The Many Ways to Deal with STING
Abstract
1. Introduction
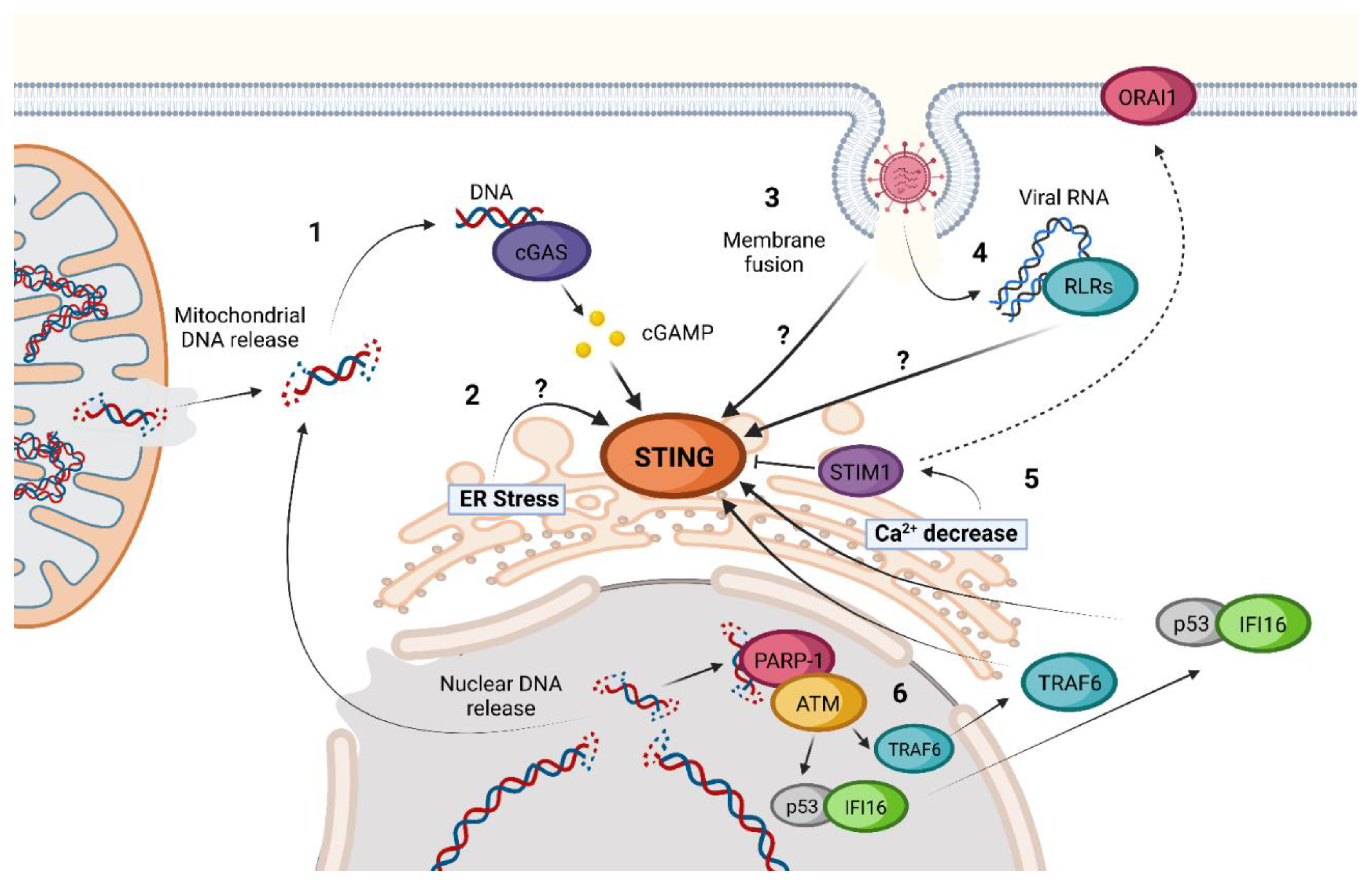
2. cGAS-STING Canonical Signaling Pathway
3. STING Non-Canonical Signaling Pathway
4. STING Regulation and Inhibition Mechanisms
| Modification | Protein Involved | Locus (Human STING) | Outcome | |
|---|---|---|---|---|
| Positive regulation | K63-linked ubiquitination | TRIM32 [91,92], TRIM56 [92], UBXN3B [107], MUL1 [43], RNF115 [98] | K20, K150, K224, K236, K289 | Increase of STING-TBK1 interaction |
| K27-linked ubiquitination | AMFR [108] | K137, K150, K224, K236 | Increase of STING-TBK1 interaction | |
| K11-linked ubiquitination | RNF26 [97] | K150 | Prevention of K48-linked ubiquitination | |
| K48-linked deubiquitination | USP20 [99,100,102], USP44 [101], CYLD [109], EIF3S5 [42] and OTUD5 [110] | K33, K236, K150, K347 | Increase of STING stability | |
| Y245 phosphorylation | EGFR [40] | Y245 | Enabling STING trafficking | |
| S358 phosphorylation | TBK1 [48] | S358 | Enabling TBK1-STING complex formation | |
| S366 phosphorylation | TBK1 [46] | S366 | Enabling STING interaction with IRF3 | |
| Palmitoylation | DHHC3, DHHC7, DHHC15 [45] | C88, C91 | Induction of STING oligomerization | |
| SUMOylation | TRIM38 [90] | K338 | Prevention of STING degradation | |
| Negative regulation | K48-linked ubiquitination | TRIM29 [93], TIM30α [94], RNF5 [95], RNF90 [96] | K370, K275, K150 | Induction of STING degradation |
| K63-linked deubiquitination | USP49 [103], USP21 [105], USP35 [106], MYSM1 [111] | K150 | Decrease of STING interaction with TBK1 | |
| K27- linked deubiquitination | USP13 [104], USP21 [105], USP35 [106] | Unknown | Decrease of STING interaction with TBK1 | |
| K6-, K11- and K29-linked deubiquitination | USP35 [106] | Unknown | Decrease of STING interaction with TBK1 | |
| S366 phosphorylation | ULK1 [88] | S366 | Inhibition of STING-dependent IRF3 activation | |
| S358 dephosphorylation | PPM1A [48] | S358 | Reduction of STING oligomerization | |
| deSUMOylation | SENP2 [90] | K338 | Induction of STING degradation |
5. Tridimensional Structure of STING and Conformational Changes upon Activation
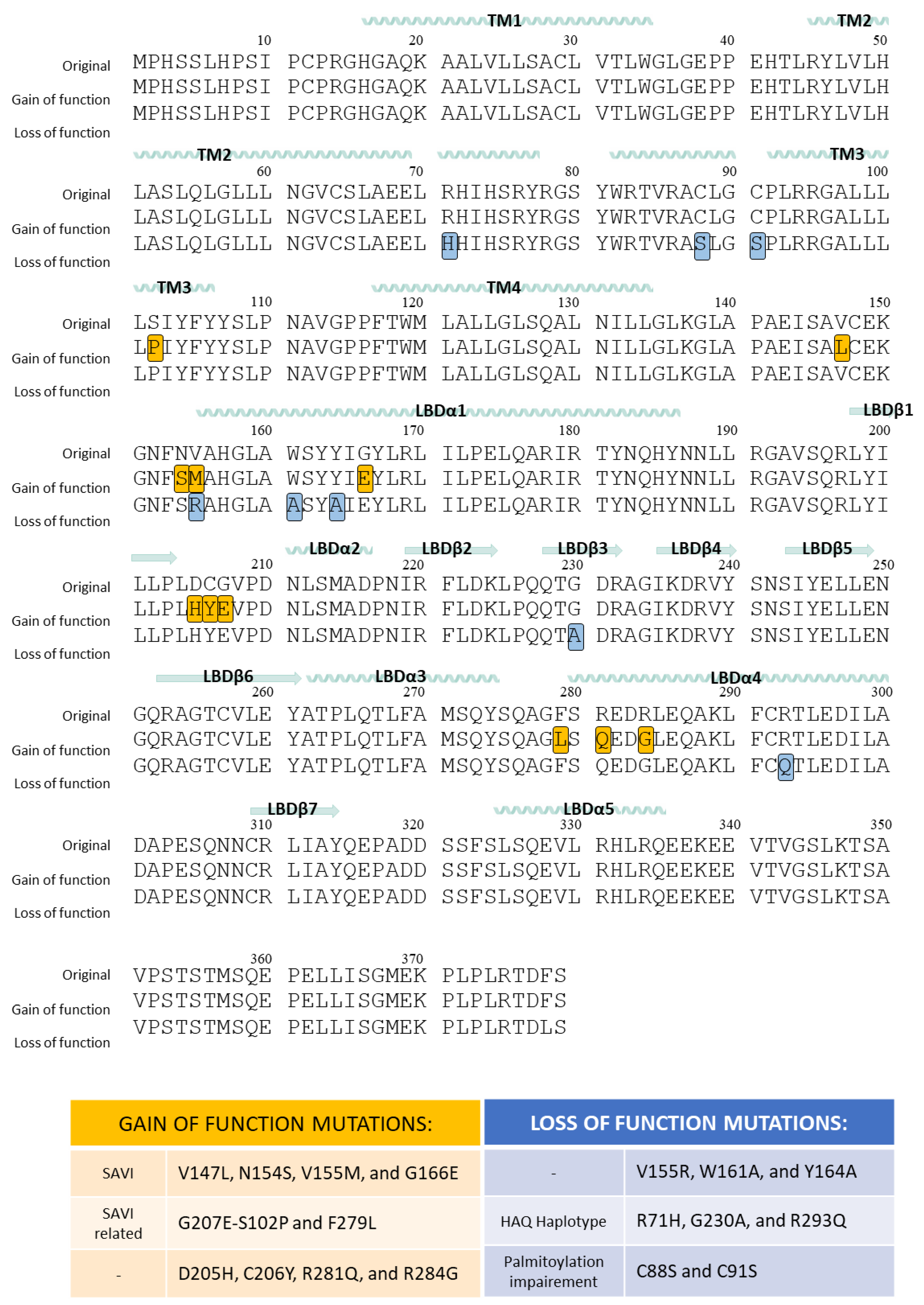
6. STING Polymorphisms
6.1. Gain-of-Function Mutations of STING
6.2. Loss-of-Function Mutations of STING
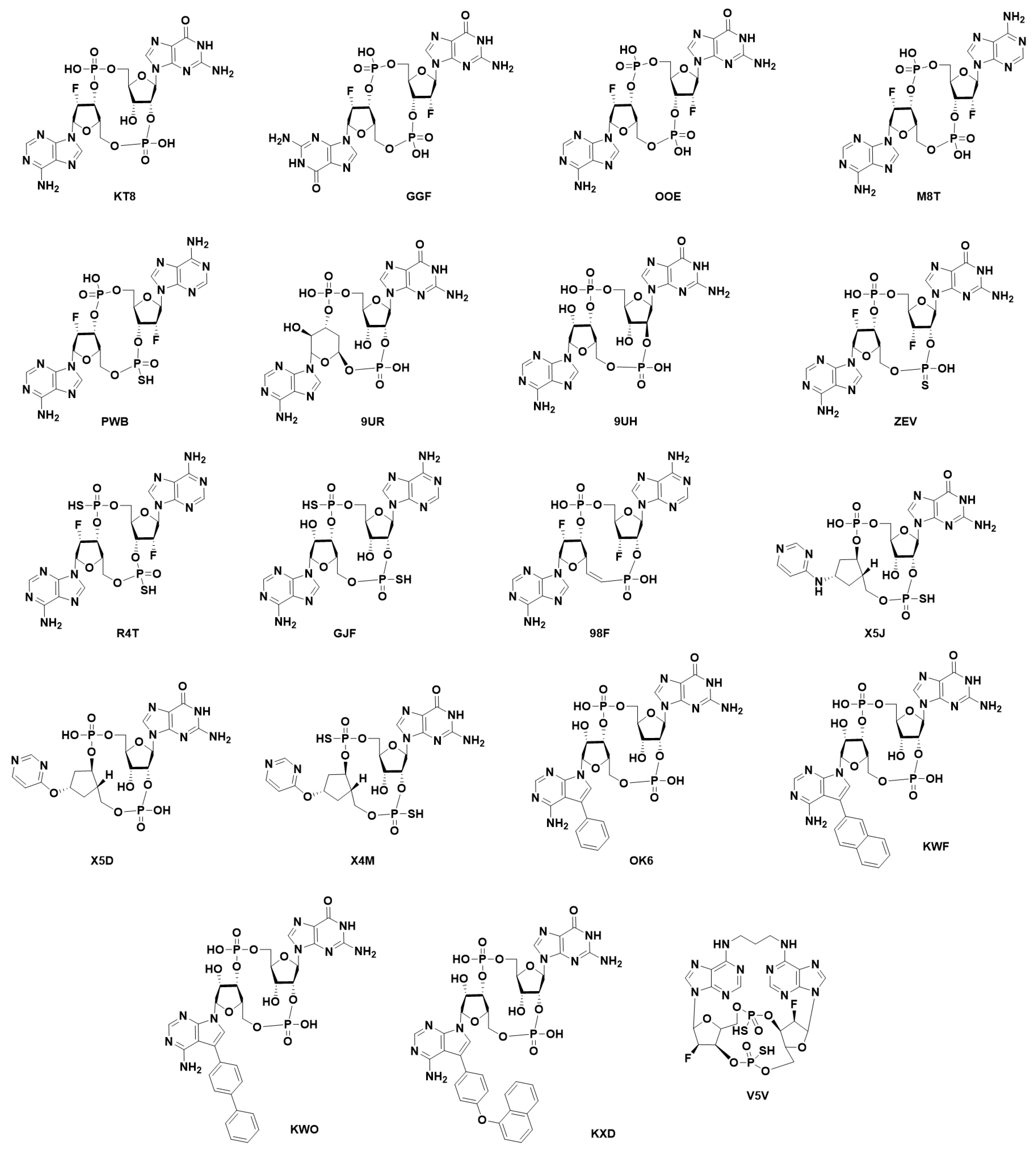
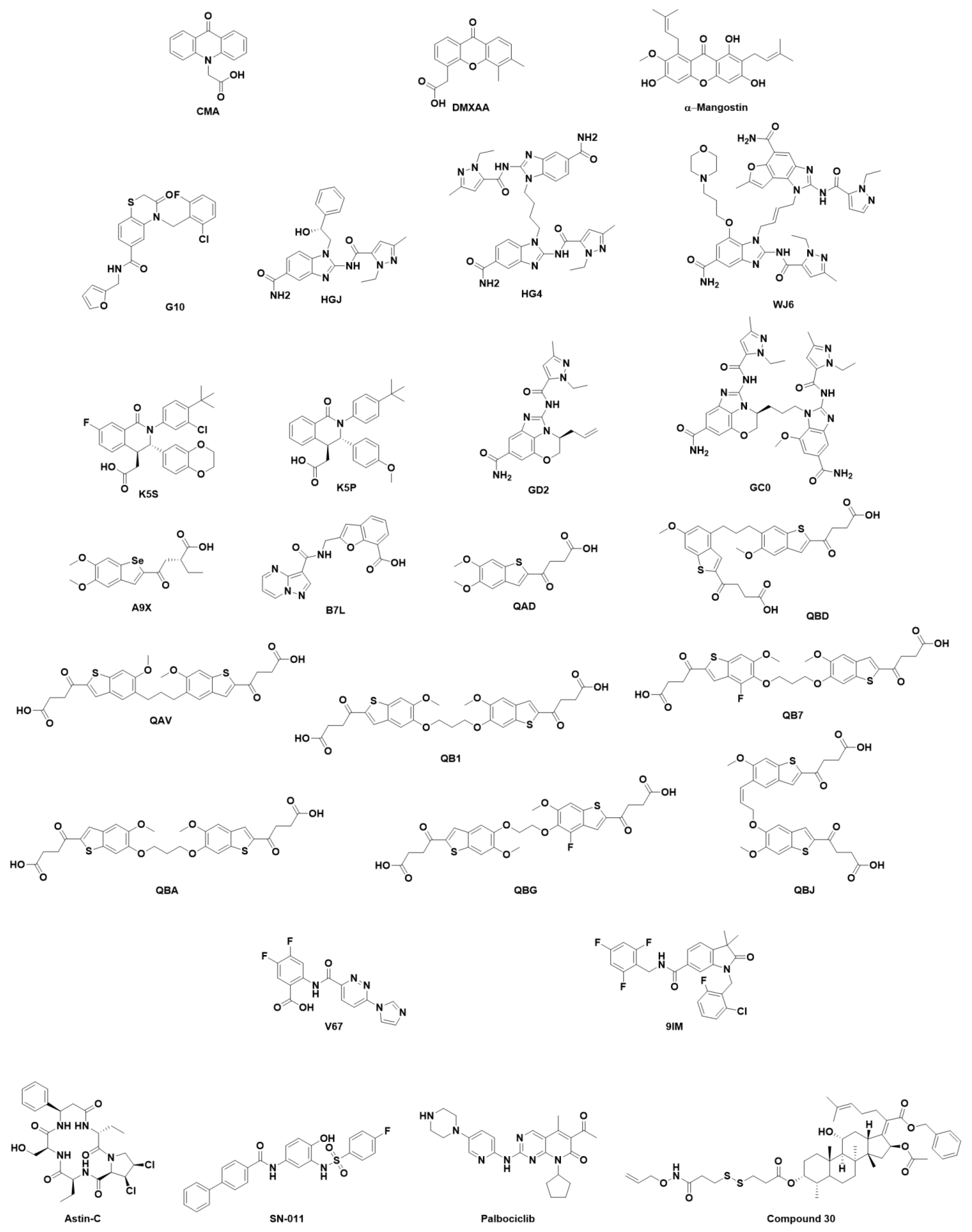
7. Structural Insight into Ligand-STING Interaction
7.1. Cyclic Di-Nucleotides as Natural STING Activators
7.2. Small Organic Molecules as Drug Candidates
8. Perspectives
Supplementary Materials
Funding
Conflicts of Interest
References
- Ishikawa, H.; Barber, G.N. STING is an Endoplasmic Reticulum Adaptor that Facilitates Innate Immune Signalling. Nature 2008, 455, 674–678. [Google Scholar] [CrossRef] [PubMed]
- Barber, G.N. STING: Infection, Inflammation, and Cancer. Nat. Rev. Immunol. 2015, 15, 760–770. [Google Scholar] [CrossRef] [PubMed]
- Cai, H.; Imler, J. cGAS-STING: Insight on the Evolution of a Primordial Antiviral Signaling Cassette. Fac. Rev. 2021, 10, 54. [Google Scholar] [CrossRef] [PubMed]
- Kranzusch, P.J. cGAS and CD-NTase Enzymes: Structure, Mechanism, and Evolution. Curr. Opin. Struct. Biol. 2019, 59, 178–187. [Google Scholar] [CrossRef]
- Morehouse, B.R.; Govande, A.A.; Millman, A.; Keszei, A.F.A.; Lowey, B.; Ofir, G.; Shao, S.; Sorek, R.; Kranzusch, P.J. STING Cyclic Dinucleotide Sensing Originated in Bacteria. Nature 2020, 586, 429–433. [Google Scholar] [CrossRef]
- Anastasiou, M.; Newton, G.A.; Kaur, K.; Carrillo-Salinas, F.J.; Smolgovsky, S.A.; Bayer, A.L.; Ilyukha, V.; Sharma, S.; Poltorak, A.; Luscinskas, F.W.; et al. Endothelial STING Controls T Cell Transmigration in an IFNI-Dependent Manner. JCI Insight. 2021, 6, e149346. [Google Scholar]
- Yu, Y.; Yang, W.; Bilotta, A.J.; Yu, Y.; Zhao, X.; Zhou, Z.; Yao, S.; Xu, J.; Zhou, J.; Dann, S.M.; et al. STING Controls Intestinal Homeostasis through Promoting Antimicrobial Peptide Expression in Epithelial Cells. FASEB J. 2020, 34, 15417–15430. [Google Scholar] [CrossRef]
- Nazmi, A.; Mukhopadhyay, R.; Dutta, K.; Basu, A. STING Mediates Neuronal Innate Immune Response Following Japanese Encephalitis Virus Infection. Sci. Rep. 2012, 2, 347. [Google Scholar] [CrossRef]
- Larkin, B.; Ilyukha, V.; Sorokin, M.; Buzdin, A.; Vannier, E.; Poltorak, A. Cutting Edge: Activation of STING in T Cells Induces Type I IFN Responses and Cell Death. J. Immunol. 2017, 199, 397–402. [Google Scholar] [CrossRef]
- Walker, M.M.; Crute, B.W.; Cambier, J.C.; Getahun, A. B Cell-Intrinsic STING Signaling Triggers Cell Activation, Synergizes with B Cell Receptor Signals, and Promotes Antibody Responses. J. Immunol. 2018, 201, 2641–2653. [Google Scholar] [CrossRef]
- Souza-Fonseca-Guimaraes, F.; Parlato, M.; de Oliveira, R.B.; Golenbock, D.; Fitzgerald, K.; Shalova, I.N.; Biswas, S.K.; Cavaillon, J.; Adib-Conquy, M. Interferon-Γ and Granulocyte/Monocyte Colony-Stimulating Factor Production by Natural Killer Cells Involves Different Signaling Pathways and the Adaptor Stimulator of Interferon Genes (STING). J. Biol. Chem. 2013, 288, 10715–10721. [Google Scholar] [CrossRef]
- Margolis, S.R.; Wilson, S.C.; Vance, R.E. Evolutionary Origins of cGAS-STING Signaling. Trends Immunol. 2017, 38, 733–743. [Google Scholar] [CrossRef] [PubMed]
- Paludan, S.R.; Bowie, A.G. Immune Sensing of DNA. Immunity 2013, 38, 870–880. [Google Scholar] [CrossRef] [PubMed]
- Yang, P.; An, H.; Liu, X.; Wen, M.; Zheng, Y.; Rui, Y.; Cao, X. The Cytosolic Nucleic Acid Sensor LRRFIP1 Mediates the Production of Type I Interferon Via a Beta-Catenin-Dependent Pathway. Nat. Immunol. 2010, 11, 487–494. [Google Scholar] [CrossRef] [PubMed]
- Lin, B.; Goldbach-Mansky, R. Pathogenic Insights from Genetic Causes of Autoinflammatory Inflammasomopathies and Interferonopathies. J. Allergy Clin. Immunol. 2022, 149, 819–832. [Google Scholar] [CrossRef] [PubMed]
- Ishikawa, H.; Ma, Z.; Barber, G.N. STING Regulates Intracellular DNA-Mediated, Type I Interferon-Dependent Innate Immunity. Nature 2009, 461, 788–792. [Google Scholar] [CrossRef]
- Li, X.; Wu, J.; Gao, D.; Wang, H.; Sun, L.; Chen, Z.J. Pivotal Roles of cGAS-cGAMP Signaling in Antiviral Defense and Immune Adjuvant Effects. Science 2013, 341, 1390–1394. [Google Scholar] [CrossRef]
- Sun, Y.; Cheng, Y. STING Or Sting: cGAS-STING-Mediated Immune Response to Protozoan Parasites. Trends Parasitol. 2020, 36, 773–784. [Google Scholar] [CrossRef]
- Webb, L.G.; Fernandez-Sesma, A. RNA Viruses, and the cGAS-STING Pathway: Reframing our Understanding of Innate Immune Sensing. Curr. Opin. Virol. 2022, 53, 101206. [Google Scholar] [CrossRef]
- Nitta, S.; Sakamoto, N.; Nakagawa, M.; Kakinuma, S.; Mishima, K.; Kusano-Kitazume, A.; Kiyohashi, K.; Murakawa, M.; Nishimura-Sakurai, Y.; Azuma, S.; et al. Hepatitis C Virus NS4B Protein Targets STING and Abrogates RIG-I-Mediated Type I Interferon-Dependent Innate Immunity. Hepatology 2013, 57, 46–58. [Google Scholar] [CrossRef]
- Rui, Y.; Su, J.; Shen, S.; Hu, Y.; Huang, D.; Zheng, W.; Lou, M.; Shi, Y.; Wang, M.; Chen, S.; et al. Unique and Complementary Suppression of cGAS-STING and RNA Sensing-Triggered Innate Immune Responses by SARS-CoV-2 Proteins. Signal Transduct. Target. Ther. 2021, 6, 123. [Google Scholar] [CrossRef] [PubMed]
- Corrales, L.; McWhirter, S.M.; Dubensky, T.W.; Gajewski, T.F. The Host STING Pathway at the Interface of Cancer and Immunity. J. Clin. Investig. 2016, 126, 2404–2411. [Google Scholar] [CrossRef] [PubMed]
- Foote, J.B.; Kok, M.; Leatherman, J.M.; Armstrong, T.D.; Marcinkowski, B.C.; Ojalvo, L.S.; Kanne, D.B.; Jaffee, E.M.; Dubensky, T.W.; Emens, L.A. A STING Agonist Given with OX40 Receptor and PD-L1 Modulators Primes Immunity and Reduces Tumor Growth in Tolerized Mice. Cancer Immunol. Res. 2017, 5, 468–479. [Google Scholar] [CrossRef] [PubMed]
- Demaria, O.; De Gassart, A.; Coso, S.; Gestermann, N.; Di Domizio, J.; Flatz, L.; Gaide, O.; Michielin, O.; Hwu, P.; Petrova, T.V.; et al. STING Activation of Tumor Endothelial Cells Initiates Spontaneous and Therapeutic Antitumor Immunity. Proc. Natl. Acad. Sci. USA 2015, 112, 15408–15413. [Google Scholar] [CrossRef]
- Yu, L.; Liu, P. Cytosolic DNA Sensing by cGAS: Regulation, Function, and Human Diseases. Signal Transduct. Target. Ther. 2021, 6, 170. [Google Scholar] [CrossRef]
- Sun, L.; Wu, J.; Du, F.; Chen, X.; Chen, Z.J. Cyclic GMP-AMP Synthase is a Cytosolic DNA Sensor that Activates the Type I Interferon Pathway. Science 2013, 339, 786–791. [Google Scholar] [CrossRef]
- Gehrke, N.; Mertens, C.; Zillinger, T.; Wenzel, J.; Bald, T.; Zahn, S.; Tüting, T.; Hartmann, G.; Barchet, W. Oxidative Damage of DNA Confers Resistance to Cytosolic Nuclease TREX1 Degradation and Potentiates STING-Dependent Immune Sensing. Immunity 2013, 39, 482–495. [Google Scholar] [CrossRef]
- Decout, A.; Katz, J.D.; Venkatraman, S.; Ablasser, A. The cGAS-STING Pathway as a Therapeutic Target in Inflammatory Diseases. Nat. Rev. Immunol. 2021, 21, 548–569. [Google Scholar] [CrossRef]
- Luecke, S.; Holleufer, A.; Christensen, M.H.; Jønsson, K.L.; Boni, G.A.; Sørensen, L.K.; Johannsen, M.; Jakobsen, M.R.; Hartmann, R.; Paludan, S.R. cGAS is Activated by DNA in a Length-Dependent Manner. EMBO Rep. 2017, 18, 1707–1715. [Google Scholar] [CrossRef]
- Wan, D.; Jiang, W.; Hao, J. Research Advances in how the cGAS-STING Pathway Controls the Cellular Inflammatory Response. Front. Immunol. 2020, 11, 615. [Google Scholar] [CrossRef]
- Burdette, D.L.; Monroe, K.M.; Sotelo-Troha, K.; Iwig, J.S.; Eckert, B.; Hyodo, M.; Hayakawa, Y.; Vance, R.E. STING is a Direct Innate Immune Sensor of Cyclic Di-GMP. Nature 2011, 478, 515–518. [Google Scholar] [CrossRef] [PubMed]
- Gürsoy, U.K.; Gürsoy, M.; Könönen, E.; Sintim, H.O. Cyclic Dinucleotides in Oral Bacteria and in Oral Biofilms. Front. Cell. Infect. Microbiol. 2017, 7, 273. [Google Scholar] [CrossRef] [PubMed]
- Devaux, L.; Sleiman, D.; Mazzuoli, M.; Gominet, M.; Lanotte, P.; Trieu-Cuot, P.; Kaminski, P.; Firon, A. Cyclic Di-AMP Regulation of Osmotic Homeostasis is Essential in Group B Streptococcus. PLoS Genet. 2018, 14, e1007342. [Google Scholar] [CrossRef] [PubMed]
- Purcell, E.B.; Tamayo, R. Cyclic Diguanylate Signaling in Gram-Positive Bacteria. FEMS Microbiol. Rev. 2016, 40, 753–773. [Google Scholar] [CrossRef] [PubMed]
- Randall, T.E.; Eckartt, K.; Kakumanu, S.; Price-Whelan, A.; Dietrich, L.E.P.; Harrison, J.J. Sensory Perception in Bacterial Cyclic Diguanylate Signal Transduction. J. Bacteriol. 2022, 204, e0043321. [Google Scholar] [CrossRef] [PubMed]
- Ha, D.; O’Toole, G.A. C-Di-GMP and its Effects on Biofilm Formation and Dispersion: A Pseudomonas Aeruginosa Review. Microbiol. Spectr. 2015, 3, MB-2014. [Google Scholar] [CrossRef]
- Cohen, D.; Melamed, S.; Millman, A.; Shulman, G.; Oppenheimer-Shaanan, Y.; Kacen, A.; Doron, S.; Amitai, G.; Sorek, R. Cyclic GMP-AMP Signalling Protects Bacteria Against Viral Infection. Nature 2019, 574, 691–695. [Google Scholar] [CrossRef]
- Ergun, S.L.; Fernandez, D.; Weiss, T.M.; Li, L. STING Polymer Structure Reveals Mechanisms for Activation, Hyperactivation, and Inhibition. Cell 2019, 178, 290–301.e10. [Google Scholar] [CrossRef]
- Shang, G.; Zhang, C.; Chen, Z.J.; Bai, X.; Zhang, X. Cryo-EM Structures of STING Reveal its Mechanism of Activation by Cyclic GMP-AMP. Nature 2019, 567, 389–393. [Google Scholar] [CrossRef]
- Wang, C.; Wang, X.; Veleeparambil, M.; Kessler, P.M.; Willard, B.; Chattopadhyay, S.; Sen, G.C. EGFR-Mediated Tyrosine Phosphorylation of STING Determines its Trafficking Route and Cellular Innate Immunity Functions. EMBO J. 2020, 39, e104106. [Google Scholar] [CrossRef]
- Dobbs, N.; Burnaevskiy, N.; Chen, D.; Gonugunta, V.K.; Alto, N.M.; Yan, N. STING Activation by Translocation from the ER is Associated with Infection and Autoinflammatory Disease. Cell Host Microbe 2015, 18, 157–168. [Google Scholar] [CrossRef] [PubMed]
- Luo, W.; Li, S.; Li, C.; Lian, H.; Yang, Q.; Zhong, B.; Shu, H. iRhom2 is Essential for Innate Immunity to DNA Viruses by Mediating Trafficking and Stability of the Adaptor STING. Nat. Immunol. 2016, 17, 1057–1066. [Google Scholar] [CrossRef] [PubMed]
- Ni, G.; Konno, H.; Barber, G.N. Ubiquitination of STING at Lysine 224 Controls IRF3 Activation. Sci. Immunol. 2017, 2, eaah7119. [Google Scholar] [CrossRef] [PubMed]
- Fang, R.; Jiang, Q.; Guan, Y.; Gao, P.; Zhang, R.; Zhao, Z.; Jiang, Z. Golgi Apparatus-Synthesized Sulfated Glycosaminoglycans Mediate Polymerization and Activation of the cGAMP Sensor STING. Immunity 2021, 54, 962–975.e8. [Google Scholar] [CrossRef] [PubMed]
- Mukai, K.; Konno, H.; Akiba, T.; Uemura, T.; Waguri, S.; Kobayashi, T.; Barber, G.N.; Arai, H.; Taguchi, T. Activation of STING Requires Palmitoylation at the Golgi. Nat. Commun. 2016, 7, 11932. [Google Scholar] [CrossRef] [PubMed]
- Zhang, C.; Shang, G.; Gui, X.; Zhang, X.; Bai, X.; Chen, Z.J. Structural Basis of STING Binding with and Phosphorylation by TBK1. Nature 2019, 567, 394–398. [Google Scholar] [CrossRef]
- Yu, Y.; Liu, J.; Liu, C.; Liu, R.; Liu, L.; Yu, Z.; Zhuang, J.; Sun, C. Post-Translational Modifications of cGAS-STING: A Critical Switch for Immune Regulation. Cells 2022, 11, 3043. [Google Scholar] [CrossRef]
- Li, Z.; Liu, G.; Sun, L.; Teng, Y.; Guo, X.; Jia, J.; Sha, J.; Yang, X.; Chen, D.; Sun, Q. PPM1A Regulates Antiviral Signaling by Antagonizing TBK1-Mediated STING Phosphorylation and Aggregation. PLoS Pathog. 2015, 11, e1004783. [Google Scholar] [CrossRef]
- Tamura, T.; Yanai, H.; Savitsky, D.; Taniguchi, T. The IRF Family Transcription Factors in Immunity and Oncogenesis. Annu. Rev. Immunol. 2008, 26, 535–584. [Google Scholar] [CrossRef]
- Balka, K.R.; Louis, C.; Saunders, T.L.; Smith, A.M.; Calleja, D.J.; D’Silva, D.B.; Moghaddas, F.; Tailler, M.; Lawlor, K.E.; Zhan, Y.; et al. TBK1 and IKKε Act Redundantly to Mediate STING-Induced NF-κB Responses in Myeloid Cells. Cell Rep. 2020, 31, 107492. [Google Scholar] [CrossRef]
- Zhu, Q.; Man, S.M.; Gurung, P.; Liu, Z.; Vogel, P.; Lamkanfi, M.; Kanneganti, T. Cutting Edge: STING Mediates Protection Against Colorectal Tumorigenesis by Governing the Magnitude of Intestinal Inflammation. J. Immunol. 2014, 193, 4779–4782. [Google Scholar] [CrossRef] [PubMed]
- Gui, X.; Yang, H.; Li, T.; Tan, X.; Shi, P.; Li, M.; Du, F.; Chen, Z.J. Autophagy Induction Via STING Trafficking is a Primordial Function of the cGAS Pathway. Nature 2019, 567, 262–266. [Google Scholar] [CrossRef] [PubMed]
- Zhang, R.; Kang, R.; Tang, D. The STING1 Network Regulates Autophagy and Cell Death. Signal Transduct. Target. Ther. 2021, 6, 208. [Google Scholar] [CrossRef]
- Liu, D.; Wu, H.; Wang, C.; Li, Y.; Tian, H.; Siraj, S.; Sehgal, S.A.; Wang, X.; Wang, J.; Shang, Y.; et al. STING Directly Activates Autophagy to Tune the Innate Immune Response. Cell Death Differ. 2019, 26, 1735–1749. [Google Scholar] [CrossRef] [PubMed]
- Tian, Y.; Wang, M.; Zhao, J. Crosstalk between Autophagy and Type I Interferon Responses in Innate Antiviral Immunity. Viruses 2019, 11, 132. [Google Scholar] [CrossRef]
- Srikanth, S.; Woo, J.S.; Wu, B.; El-Sherbiny, Y.M.; Leung, J.; Chupradit, K.; Rice, L.; Seo, G.J.; Calmettes, G.; Ramakrishna, C.; et al. The Ca2+ Sensor STIM1 Regulates the Type I Interferon Response by Retaining the Signaling Adaptor STING at the Endoplasmic Reticulum. Nat. Immunol. 2019, 20, 152–162. [Google Scholar] [CrossRef] [PubMed]
- Petrasek, J.; Iracheta-Vellve, A.; Csak, T.; Satishchandran, A.; Kodys, K.; Kurt-Jones, E.A.; Fitzgerald, K.A.; Szabo, G. STING-IRF3 Pathway Links Endoplasmic Reticulum Stress with Hepatocyte Apoptosis in Early Alcoholic Liver Disease. Proc. Natl. Acad. Sci. USA 2013, 110, 16544–16549. [Google Scholar] [CrossRef]
- Dunphy, G.; Flannery, S.M.; Almine, J.F.; Connolly, D.J.; Paulus, C.; Jønsson, K.L.; Jakobsen, M.R.; Nevels, M.M.; Bowie, A.G.; Unterholzner, L. Non-Canonical Activation of the DNA Sensing Adaptor STING by ATM and IFI16 Mediates NF-κB Signaling After Nuclear DNA Damage. Mol. Cell 2018, 71, 745–760.e5. [Google Scholar] [CrossRef]
- Jønsson, K.L.; Laustsen, A.; Krapp, C.; Skipper, K.A.; Thavachelvam, K.; Hotter, D.; Egedal, J.H.; Kjolby, M.; Mohammadi, P.; Prabakaran, T.; et al. IFI16 is Required for DNA Sensing in Human Macrophages by Promoting Production and Function of cGAMP. Nat. Commun. 2017, 8, 14391. [Google Scholar] [CrossRef]
- Li, S.; Kong, L.; Yu, X. The Expanding Roles of Endoplasmic Reticulum Stress in Virus Replication and Pathogenesis. Crit. Rev. Microbiol. 2015, 41, 150–164. [Google Scholar] [CrossRef]
- Choi, J.; Song, C. Insights into the Role of Endoplasmic Reticulum Stress in Infectious Diseases. Front. Immunol. 2019, 10, 3147. [Google Scholar] [CrossRef] [PubMed]
- McGuckin Wuertz, K.; Treuting, P.M.; Hemann, E.A.; Esser-Nobis, K.; Snyder, A.G.; Graham, J.B.; Daniels, B.P.; Wilkins, C.; Snyder, J.M.; Voss, K.M.; et al. STING is Required for Host Defense Against Neuropathological West Nile Virus Infection. PLoS Pathog. 2019, 15, e1007899. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Chen, Y.; Chen, Y.; Chiu, W.; Shen, M. The STIM1-Orai1 Pathway of Store-Operated Ca2+ Entry Controls the Checkpoint in Cell Cycle G1/S Transition. Sci. Rep. 2016, 6, 22142. [Google Scholar] [CrossRef] [PubMed]
- Mekahli, D.; Bultynck, G.; Parys, J.B.; De Smedt, H.; Missiaen, L. Endoplasmic-Reticulum Calcium Depletion and Disease. Cold Spring Harb. Perspect. Biol. 2011, 3, a004317. [Google Scholar] [CrossRef]
- Wrighton, K. The STING Behind Dengue Virus Infection. Nat. Rev. Microbiol. 2018, 16, 330. [Google Scholar] [CrossRef]
- Ni, G.; Ma, Z.; Damania, B. cGAS and STING: At the Intersection of DNA and RNA Virus-Sensing Networks. PLoS Pathog. 2018, 14, e1007148. [Google Scholar] [CrossRef]
- Franz, K.M.; Neidermyer, W.J.; Tan, Y.; Whelan, S.P.J.; Kagan, J.C. STING-Dependent Translation Inhibition Restricts RNA Virus Replication. Proc. Natl. Acad. Sci. USA 2018, 115, E2058–E2067. [Google Scholar] [CrossRef]
- Webb, L.G.; Veloz, J.; Pintado-Silva, J.; Zhu, T.; Rangel, M.V.; Mutetwa, T.; Zhang, L.; Bernal-Rubio, D.; Figueroa, D.; Carrau, L.; et al. Chikungunya Virus Antagonizes cGAS-STING Mediated Type-I Interferon Responses by Degrading cGAS. PLoS Pathog. 2020, 16, e1008999. [Google Scholar] [CrossRef]
- Aguirre, S.; Maestre, A.M.; Pagni, S.; Patel, J.R.; Savage, T.; Gutman, D.; Maringer, K.; Bernal-Rubio, D.; Shabman, R.S.; Simon, V.; et al. DENV Inhibits Type I IFN Production in Infected Cells by Cleaving Human STING. PLoS Pathog. 2012, 8, e1002934. [Google Scholar] [CrossRef]
- Ding, Q.; Cao, X.; Lu, J.; Huang, B.; Liu, Y.; Kato, N.; Shu, H.; Zhong, J. Hepatitis C Virus NS4B Blocks the Interaction of STING and TBK1 to Evade Host Innate Immunity. J. Hepatol. 2013, 59, 52–58. [Google Scholar] [CrossRef]
- Sun, L.; Xing, Y.; Chen, X.; Zheng, Y.; Yang, Y.; Nichols, D.B.; Clementz, M.A.; Banach, B.S.; Li, K.; Baker, S.C.; et al. Coronavirus Papain-Like Proteases Negatively Regulate Antiviral Innate Immune Response through Disruption of STING-Mediated Signaling. PLoS ONE 2012, 7, e30802. [Google Scholar] [CrossRef] [PubMed]
- Aguirre, S.; Luthra, P.; Sanchez-Aparicio, M.T.; Maestre, A.M.; Patel, J.; Lamothe, F.; Fredericks, A.C.; Tripathi, S.; Zhu, T.; Pintado-Silva, J.; et al. Dengue Virus NS2B Protein Targets cGAS for Degradation and Prevents Mitochondrial DNA Sensing during Infection. Nat. Microbiol. 2017, 2, 17037. [Google Scholar] [CrossRef] [PubMed]
- Lai, J.; Wang, M.; Huang, C.; Wu, C.; Hung, L.; Yang, C.; Ke, P.; Luo, S.; Liu, S.; Ho, L. Infection with the Dengue RNA Virus Activates TLR9 Signaling in Human Dendritic Cells. EMBO Rep. 2018, 19, e46182. [Google Scholar] [CrossRef] [PubMed]
- Sun, B.; Sundström, K.B.; Chew, J.J.; Bist, P.; Gan, E.S.; Tan, H.C.; Goh, K.C.; Chawla, T.; Tang, C.K.; Ooi, E.E. Dengue Virus Activates cGAS through the Release of Mitochondrial DNA. Sci. Rep. 2017, 7, 3594. [Google Scholar] [CrossRef]
- Liu, X.; Wei, L.; Xu, F.; Zhao, F.; Huang, Y.; Fan, Z.; Mei, S.; Hu, Y.; Zhai, L.; Guo, J.; et al. SARS-CoV-2 Spike Protein-Induced Cell Fusion Activates the cGAS-STING Pathway and the Interferon Response. Sci. Signal. 2022, 15, eabg8744. [Google Scholar] [CrossRef]
- Holm, C.K.; Jensen, S.B.; Jakobsen, M.R.; Cheshenko, N.; Horan, K.A.; Moeller, H.B.; Gonzalez-Dosal, R.; Rasmussen, S.B.; Christensen, M.H.; Yarovinsky, T.O.; et al. Virus-Cell Fusion as a Trigger of Innate Immunity Dependent on the Adaptor STING. Nat. Immunol. 2012, 13, 737–743. [Google Scholar] [CrossRef]
- Holm, C.K.; Rahbek, S.H.; Gad, H.H.; Bak, R.O.; Jakobsen, M.R.; Jiang, Z.; Hansen, A.L.; Jensen, S.K.; Sun, C.; Thomsen, M.K.; et al. Influenza A Virus Targets a cGAS-Independent STING Pathway that Controls Enveloped RNA Viruses. Nat. Commun. 2016, 7, 10680. [Google Scholar] [CrossRef]
- Liu, Y.; Goulet, M.; Sze, A.; Hadj, S.B.; Belgnaoui, S.M.; Lababidi, R.R.; Zheng, C.; Fritz, J.H.; Olagnier, D.; Lin, R. RIG-I-Mediated STING Upregulation Restricts Herpes Simplex Virus 1 Infection. J. Virol. 2016, 90, 9406–9419. [Google Scholar] [CrossRef]
- Pathare, G.R.; Decout, A.; Glück, S.; Cavadini, S.; Makasheva, K.; Hovius, R.; Kempf, G.; Weiss, J.; Kozicka, Z.; Guey, B.; et al. Structural Mechanism of cGAS Inhibition by The nucleosome. Nature 2020, 587, 668–672. [Google Scholar] [CrossRef]
- Guey, B.; Wischnewski, M.; Decout, A.; Makasheva, K.; Kaynak, M.; Sakar, M.S.; Fierz, B.; Ablasser, A. BAF Restricts cGAS on Nuclear DNA to Prevent Innate Immune Activation. Science 2020, 369, 823–828. [Google Scholar] [CrossRef]
- Xia, P.; Wang, S.; Ye, B.; Du, Y.; Li, C.; Xiong, Z.; Qu, Y.; Fan, Z. A Circular RNA Protects Dormant Hematopoietic Stem Cells from DNA Sensor cGAS-Mediated Exhaustion. Immunity 2018, 48, 688–701.e7. [Google Scholar] [CrossRef]
- Sun, X.; Liu, T.; Zhao, J.; Xia, H.; Xie, J.; Guo, Y.; Zhong, L.; Li, M.; Yang, Q.; Peng, C.; et al. DNA-PK Deficiency Potentiates cGAS-Mediated Antiviral Innate Immunity. Nat. Commun. 2020, 11, 6182. [Google Scholar] [CrossRef] [PubMed]
- Liang, J.; Hong, Z.; Sun, B.; Guo, Z.; Wang, C.; Zhu, J. The Alternatively Spliced Isoforms of Key Molecules in the cGAS-STING Signaling Pathway. Front. Immunol. 2021, 12, 771744. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Li, M.; Li, L.; Qian, G.; Wang, Y.; Chen, Z.; Liu, J.; Fang, C.; Huang, F.; Guo, D.; et al. Β-Arrestin 2 as an Activator of cGAS-STING Signaling and Target of Viral Immune Evasion. Nat. Commun. 2020, 11, 6000. [Google Scholar] [CrossRef] [PubMed]
- Prabakaran, T.; Bodda, C.; Krapp, C.; Zhang, B.; Christensen, M.H.; Sun, C.; Reinert, L.; Cai, Y.; Jensen, S.B.; Skouboe, M.K.; et al. Attenuation of cGAS-STING Signaling is Mediated by a p62/SQSTM1-Dependent Autophagy Pathway Activated by TBK1. EMBO J. 2018, 37, e97858. [Google Scholar] [CrossRef]
- Gonugunta, V.K.; Sakai, T.; Pokatayev, V.; Yang, K.; Wu, J.; Dobbs, N.; Yan, N. Trafficking-Mediated STING Degradation Requires Sorting to Acidified Endolysosomes and can be Targeted to Enhance Anti-Tumor Response. Cell Rep. 2017, 21, 3234–3242. [Google Scholar] [CrossRef]
- Saitoh, T.; Fujita, N.; Hayashi, T.; Takahara, K.; Satoh, T.; Lee, H.; Matsunaga, K.; Kageyama, S.; Omori, H.; Noda, T.; et al. Atg9a Controls dsDNA-Driven Dynamic Translocation of STING and the Innate Immune Response. Proc. Natl. Acad. Sci. USA 2009, 106, 20842–20846. [Google Scholar] [CrossRef]
- Konno, H.; Konno, K.; Barber, G.N. Cyclic Dinucleotides Trigger ULK1 (ATG1) Phosphorylation of STING to Prevent Sustained Innate Immune Signaling. Cell 2013, 155, 688–698. [Google Scholar] [CrossRef]
- van Gent, M.; Sparrer, K.M.J.; Gack, M.U. TRIM Proteins and their Roles in Antiviral Host Defenses. Annu. Rev. Virol. 2018, 5, 385–405. [Google Scholar] [CrossRef]
- Hu, M.; Yang, Q.; Xie, X.; Liao, C.; Lin, H.; Liu, T.; Yin, L.; Shu, H. Sumoylation Promotes the Stability of the DNA Sensor cGAS and the Adaptor STING to Regulate the Kinetics of Response to DNA Virus. Immunity 2016, 45, 555–569. [Google Scholar] [CrossRef]
- Zhang, J.; Hu, M.; Wang, Y.; Shu, H. TRIM32 Protein Modulates Type I Interferon Induction and Cellular Antiviral Response by Targeting MITA/STING Protein for K63-Linked Ubiquitination. J. Biol. Chem. 2012, 287, 28646–28655. [Google Scholar] [CrossRef] [PubMed]
- Tsuchida, T.; Zou, J.; Saitoh, T.; Kumar, H.; Abe, T.; Matsuura, Y.; Kawai, T.; Akira, S. The Ubiquitin Ligase TRIM56 Regulates Innate Immune Responses to Intracellular Double-Stranded DNA. Immunity 2010, 33, 765–776. [Google Scholar] [CrossRef] [PubMed]
- Xing, J.; Zhang, A.; Zhang, H.; Wang, J.; Li, X.C.; Zeng, M.; Zhang, Z. TRIM29 Promotes DNA Virus Infections by Inhibiting Innate Immune Response. Nat. Commun. 2017, 8, 945. [Google Scholar] [CrossRef]
- Wang, Y.; Lian, Q.; Yang, B.; Yan, S.; Zhou, H.; He, L.; Lin, G.; Lian, Z.; Jiang, Z.; Sun, B. TRIM30α is a Negative-Feedback Regulator of the Intracellular DNA and DNA Virus-Triggered Response by Targeting STING. PLoS Pathog. 2015, 11, e1005012. [Google Scholar] [CrossRef]
- Zhong, B.; Zhang, L.; Lei, C.; Li, Y.; Mao, A.; Yang, Y.; Wang, Y.; Zhang, X.; Shu, H. The Ubiquitin Ligase RNF5 Regulates Antiviral Responses by Mediating Degradation of the Adaptor Protein MITA. Immunity 2009, 30, 397–407. [Google Scholar] [CrossRef]
- Yang, B.; Liu, Y.; Cui, Y.; Song, D.; Zhang, G.; Ma, S.; Liu, Y.; Chen, M.; Chen, F.; Wang, H.; et al. RNF90 Negatively Regulates Cellular Antiviral Responses by Targeting MITA for Degradation. PLoS Pathog. 2020, 16, e1008387. [Google Scholar] [CrossRef] [PubMed]
- Qin, Y.; Zhou, M.; Hu, M.; Hu, Y.; Zhang, J.; Guo, L.; Zhong, B.; Shu, H. RNF26 Temporally Regulates Virus-Triggered Type I Interferon Induction by Two Distinct Mechanisms. PLoS Pathog. 2014, 10, e1004358. [Google Scholar] [CrossRef]
- Zhang, Z.; Xiong, T.; Yao, S.; Wei, M.; Chen, M.; Lin, D.; Zhong, B. RNF115 Plays Dual Roles in Innate Antiviral Responses by Catalyzing Distinct Ubiquitination of MAVS and MITA. Nat. Commun. 2020, 11, 5536. [Google Scholar] [CrossRef]
- Zhang, M.; Zhang, M.; Zhang, Q.; Zhu, G.; Yuan, L.; Zhang, D.; Zhu, Q.; Yao, J.; Shu, H.; Zhong, B. USP18 Recruits USP20 to Promote Innate Antiviral Response through Deubiquitinating STING/MITA. Cell. Res. 2016, 26, 1302–1319. [Google Scholar] [CrossRef]
- Zhang, M.; Cai, Z.; Zhang, M.; Wang, X.; Wang, Y.; Zhao, F.; Zhou, J.; Luo, M.; Zhu, Q.; Xu, Z.; et al. USP20 Promotes Cellular Antiviral Responses Via Deconjugating K48-Linked Ubiquitination of MITA. J. Immunol. 2019, 202, 2397–2406. [Google Scholar] [CrossRef]
- Zhang, H.; Liao, B.; Xu, Z.; Ran, Y.; Wang, D.; Yang, Y.; Luo, W.; Wang, Y. USP44 Positively Regulates Innate Immune Response to DNA Viruses through Deubiquitinating MITA. PLoS Pathog. 2020, 16, e1008178. [Google Scholar] [CrossRef] [PubMed]
- Schneider, W.M.; Chevillotte, M.D.; Rice, C.M. Interferon-Stimulated Genes: A Complex Web of Host Defenses. Annu. Rev. Immunol. 2014, 32, 513–545. [Google Scholar] [CrossRef] [PubMed]
- Ye, L.; Zhang, Q.; Liuyu, T.; Xu, Z.; Zhang, M.; Luo, M.; Zeng, W.; Zhu, Q.; Lin, D.; Zhong, B. USP49 Negatively Regulates Cellular Antiviral Responses Via Deconjugating K63-Linked Ubiquitination of MITA. PLoS Pathog. 2019, 15, e1007680. [Google Scholar] [CrossRef]
- Sun, H.; Zhang, Q.; Jing, Y.; Zhang, M.; Wang, H.; Cai, Z.; Liuyu, T.; Zhang, Z.; Xiong, T.; Wu, Y.; et al. USP13 Negatively Regulates Antiviral Responses by Deubiquitinating STING. Nat. Commun. 2017, 8, 15534. [Google Scholar] [CrossRef]
- Chen, Y.; Wang, L.; Jin, J.; Luan, Y.; Chen, C.; Li, Y.; Chu, H.; Wang, X.; Liao, G.; Yu, Y.; et al. p38 Inhibition Provides Anti-DNA Virus Immunity by Regulation of USP21 Phosphorylation and STING Activation. J. Exp. Med. 2017, 214, 991–1010. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Chen, Y.; Chen, X.; Zhang, W.; Zhao, L.; Weng, L.; Tian, H.; Wu, Z.; Tan, X.; Ge, X.; et al. Deubiquitinase USP35 Restrains STING-Mediated Interferon Signaling in Ovarian Cancer. Cell Death Differ. 2021, 28, 139–155. [Google Scholar] [CrossRef]
- Yang, L.; Wang, L.; Ketkar, H.; Ma, J.; Yang, G.; Cui, S.; Geng, T.; Mordue, D.G.; Fujimoto, T.; Cheng, G.; et al. UBXN3B Positively Regulates STING-Mediated Antiviral Immune Responses. Nat. Commun. 2018, 9, 2329. [Google Scholar] [CrossRef] [PubMed]
- Wang, Q.; Liu, X.; Cui, Y.; Tang, Y.; Chen, W.; Li, S.; Yu, H.; Pan, Y.; Wang, C. The E3 Ubiquitin Ligase AMFR and INSIG1 Bridge the Activation of TBK1 Kinase by Modifying the Adaptor STING. Immunity 2014, 41, 919–933. [Google Scholar] [CrossRef]
- Zhang, L.; Wei, N.; Cui, Y.; Hong, Z.; Liu, X.; Wang, Q.; Li, S.; Liu, H.; Yu, H.; Cai, Y.; et al. The Deubiquitinase CYLD is a Specific Checkpoint of the STING Antiviral Signaling Pathway. PLoS Pathog. 2018, 14, e1007435. [Google Scholar] [CrossRef]
- Guo, Y.; Jiang, F.; Kong, L.; Wu, H.; Zhang, H.; Chen, X.; Zhao, J.; Cai, B.; Li, Y.; Ma, C.; et al. OTUD5 Promotes Innate Antiviral and Antitumor Immunity through Deubiquitinating and Stabilizing STING. Cell. Mol. Immunol. 2021, 18, 1945–1955. [Google Scholar] [CrossRef]
- Tian, M.; Liu, W.; Zhang, Q.; Huang, Y.; Li, W.; Wang, W.; Zhao, P.; Huang, S.; Song, Y.; Shereen, M.A.; et al. MYSM1 Represses Innate Immunity and Autoimmunity through Suppressing the cGAS-STING Pathway. Cell Rep. 2020, 33, 108297. [Google Scholar] [CrossRef]
- Landman, S.L.; Ressing, M.E.; van der Veen, A.G. Balancing STING in Antimicrobial Defense and Autoinflammation. Cytokine Growth Factor Rev. 2020, 55, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Hertzog, J.; Rehwinkel, J. Regulation and Inhibition of the DNA Sensor cGAS. EMBO Rep. 2020, 21, e51345. [Google Scholar] [CrossRef] [PubMed]
- Eaglesham, J.B.; Pan, Y.; Kupper, T.S.; Kranzusch, P.J. Viral and Metazoan Poxins are cGAMP-Specific Nucleases that Restrict cGAS-STING Signalling. Nature 2019, 566, 259–263. [Google Scholar] [CrossRef]
- Eaglesham, J.B.; McCarty, K.L.; Kranzusch, P.J. Structures of Diverse Poxin cGAMP Nucleases Reveal a Widespread Role for cGAS-STING Evasion in Host-Pathogen Conflict. eLife 2020, 9, e59753. [Google Scholar] [CrossRef]
- Zou, H.; Huang, Z.; Yang, Y.; Luo, W.; Wang, S.; Luo, M.; Fu, Y.; Wang, Y. Human Cytomegalovirus Protein UL94 Targets MITA to Evade the Antiviral Immune Response. J. Virol. 2020, 94, 22. [Google Scholar] [CrossRef] [PubMed]
- Fu, Y.; Su, S.; Gao, Y.; Wang, P.; Huang, Z.; Hu, M.; Luo, W.; Li, S.; Luo, M.; Wang, Y.; et al. Human Cytomegalovirus Tegument Protein UL82 Inhibits STING-Mediated Signaling to Evade Antiviral Immunity. Cell Host Microbe 2017, 21, 231–243. [Google Scholar] [CrossRef] [PubMed]
- Pan, S.; Liu, X.; Ma, Y.; Cao, Y.; He, B. Herpes Simplex Virus 1 γ134.5 Protein Inhibits STING Activation that Restricts Viral Replication. J. Virol. 2018, 92, 1015. [Google Scholar] [CrossRef]
- Wang, J.; Yang, S.; Liu, L.; Wang, H.; Yang, B. HTLV-1 Tax Impairs K63-Linked Ubiquitination of STING to Evade Host Innate Immunity. Virus Res. 2017, 232, 13–21. [Google Scholar] [CrossRef]
- Chen, X.; Yang, X.; Zheng, Y.; Yang, Y.; Xing, Y.; Chen, Z. SARS Coronavirus Papain-Like Protease Inhibits the Type I Interferon Signaling Pathway through Interaction with the STING-TRAF3-TBK1 Complex. Protein Cell 2014, 5, 369–381. [Google Scholar] [CrossRef]
- Liu, Y.; Li, J.; Chen, J.; Li, Y.; Wang, W.; Du, X.; Song, W.; Zhang, W.; Lin, L.; Yuan, Z. Hepatitis B Virus Polymerase Disrupts K63-Linked Ubiquitination of STING to Block Innate Cytosolic DNA-Sensing Pathways. J. Virol. 2015, 89, 2287–2300. [Google Scholar] [CrossRef] [PubMed]
- Bodda, C.; Reinert, L.S.; Fruhwürth, S.; Richardo, T.; Sun, C.; Zhang, B.; Kalamvoki, M.; Pohlmann, A.; Mogensen, T.H.; Bergström, P.; et al. HSV1 VP1-2 Deubiquitinates STING to Block Type I Interferon Expression and Promote Brain Infection. J. Exp. Med. 2020, 217, e20191422. [Google Scholar] [CrossRef] [PubMed]
- Xing, Y.; Chen, J.; Tu, J.; Zhang, B.; Chen, X.; Shi, H.; Baker, S.C.; Feng, L.; Chen, Z. The Papain-Like Protease of Porcine Epidemic Diarrhea Virus Negatively Regulates Type I Interferon Pathway by Acting as a Viral Deubiquitinase. J. Gen. Virol. 2013, 94, 1554–1567. [Google Scholar] [CrossRef] [PubMed]
- Deschamps, T.; Kalamvoki, M. Evasion of the STING DNA-Sensing Pathway by VP11/12 of Herpes Simplex Virus 1. J. Virol. 2017, 91, 535. [Google Scholar] [CrossRef]
- Christensen, M.H.; Jensen, S.B.; Miettinen, J.J.; Luecke, S.; Prabakaran, T.; Reinert, L.S.; Mettenleiter, T.; Chen, Z.J.; Knipe, D.M.; Sandri-Goldin, R.M.; et al. HSV-1 ICP27 Targets the TBK1-Activated STING Signalsome to Inhibit Virus-Induced Type I IFN expression. EMBO J. 2016, 35, 1385–1399. [Google Scholar] [CrossRef]
- Su, J.; Rui, Y.; Lou, M.; Yin, L.; Xiong, H.; Zhou, Z.; Shen, S.; Chen, T.; Zhang, Z.; Zhao, N.; et al. HIV-2/SIV Vpx Targets a Novel Functional Domain of STING to Selectively Inhibit cGAS-STING-Mediated NF-κB Signalling. Nat. Microbiol. 2019, 4, 2552–2564. [Google Scholar] [CrossRef]
- Anukanth, A.; Ponnuswamy, P.K. Conformational Characteristics of Mixed Sugar Puckered Deoxytetranucleoside Triphosphate D-GpCpGpC from Energy Minimization Studies. J. Biomol. Struct. Dyn. 1989, 6, 801–814. [Google Scholar] [CrossRef]
- Ni, G.; Ma, Z.; Wong, J.P.; Zhang, Z.; Cousins, E.; Major, M.B.; Damania, B. PPP6C Negatively Regulates STING-Dependent Innate Immune Responses. mBio 2020, 11, 1728. [Google Scholar] [CrossRef]
- Yu, K.; Tian, H.; Deng, H. PPM1G Restricts Innate Immune Signaling Mediated by STING and MAVS and is Hijacked by KSHV for Immune Evasion. Sci. Adv. 2020, 6, eabd0276. [Google Scholar] [CrossRef]
- Zhang, W.; Jiang, B.; Zeng, M.; Duan, Y.; Wu, Z.; Wu, Y.; Wang, T.; Wang, M.; Jia, R.; Zhu, D.; et al. Binding of Duck Tembusu Virus Nonstructural Protein 2A to Duck STING Disrupts Induction of its Signal Transduction Cascade to Inhibit Beta Interferon Induction. J. Virol. 2020, 94, 1850. [Google Scholar] [CrossRef]
- Ding, Q.; Gaska, J.M.; Douam, F.; Wei, L.; Kim, D.; Balev, M.; Heller, B.; Ploss, A. Species-Specific Disruption of STING-Dependent Antiviral Cellular Defenses by the Zika Virus NS2B3 Protease. Proc. Natl. Acad. Sci. USA 2018, 115, E6310–E6318. [Google Scholar] [CrossRef] [PubMed]
- Wu, Z.; Zhang, W.; Wu, Y.; Wang, T.; Wu, S.; Wang, M.; Jia, R.; Zhu, D.; Liu, M.; Zhao, X.; et al. Binding of the Duck Tembusu Virus Protease to STING is Mediated by NS2B and is Crucial for STING Cleavage and for Impaired Induction of IFN-B. J. Immunol. 2019, 203, 3374–3385. [Google Scholar] [CrossRef] [PubMed]
- Rodríguez-García, E.; Olagüe, C.; Ríus-Rocabert, S.; Ferrero, R.; Llorens, C.; Larrea, E.; Fortes, P.; Prieto, J.; González-Aseguinolaza, G.; Nistal-Villan, E. TMEM173 Alternative Spliced Isoforms Modulate Viral Replication through the STING Pathway. Immunohorizons 2018, 2, 363–376. [Google Scholar] [CrossRef] [PubMed]
- Lu, D.; Shang, G.; Li, J.; Lu, Y.; Bai, X.; Zhang, X. Activation of STING by Targeting a Pocket in the Transmembrane Domain. Nature 2022, 604, 557–562. [Google Scholar] [CrossRef]
- Shu, C.; Yi, G.; Watts, T.; Kao, C.C.; Li, P. Structure of STING Bound to Cyclic Di-GMP Reveals the Mechanism of Cyclic Dinucleotide Recognition by the Immune System. Nat. Struct. Mol. Biol. 2012, 19, 722–724. [Google Scholar] [CrossRef] [PubMed]
- Gao, P.; Zillinger, T.; Wang, W.; Ascano, M.; Dai, P.; Hartmann, G.; Tuschl, T.; Deng, L.; Barchet, W.; Patel, D.J. Binding-Pocket and Lid-Region Substitutions Render Human STING Sensitive to the Species-Specific Drug DMXAA. Cell Rep. 2014, 8, 1668–1676. [Google Scholar] [CrossRef] [PubMed]
- Ruiz-Moreno, J.S.; Hamann, L.; Shah, J.A.; Verbon, A.; Mockenhaupt, F.P.; Puzianowska-Kuznicka, M.; Naujoks, J.; Sander, L.E.; Witzenrath, M.; Cambier, J.C.; et al. The Common HAQ STING Variant Impairs cGAS-Dependent Antibacterial Responses and is Associated with Susceptibility to Legionnaires’ Disease in Humans. PLoS Pathog. 2018, 14, e1006829. [Google Scholar] [CrossRef] [PubMed]
- Yi, G.; Brendel, V.P.; Shu, C.; Li, P.; Palanathan, S.; Cheng Kao, C. Single Nucleotide Polymorphisms of Human STING can Affect Innate Immune Response to Cyclic Dinucleotides. PLoS ONE 2013, 8, e77846. [Google Scholar] [CrossRef]
- Patel, S.; Jin, L. TMEM173 Variants and Potential Importance to Human Biology and Disease. Genes Immun. 2019, 20, 82–89. [Google Scholar] [CrossRef]
- Xiao, T.S.; Fitzgerald, K.A. The cGAS-STING Pathway for DNA Sensing. Mol. Cell 2013, 51, 135–139. [Google Scholar] [CrossRef]
- Liu, Y.; Jesus, A.A.; Marrero, B.; Yang, D.; Ramsey, S.E.; Sanchez, G.A.M.; Tenbrock, K.; Wittkowski, H.; Jones, O.Y.; Kuehn, H.S.; et al. Activated STING in a Vascular and Pulmonary Syndrome. N. Engl. J. Med. 2014, 371, 507–518. [Google Scholar] [CrossRef] [PubMed]
- Ouyang, S.; Song, X.; Wang, Y.; Ru, H.; Shaw, N.; Jiang, Y.; Niu, F.; Zhu, Y.; Qiu, W.; Parvatiyar, K.; et al. Structural Analysis of the STING Adaptor Protein Reveals a Hydrophobic Dimer Interface and Mode of Cyclic Di-GMP Binding. Immunity 2012, 36, 1073–1086. [Google Scholar] [CrossRef] [PubMed]
- Jeremiah, N.; Neven, B.; Gentili, M.; Callebaut, I.; Maschalidi, S.; Stolzenberg, M.; Goudin, N.; Frémond, M.; Nitschke, P.; Molina, T.J.; et al. Inherited STING-Activating Mutation Underlies a Familial Inflammatory Syndrome with Lupus-Like Manifestations. J. Clin. Investig. 2014, 124, 5516–5520. [Google Scholar] [CrossRef] [PubMed]
- Melki, I.; Rose, Y.; Uggenti, C.; Van Eyck, L.; Frémond, M.; Kitabayashi, N.; Rice, G.I.; Jenkinson, E.M.; Boulai, A.; Jeremiah, N.; et al. Disease-Associated Mutations Identify a Novel Region in Human STING Necessary for the Control of Type I Interferon Signaling. J. Allergy Clin. Immunol. 2017, 140, 543–552.e5. [Google Scholar] [CrossRef] [PubMed]
- Keskitalo, S.; Haapaniemi, E.; Einarsdottir, E.; Rajamäki, K.; Heikkilä, H.; Ilander, M.; Pöyhönen, M.; Morgunova, E.; Hokynar, K.; Lagström, S.; et al. Novel TMEM173 Mutation and the Role of Disease Modifying Alleles. Front. Immunol. 2019, 10, 2770. [Google Scholar] [CrossRef] [PubMed]
- Seo, J.; Kang, J.; Suh, D.I.; Park, E.; Lee, C.; Choi, S.A.; Kim, S.Y.; Kim, Y.; Park, S.; Ye, M.; et al. Tofacitinib Relieves Symptoms of Stimulator of Interferon Genes (STING)-Associated Vasculopathy with Onset in Infancy Caused by 2 De Novo Variants in TMEM173. J. Allergy Clin. Immunol. 2017, 139, 1396–1399.e12. [Google Scholar] [CrossRef] [PubMed]
- Jin, L.; Xu, L.; Yang, I.V.; Davidson, E.J.; Schwartz, D.A.; Wurfel, M.M.; Cambier, J.C. Identification and Characterization of a Loss-of-Function Human MPYS Variant. Genes Immun. 2011, 12, 263–269. [Google Scholar] [CrossRef]
- Hansen, A.L.; Mukai, K.; Schopfer, F.J.; Taguchi, T.; Holm, C.K. STING Palmitoylation as a Therapeutic Target. Cell. Mol. Immunol. 2019, 16, 236–241. [Google Scholar] [CrossRef] [PubMed]
- Haag, S.M.; Gulen, M.F.; Reymond, L.; Gibelin, A.; Abrami, L.; Decout, A.; Heymann, M.; van der Goot, F.G.; Turcatti, G.; Behrendt, R.; et al. Targeting STING with Covalent Small-Molecule Inhibitors. Nature 2018, 559, 269–273. [Google Scholar] [CrossRef]
- Donnelly, C.R.; Jiang, C.; Andriessen, A.S.; Wang, K.; Wang, Z.; Ding, H.; Zhao, J.; Luo, X.; Lee, M.S.; Lei, Y.L.; et al. STING Controls Nociception Via Type I Interferon Signalling in Sensory Neurons. Nature 2021, 591, 275–280. [Google Scholar] [CrossRef]
- Davies, B.W.; Bogard, R.W.; Young, T.S.; Mekalanos, J.J. Coordinated Regulation of Accessory Genetic Elements Produces Cyclic Di-Nucleotides for V. Cholerae Virulence. Cell 2012, 149, 358–370. [Google Scholar] [CrossRef]
- Wu, J.; Sun, L.; Chen, X.; Du, F.; Shi, H.; Chen, C.; Chen, Z.J. Cyclic GMP-AMP is an Endogenous Second Messenger in Innate Immune Signaling by Cytosolic DNA. Science 2013, 339, 826–830. [Google Scholar] [CrossRef] [PubMed]
- Kranzusch, P.J.; Wilson, S.C.; Lee, A.S.Y.; Berger, J.M.; Doudna, J.A.; Vance, R.E. Ancient Origin of cGAS-STING Reveals Mechanism of Universal 2′,3′ cGAMP Signaling. Mol. Cell 2015, 59, 891–903. [Google Scholar] [CrossRef] [PubMed]
- Gao, P.; Ascano, M.; Zillinger, T.; Wang, W.; Dai, P.; Serganov, A.A.; Gaffney, B.L.; Shuman, S.; Jones, R.A.; Deng, L.; et al. Structure-Function Analysis of STING Activation by C[G(2′,5′)pA(3′,5′)P] and Targeting by Antiviral DMXAA. Cell 2013, 154, 748–762. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Shi, H.; Wu, J.; Zhang, X.; Sun, L.; Chen, C.; Chen, Z.J. Cyclic GMP-AMP Containing Mixed Phosphodiester Linkages is an Endogenous High-Affinity Ligand for STING. Mol. Cell 2013, 51, 226–235. [Google Scholar] [CrossRef]
- Shi, H.; Wu, J.; Chen, Z.J.; Chen, C. Molecular Basis for the Specific Recognition of the Metazoan Cyclic GMP-AMP by the Innate Immune Adaptor Protein STING. Proc. Natl. Acad. Sci. USA 2015, 112, 8947–8952. [Google Scholar] [CrossRef]
- Smola, M.; Gutten, O.; Dejmek, M.; Kožíšek, M.; Evangelidis, T.; Tehrani, Z.A.; Novotná, B.; Nencka, R.; Birkuš, G.; Rulíšek, L.; et al. Ligand Strain and its Conformational Complexity is a Major Factor in the Binding of Cyclic Dinucleotides to STING Protein. Angew. Chem. Int. Ed. Engl. 2021, 60, 10172–10178. [Google Scholar] [CrossRef] [PubMed]
- Morehouse, B.R.; Yip, M.C.J.; Keszei, A.F.A.; McNamara-Bordewick, N.K.; Shao, S.; Kranzusch, P.J. Cryo-EM Structure of an Active Bacterial TIR-STING Filament Complex. Nature 2022, 608, 803–807. [Google Scholar] [CrossRef]
- Ko, T.; Wang, Y.; Yang, C.; Hou, M.; Chen, C.; Chiu, Y.; Chen, Y. Crystal Structure and Functional Implication of Bacterial STING. Nat. Commun. 2022, 13, 26. [Google Scholar] [CrossRef]
- Yin, Q.; Tian, Y.; Kabaleeswaran, V.; Jiang, X.; Tu, D.; Eck, M.J.; Chen, Z.J.; Wu, H. Cyclic Di-GMP Sensing Via the Innate Immune Signaling Protein STING. Mol. Cell 2012, 46, 735–745. [Google Scholar] [CrossRef]
- Li, Q.; Tian, S.; Liang, J.; Fan, J.; Lai, J.; Chen, Q. Therapeutic Development by Targeting the cGAS-STING Pathway in Autoimmune Disease and Cancer. Front. Pharmacol. 2021, 12, 779425. [Google Scholar] [CrossRef] [PubMed]
- Gray, P.M.; Forrest, G.; Wisniewski, T.; Porter, G.; Freed, D.C.; DeMartino, J.A.; Zaller, D.M.; Guo, Z.; Leone, J.; Fu, T.; et al. Evidence for Cyclic Diguanylate as a Vaccine Adjuvant with Novel Immunostimulatory Activities. Cell. Immunol. 2012, 278, 113–119. [Google Scholar] [CrossRef] [PubMed]
- Libanova, R.; Becker, P.D.; Guzmán, C.A. Cyclic Di-Nucleotides: New Era for Small Molecules as Adjuvants. Microb. Biotechnol. 2012, 5, 168–176. [Google Scholar] [CrossRef]
- Van Herck, S.; Feng, B.; Tang, L. Delivery of STING Agonists for Adjuvanting Subunit Vaccines. Adv. Drug Deliv. Rev. 2021, 179, 114020. [Google Scholar] [CrossRef] [PubMed]
- Padron-Regalado, E.; Ulaszewska, M.; Douglas, A.D.; Hill, A.V.S.; Spencer, A.J. STING-Pathway Modulation to Enhance the Immunogenicity of Adenoviral-Vectored Vaccines. Sci. Rep. 2022, 12, 14464. [Google Scholar] [CrossRef] [PubMed]
- Li, L.; Yin, Q.; Kuss, P.; Maliga, Z.; Millán, J.L.; Wu, H.; Mitchison, T.J. Hydrolysis of 2′3′-cGAMP by ENPP1 and Design of Nonhydrolyzable Analogs. Nat. Chem. Biol. 2014, 10, 1043–1048. [Google Scholar] [CrossRef] [PubMed]
- Kong, X.; Zuo, H.; Huang, H.; Zhang, Q.; Chen, J.; He, C.; Hu, Y. STING as an Emerging Therapeutic Target for Drug Discovery: Perspectives from the Global Patent Landscape. J. Adv. Res. 2023, 44, 119–133. [Google Scholar] [CrossRef] [PubMed]
- Novotná, B.; Vaneková, L.; Zavřel, M.; Buděšínský, M.; Dejmek, M.; Smola, M.; Gutten, O.; Tehrani, Z.A.; Pimková Polidarová, M.; Brázdová, A.; et al. Enzymatic Preparation of 2′-5′,3′-5′-Cyclic Dinucleotides, their Binding Properties to Stimulator of Interferon Genes Adaptor Protein, and Structure/Activity Correlations. J. Med. Chem. 2019, 62, 10676–10690. [Google Scholar] [CrossRef]
- Zhang, H.; You, Q.; Xu, X. Targeting Stimulator of Interferon Genes (STING): A Medicinal Chemistry Perspective. J. Med. Chem. 2020, 63, 3785–3816. [Google Scholar] [CrossRef]
- Vyskocil, S.; Cardin, D.; Ciavarri, J.; Conlon, J.; Cullis, C.; England, D.; Gershman, R.; Gigstad, K.; Gipson, K.; Gould, A.; et al. Identification of Novel Carbocyclic Pyrimidine Cyclic Dinucleotide STING Agonists for Antitumor Immunotherapy using Systemic Intravenous Route. J. Med. Chem. 2021, 64, 6902–6923. [Google Scholar] [CrossRef]
- Kozlova, A.; Thabault, L.; Liberelle, M.; Klaessens, S.; Prévost, J.R.C.; Mathieu, C.; Pilotte, L.; Stroobant, V.; Van den Eynde, B.; Frédérick, R. Rational Design of Original Fused-Cycle Selective Inhibitors of Tryptophan 2,3-Dioxygenase. J. Med. Chem. 2021, 64, 10967–10980. [Google Scholar] [CrossRef] [PubMed]
- Kim, D.; Endo, A.; Fang, F.G.; Huang, K.; Bao, X.; Choi, H.; Majumder, U.; Shen, Y.Y.; Mathieu, S.; Zhu, X.; et al. E7766, a Macrocycle-Bridged Stimulator of Interferon Genes (STING) Agonist with Potent Pan-Genotypic Activity. ChemMedChem 2021, 16, 1740–1743. [Google Scholar] [CrossRef] [PubMed]
- McIntosh, J.A.; Liu, Z.; Andresen, B.M.; Marzijarani, N.S.; Moore, J.C.; Marshall, N.M.; Borra-Garske, M.; Obligacion, J.V.; Fier, P.S.; Peng, F.; et al. A Kinase-cGAS Cascade to Synthesize a Therapeutic STING Activator. Nature 2022, 603, 439–444. [Google Scholar] [CrossRef] [PubMed]
- Ding, C.; Song, Z.; Shen, A.; Chen, T.; Zhang, A. Small Molecules Targeting the Innate Immune cGAS-STING-TBK1 Signaling Pathway. Acta Pharm. Sin. B 2020, 10, 2272–2298. [Google Scholar] [CrossRef]
- Meric-Bernstam, F.; Sweis, R.F.; Hodi, F.S.; Messersmith, W.A.; Andtbacka, R.H.I.; Ingham, M.; Lewis, N.; Chen, X.; Pelletier, M.; Chen, X.; et al. Phase I Dose-Escalation Trial of MIW815 (ADU-S100), an Intratumoral STING Agonist, in Patients with Advanced/Metastatic Solid Tumors Or Lymphomas. Clin. Cancer Res. 2022, 28, 677–688. [Google Scholar] [CrossRef]
- Cavlar, T.; Deimling, T.; Ablasser, A.; Hopfner, K.; Hornung, V. Species-Specific Detection of the Antiviral Small-Molecule Compound CMA by STING. EMBO J. 2013, 32, 1440–1450. [Google Scholar] [CrossRef]
- Le Naour, J.; Zitvogel, L.; Galluzzi, L.; Vacchelli, E.; Kroemer, G. Trial Watch: STING Agonists in Cancer Therapy. Oncoimmunology 2020, 9, 1777624. [Google Scholar] [CrossRef]
- Conlon, J.; Burdette, D.L.; Sharma, S.; Bhat, N.; Thompson, M.; Jiang, Z.; Rathinam, V.A.K.; Monks, B.; Jin, T.; Xiao, T.S.; et al. Mouse, but Not Human STING, Binds and Signals in Response to the Vascular Disrupting Agent 5,6-Dimethylxanthenone-4-Acetic Acid. J. Immunol. 2013, 190, 5216–5225. [Google Scholar] [CrossRef]
- Kim, S.; Li, L.; Maliga, Z.; Yin, Q.; Wu, H.; Mitchison, T.J. Anticancer Flavonoids are Mouse-Selective STING Agonists. ACS Chem. Biol. 2013, 8, 1396–1401. [Google Scholar] [CrossRef]
- Tijono, S.M.; Guo, K.; Henare, K.; Palmer, B.D.; Wang, L.-S.; Albelda, S.M.; Ching, L. Identification of Human-Selective Analogues of the Vascular-Disrupting Agent 5,6-Dimethylxanthenone-4-Acetic Acid (DMXAA). Br. J. Cancer 2013, 108, 1306–1315. [Google Scholar] [CrossRef]
- Gobbi, S.; Belluti, F.; Rampa, A.; Bisi, A. Flavonoid-Inspired Vascular Disrupting Agents: Exploring Flavone-8-Acetic Acid and Derivatives in the New Century. Molecules 2021, 26, 4228. [Google Scholar] [CrossRef] [PubMed]
- Feng, X.; Liu, D.; Li, Z.; Bian, J. Bioactive Modulators Targeting STING Adaptor in cGAS-STING Pathway. Drug Discov. Today 2020, 25, 230–237. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Sun, Z.; Pei, J.; Luo, Q.; Zeng, X.; Li, Q.; Yang, Z.; Quan, J. Identification of A-Mangostin as an Agonist of Human STING. ChemMedChem 2018, 13, 2057–2064. [Google Scholar] [CrossRef] [PubMed]
- Zhong, S.; Li, W.; Bai, Y.; Wu, B.; Wang, X.; Jiang, S.; Zhao, Y.; Ren, J.; Li, H.; Jin, R. Computational Study on New Natural Compound Agonists of Stimulator of Interferon Genes (STING). PLoS ONE 2019, 14, e0216678. [Google Scholar] [CrossRef]
- Sali, T.M.; Pryke, K.M.; Abraham, J.; Liu, A.; Archer, I.; Broeckel, R.; Staverosky, J.A.; Smith, J.L.; Al-Shammari, A.; Amsler, L.; et al. Characterization of a Novel Human-Specific STING Agonist that Elicits Antiviral Activity Against Emerging Alphaviruses. PLoS Pathog. 2015, 11, e1005324. [Google Scholar] [CrossRef]
- Banerjee, M.; Middya, S.; Shrivastava, R.; Basu, S.; Ghosh, R.; Pryde, D.C.; Yadav, D.B.; Surya, A. G10 is a Direct Activator of Human STING. PLoS ONE 2020, 15, e0237743. [Google Scholar] [CrossRef]
- Xie, Z.; Wang, Z.; Fan, F.; Zhou, J.; Hu, Z.; Wang, Q.; Wang, X.; Zeng, Q.; Zhang, Y.; Qiu, J.; et al. Structural Insights into a Shared Mechanism of Human STING Activation by a Potent Agonist and an Autoimmune Disease-Associated Mutation. Cell Discov. 2022, 8, 133. [Google Scholar] [CrossRef]
- Ramanjulu, J.M.; Pesiridis, G.S.; Yang, J.; Concha, N.; Singhaus, R.; Zhang, S.; Tran, J.; Moore, P.; Lehmann, S.; Eberl, H.C.; et al. Design of Amidobenzimidazole STING Receptor Agonists with Systemic Activity. Nature 2018, 564, 439–443. [Google Scholar] [CrossRef]
- Siu, T.; Altman, M.D.; Baltus, G.A.; Childers, M.; Ellis, J.M.; Gunaydin, H.; Hatch, H.; Ho, T.; Jewell, J.; Lacey, B.M.; et al. Discovery of a Novel cGAMP Competitive Ligand of the Inactive Form of STING. ACS Med. Chem. Lett. 2019, 10, 92–97. [Google Scholar] [CrossRef]
- Cherney, E.C.; Zhang, L.; Lo, J.; Huynh, T.; Wei, D.; Ahuja, V.; Quesnelle, C.; Schieven, G.L.; Futran, A.; Locke, G.A.; et al. Discovery of Non-Nucleotide Small-Molecule STING Agonists Via Chemotype Hybridization. J. Med. Chem. 2022, 65, 3518–3538. [Google Scholar] [CrossRef]
- Song, C.; Liu, D.; Liu, S.; Li, D.; Horecny, I.; Zhang, X.; Li, P.; Chen, L.; Miller, M.; Chowdhury, R.; et al. SHR1032, a Novel STING Agonist, Stimulates Anti-Tumor Immunity and Directly Induces AML Apoptosis. Sci. Rep. 2022, 12, 8579. [Google Scholar] [CrossRef] [PubMed]
- Pan, B.; Perera, S.A.; Piesvaux, J.A.; Presland, J.P.; Schroeder, G.K.; Cumming, J.N.; Trotter, B.W.; Altman, M.D.; Buevich, A.V.; Cash, B.; et al. An Orally Available Non-Nucleotide STING Agonist with Antitumor Activity. Science 2020, 369, eaba6098. [Google Scholar] [CrossRef] [PubMed]
- Feng, X.; Pan, L.; Qian, Z.; Liu, D.; Guan, X.; Feng, L.; Song, B.; Xu, X.; Tan, N.; Ma, Y.; et al. Discovery of Selenium-Containing STING Agonists as Orally Available Antitumor Agents. J. Med. Chem. 2022, 65, 15048–15065. [Google Scholar] [CrossRef] [PubMed]
- Chin, E.N.; Yu, C.; Vartabedian, V.F.; Jia, Y.; Kumar, M.; Gamo, A.M.; Vernier, W.; Ali, S.H.; Kissai, M.; Lazar, D.C.; et al. Antitumor Activity of a Systemic STING-Activating Non-Nucleotide cGAMP Mimetic. Science 2020, 369, 993–999. [Google Scholar] [CrossRef]
- Hong, Z.; Mei, J.; Li, C.; Bai, G.; Maimaiti, M.; Hu, H.; Yu, W.; Sun, L.; Zhang, L.; Cheng, D.; et al. STING Inhibitors Target the Cyclic Dinucleotide Binding Pocket. Proc. Natl. Acad. Sci. USA 2021, 118, e2105465118. [Google Scholar] [CrossRef]
- Long, J.; Ying, T.; Zhang, L.; Yu, T.; Wu, J.; Liu, Y.; Li, X.; You, G.; Zhang, L.; Bi, Y. Discovery of Fusidic Acid Derivatives as Novel STING Inhibitors for Treatment of Sepsis. Eur. J. Med. Chem. 2022, 244, 114814. [Google Scholar] [CrossRef]
- Li, S.; Hong, Z.; Wang, Z.; Li, F.; Mei, J.; Huang, L.; Lou, X.; Zhao, S.; Song, L.; Chen, W.; et al. The Cyclopeptide Astin C Specifically Inhibits the Innate Immune CDN Sensor STING. Cell Rep. 2018, 25, 3405–3421.e7. [Google Scholar] [CrossRef]
- Gao, J.; Zheng, M.; Wu, X.; Zhang, H.; Su, H.; Dang, Y.; Ma, M.; Wang, F.; Xu, J.; Chen, L.; et al. CDK Inhibitor Palbociclib Targets STING to Alleviate Autoinflammation. EMBO Rep. 2022, 23, e53932. [Google Scholar] [CrossRef]
- Kang, J.; Wu, J.; Liu, Q.; Wu, X.; Zhao, Y.; Ren, J. Post-Translational Modifications of STING: A Potential Therapeutic Target. Front. Immunol. 2022, 13, 888147. [Google Scholar] [CrossRef]
- Cui, T.; Schopfer, F.J.; Zhang, J.; Chen, K.; Ichikawa, T.; Baker, P.R.S.; Batthyany, C.; Chacko, B.K.; Feng, X.; Patel, R.P.; et al. Nitrated Fatty Acids: Endogenous Anti-Inflammatory Signaling Mediators. J. Biol. Chem. 2006, 281, 35686–35698. [Google Scholar] [CrossRef]
- Vinogradova, E.V.; Zhang, X.; Remillard, D.; Lazar, D.C.; Suciu, R.M.; Wang, Y.; Bianco, G.; Yamashita, Y.; Crowley, V.M.; Schafroth, M.A.; et al. An Activity-Guided Map of Electrophile-Cysteine Interactions in Primary Human T Cells. Cell 2020, 182, 1009–1026.e29. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Yuan, L.; Ruan, Y.; Deng, B.; Yang, Z.; Ren, Y.; Li, L.; Liu, T.; Zhao, H.; Mai, R.; et al. Novel CRBN-Recruiting Proteolysis-Targeting Chimeras as Degraders of Stimulator of Interferon Genes with in Vivo Anti-Inflammatory Efficacy. J. Med. Chem. 2022, 65, 6593–6611. [Google Scholar] [CrossRef] [PubMed]
- Long, Z.; Wang, J.; Xu, J.; Lei, X.; Liu, Q. cGAS/STING Cross-Talks with Cell Cycle and Potentiates Cancer Immunotherapy. Mol. Ther. 2022, 30, 1006–1017. [Google Scholar] [CrossRef] [PubMed]



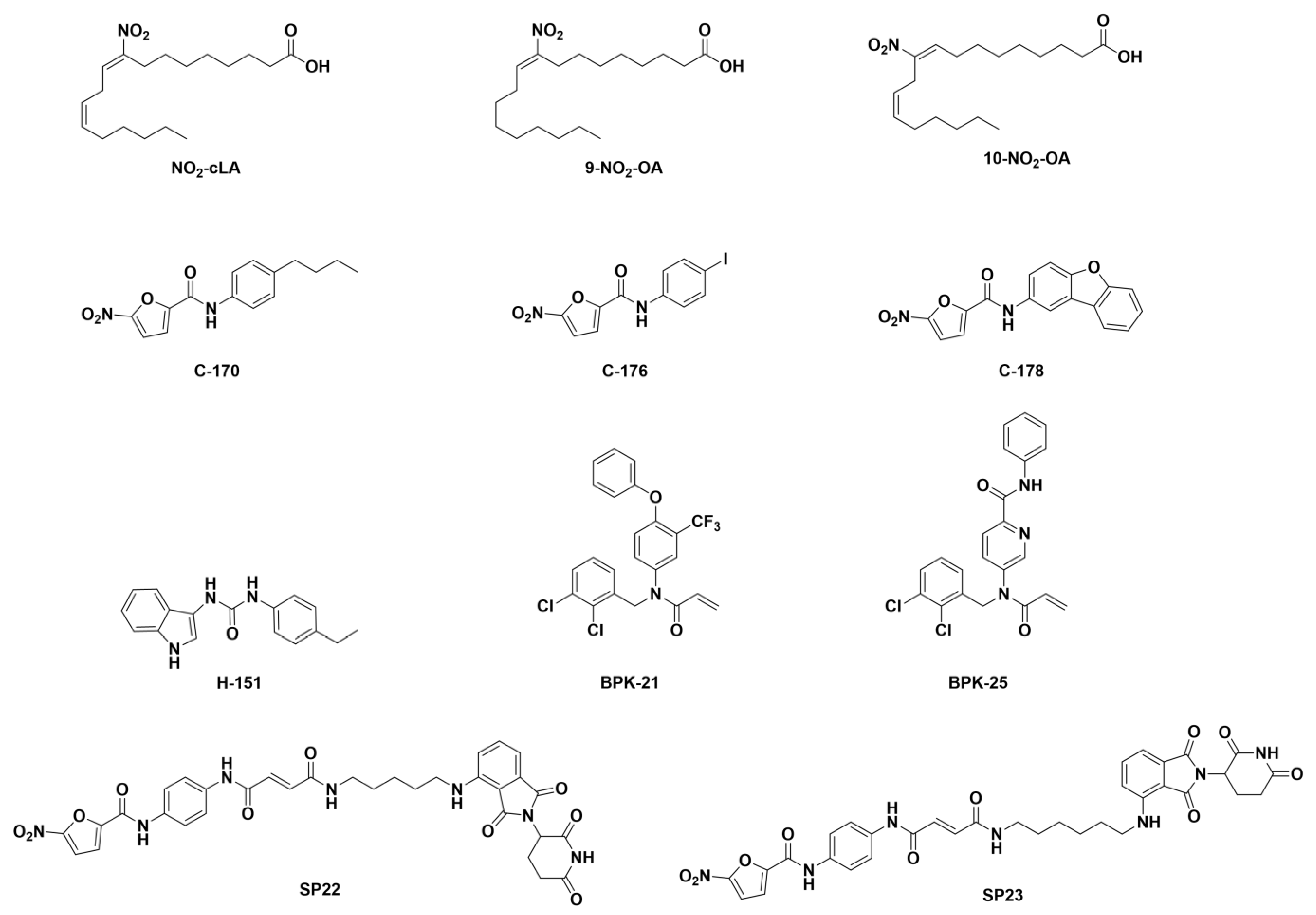
| 2′3′-cGAMP | 3′3′-cGAMP | ||
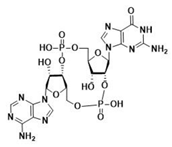 |  |  |  |
| cdiGMP | cdiAMP | ||
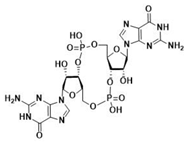 |  | 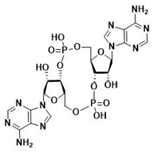 |  |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Coderch, C.; Arranz-Herrero, J.; Nistal-Villan, E.; de Pascual-Teresa, B.; Rius-Rocabert, S. The Many Ways to Deal with STING. Int. J. Mol. Sci. 2023, 24, 9032. https://doi.org/10.3390/ijms24109032
Coderch C, Arranz-Herrero J, Nistal-Villan E, de Pascual-Teresa B, Rius-Rocabert S. The Many Ways to Deal with STING. International Journal of Molecular Sciences. 2023; 24(10):9032. https://doi.org/10.3390/ijms24109032
Chicago/Turabian StyleCoderch, Claire, Javier Arranz-Herrero, Estanislao Nistal-Villan, Beatriz de Pascual-Teresa, and Sergio Rius-Rocabert. 2023. "The Many Ways to Deal with STING" International Journal of Molecular Sciences 24, no. 10: 9032. https://doi.org/10.3390/ijms24109032
APA StyleCoderch, C., Arranz-Herrero, J., Nistal-Villan, E., de Pascual-Teresa, B., & Rius-Rocabert, S. (2023). The Many Ways to Deal with STING. International Journal of Molecular Sciences, 24(10), 9032. https://doi.org/10.3390/ijms24109032







