https://doi.org/10.3390/ijms24108953







