Figure 1.
Mechanical allodynia relieved by single dose of Celecoxib theranostic nanoemulsion in both sexes. (a) Timeline of animal procedures, dosage treatment, and tissue collection for male and female cohorts. Estrus cycle tracked on days 8–12 and the females were found to be asynchronous with no apparent effect on behavior. (b) Male mechanical allodynia observed post-surgery becomes hypersensitive by day 2 and is significantly relieved following 10X CXB-NE on day 8 when compared to CCI and DF-NE groups. (Number of animals included are CCI n = 4, DF-NE n = 7, and 10X CXB-NE n = 7). (c) Female von Frey behavioral data shows post-surgery hypersensitivity occurring by day 2 aligning with the same trajectory and the same degree of hypersensitivity as the males. Following theranostic nanoemulsion treatment on day 8, significant relief from mechanical allodynia is evident when compared to CCI and DF-NE groups (Number of animals included are CCI n = 6, DF-NE n = 7, and 10X CXB-NE n = 7). Data displayed as averaged ± SEM. * ≤0.05, **** ≤0.0001. Male and female behavioral data was compared using a two-way ANOVA with a Tukey’s post hoc analysis. (a) created with BioRender.com (accessed on 6 October 2022), and (b,c) created using Prism v9.4.1.
Figure 1.
Mechanical allodynia relieved by single dose of Celecoxib theranostic nanoemulsion in both sexes. (a) Timeline of animal procedures, dosage treatment, and tissue collection for male and female cohorts. Estrus cycle tracked on days 8–12 and the females were found to be asynchronous with no apparent effect on behavior. (b) Male mechanical allodynia observed post-surgery becomes hypersensitive by day 2 and is significantly relieved following 10X CXB-NE on day 8 when compared to CCI and DF-NE groups. (Number of animals included are CCI n = 4, DF-NE n = 7, and 10X CXB-NE n = 7). (c) Female von Frey behavioral data shows post-surgery hypersensitivity occurring by day 2 aligning with the same trajectory and the same degree of hypersensitivity as the males. Following theranostic nanoemulsion treatment on day 8, significant relief from mechanical allodynia is evident when compared to CCI and DF-NE groups (Number of animals included are CCI n = 6, DF-NE n = 7, and 10X CXB-NE n = 7). Data displayed as averaged ± SEM. * ≤0.05, **** ≤0.0001. Male and female behavioral data was compared using a two-way ANOVA with a Tukey’s post hoc analysis. (a) created with BioRender.com (accessed on 6 October 2022), and (b,c) created using Prism v9.4.1.
![Ijms 24 09163 g001]()
Figure 2.
Celecoxib theranostic nanoemulsion (10X CXB-NE) reduces the infiltration of CD68 positive macrophages in the injured sciatic nerve equally for both sexes. (a) The non-surgical naïve female sciatic nerve exhibits few if any CD68 positive macrophages. (b) The non-surgical naïve male sciatic nerve typically exhibits few CD68 positive macrophages. (c) The female experiencing CCI exhibits a significant number of infiltrating CD68 positive macrophages in the injured nerve. (d) The CCI male exhibits a significant number of infiltrating CD68 positive macrophages in the injured nerve. (e) The female CCI sciatic nerve treated with drug-free nanoemulsion (DF-NE) exhibits a significant number of CD68 cells within the fasciculated nerve. (f) The male CCI sciatic nerve similarly treated with DF-NE exhibits numerous CD68 positive cells within the fasciculated nerve. (g) The female CCI sciatic nerve treated with 10X CXB-NE exhibits a significant reduction in the number of CD68 cells as compared to its CCI and DF-NE counterparts. (h) The male CCI sciatic nerve treated with 10X CXB-NE exhibits a reduced number of CD68 positive cells within the nerve as compared with its CCI and DF-NE counterparts. (i) For each animal, the number of CD68 positive cells were counted in 8 regions of interest (ROI) of equal size (140 µm × 140 µm). The average number of cells per ROI for each animal is plotted as a dot. (Naive: Male n = 2, Female n = 3. CCI: Male n = 3, Female n = 3. DFNE: Male n = 3, Female n = 4. CXBNE: Male n = 4, Female n = 5). In order to assess treatment effect per sex, a one-way ANOVA was performed with a Tukey’s post-hoc analysis. Data is displayed as averages ±SEM. * ≤0.05. Bar = 100 µm.
Figure 2.
Celecoxib theranostic nanoemulsion (10X CXB-NE) reduces the infiltration of CD68 positive macrophages in the injured sciatic nerve equally for both sexes. (a) The non-surgical naïve female sciatic nerve exhibits few if any CD68 positive macrophages. (b) The non-surgical naïve male sciatic nerve typically exhibits few CD68 positive macrophages. (c) The female experiencing CCI exhibits a significant number of infiltrating CD68 positive macrophages in the injured nerve. (d) The CCI male exhibits a significant number of infiltrating CD68 positive macrophages in the injured nerve. (e) The female CCI sciatic nerve treated with drug-free nanoemulsion (DF-NE) exhibits a significant number of CD68 cells within the fasciculated nerve. (f) The male CCI sciatic nerve similarly treated with DF-NE exhibits numerous CD68 positive cells within the fasciculated nerve. (g) The female CCI sciatic nerve treated with 10X CXB-NE exhibits a significant reduction in the number of CD68 cells as compared to its CCI and DF-NE counterparts. (h) The male CCI sciatic nerve treated with 10X CXB-NE exhibits a reduced number of CD68 positive cells within the nerve as compared with its CCI and DF-NE counterparts. (i) For each animal, the number of CD68 positive cells were counted in 8 regions of interest (ROI) of equal size (140 µm × 140 µm). The average number of cells per ROI for each animal is plotted as a dot. (Naive: Male n = 2, Female n = 3. CCI: Male n = 3, Female n = 3. DFNE: Male n = 3, Female n = 4. CXBNE: Male n = 4, Female n = 5). In order to assess treatment effect per sex, a one-way ANOVA was performed with a Tukey’s post-hoc analysis. Data is displayed as averages ±SEM. * ≤0.05. Bar = 100 µm.
![Ijms 24 09163 g002]()
Figure 3.
To reveal whether the treatments effect the DRG transcriptomes more in one group or sex than another, heatmaps displaying the top 90 genes with the highest coefficients of variation in male and female DRGs were compared. (a) Euclidean linkage was used to display the normalized log counts per million (CPM) values to show the similarities and differences in male individual animals (male naïve n = 3, male CCI n = 3, male DF-NE n = 3, male 10X CXB-NE n = 3). (b) Female treated individual animals displayed along the x-axis separated by their Euclidean distance of their gene’s normalized log CPM values (female naïve n = 3, female CCI n = 2, female DF-NE n = 2, female 10X CXB-NE n = 2). Heatmaps made in CLC Genomics Workbench 22.0.2.
Figure 3.
To reveal whether the treatments effect the DRG transcriptomes more in one group or sex than another, heatmaps displaying the top 90 genes with the highest coefficients of variation in male and female DRGs were compared. (a) Euclidean linkage was used to display the normalized log counts per million (CPM) values to show the similarities and differences in male individual animals (male naïve n = 3, male CCI n = 3, male DF-NE n = 3, male 10X CXB-NE n = 3). (b) Female treated individual animals displayed along the x-axis separated by their Euclidean distance of their gene’s normalized log CPM values (female naïve n = 3, female CCI n = 2, female DF-NE n = 2, female 10X CXB-NE n = 2). Heatmaps made in CLC Genomics Workbench 22.0.2.
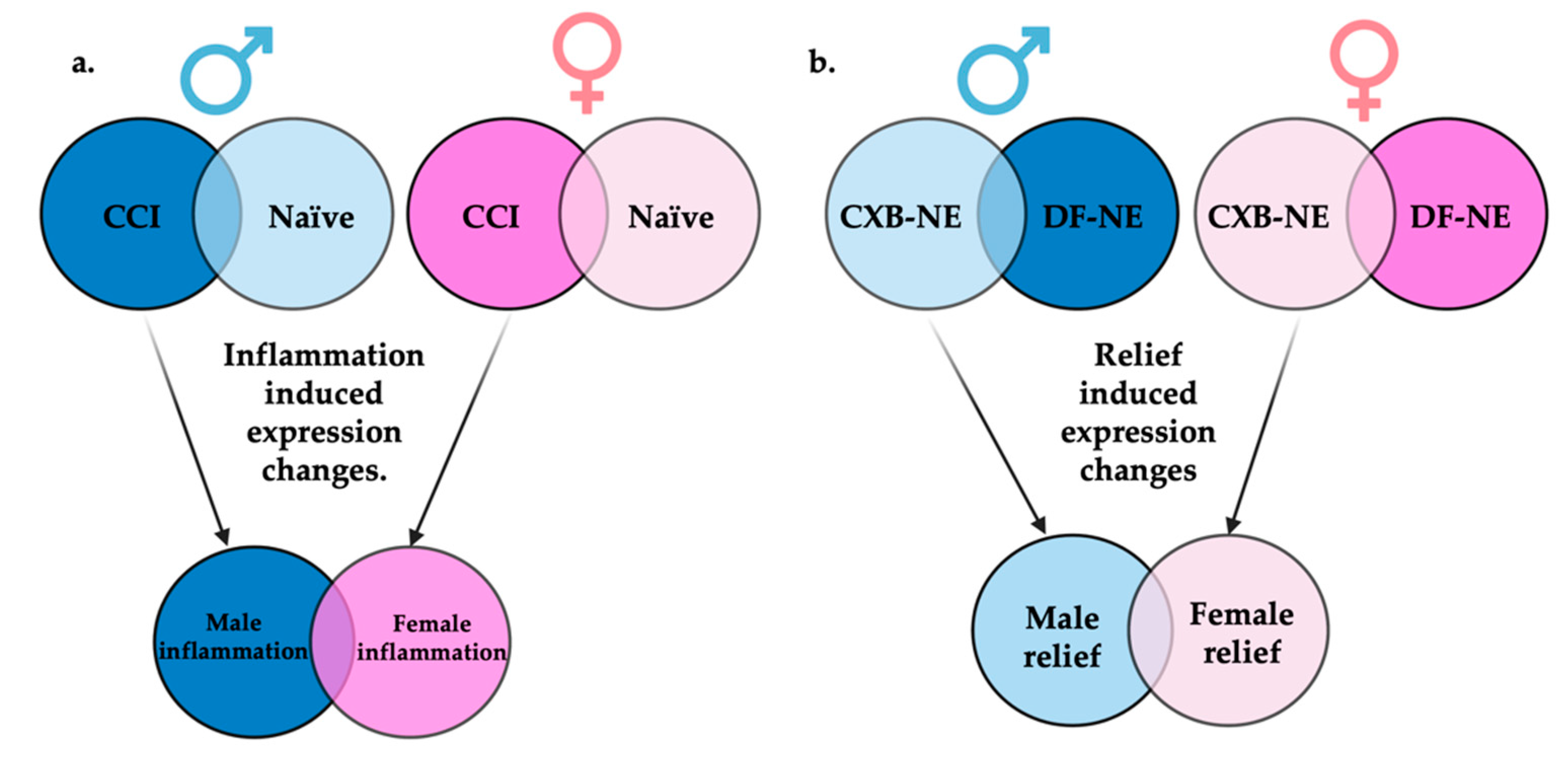
Figure 4.
Display of comparisons made between RNA-seq generated gene sets. (a) Genes differentially expressed between CCI and naïve of the same sex will be termed inflammatory induced gene expression changes. When these inflammatory gene sets are compared between sexes, genes uniquely differentially expressed in males or females can be identified. (b) Genes differentially expressed between 10X CXB-NE and DF-NE of the same sex will be termed relief induced gene expression changes. When these gene sets are compared between sexes, genes uniquely differentially expressed during relief in males or females associated can be identified. Figure made with BioRender.com (accessed on 6 October 2022).
Figure 4.
Display of comparisons made between RNA-seq generated gene sets. (a) Genes differentially expressed between CCI and naïve of the same sex will be termed inflammatory induced gene expression changes. When these inflammatory gene sets are compared between sexes, genes uniquely differentially expressed in males or females can be identified. (b) Genes differentially expressed between 10X CXB-NE and DF-NE of the same sex will be termed relief induced gene expression changes. When these gene sets are compared between sexes, genes uniquely differentially expressed during relief in males or females associated can be identified. Figure made with BioRender.com (accessed on 6 October 2022).
Figure 5.
Neuroinflammation in females causes differential gene expression in the affected DRG. (a) For visual clarity, the volcano plot features all of the differentially expressed genes due to neuroinflammation in the female DRG and highlights the top 10 significantly up-regulated (red) and top 10 significantly down-regulated genes (black). (b) Metascape enrichment analysis of the top 20 up-regulated biological pathways following induction of neuroinflammation. (c) Metascape enrichment analysis of the top 20 down-regulated biological pathways following induction of neuroinflammation.
Figure 5.
Neuroinflammation in females causes differential gene expression in the affected DRG. (a) For visual clarity, the volcano plot features all of the differentially expressed genes due to neuroinflammation in the female DRG and highlights the top 10 significantly up-regulated (red) and top 10 significantly down-regulated genes (black). (b) Metascape enrichment analysis of the top 20 up-regulated biological pathways following induction of neuroinflammation. (c) Metascape enrichment analysis of the top 20 down-regulated biological pathways following induction of neuroinflammation.
Figure 6.
Neuroinflammation in males causes differential gene expression in the affected DRG. (a) For visual clarity, the volcano plot features all the differentially expressed genes due to neuroinflammation in the male DRG with a highlight of the top 10 significantly up-regulated (red) and top 10 significantly down-regulated genes (black). (b) Metascape enrichment analysis of the top 20 up-regulated biological pathways following induction of neuroinflammation. (c) Metascape enrichment analysis of the top 20 down-regulated biological pathways following induction of neuroinflammation.
Figure 6.
Neuroinflammation in males causes differential gene expression in the affected DRG. (a) For visual clarity, the volcano plot features all the differentially expressed genes due to neuroinflammation in the male DRG with a highlight of the top 10 significantly up-regulated (red) and top 10 significantly down-regulated genes (black). (b) Metascape enrichment analysis of the top 20 up-regulated biological pathways following induction of neuroinflammation. (c) Metascape enrichment analysis of the top 20 down-regulated biological pathways following induction of neuroinflammation.
Figure 7.
Relief from neuroinflammation from COX-2 inhibiting theranostic nanoemulsion in females causes differential gene expression in the affected DRG. (a) For visual clarity, the volcano plot features all of the differentially expressed genes due to relief of neuroinflammation in the female DRG and highlights the top 6 significantly up-regulated (red) and top 10 significantly down-regulated genes (black). (b) Metascape enrichment analysis of the down-regulated biological pathways following reduction of neuroinflammation resulting from COX-2 inhibition.
Figure 7.
Relief from neuroinflammation from COX-2 inhibiting theranostic nanoemulsion in females causes differential gene expression in the affected DRG. (a) For visual clarity, the volcano plot features all of the differentially expressed genes due to relief of neuroinflammation in the female DRG and highlights the top 6 significantly up-regulated (red) and top 10 significantly down-regulated genes (black). (b) Metascape enrichment analysis of the down-regulated biological pathways following reduction of neuroinflammation resulting from COX-2 inhibition.
Figure 8.
Relief from neuroinflammation by COX-2 inhibiting theranostic nanoemulsion in males causes differential gene expression in the affected DRG. (a) For visual clarity, the volcano plot features all the differentially expressed genes due to neuroinflammatory relief in the male DRG with a highlight of the top 10 significantly up-regulated (red) and top 10 significantly down-regulated genes (black). (b) Metascape enrichment analysis of the top 20 up-regulated biological pathways following neuroinflammatory reduction via COX-2 inhibition. (c) Metascape enrichment analysis of the 9 down-regulated biological pathways following reduction of neuroinflammation resulting from COX-2 inhibition.
Figure 8.
Relief from neuroinflammation by COX-2 inhibiting theranostic nanoemulsion in males causes differential gene expression in the affected DRG. (a) For visual clarity, the volcano plot features all the differentially expressed genes due to neuroinflammatory relief in the male DRG with a highlight of the top 10 significantly up-regulated (red) and top 10 significantly down-regulated genes (black). (b) Metascape enrichment analysis of the top 20 up-regulated biological pathways following neuroinflammatory reduction via COX-2 inhibition. (c) Metascape enrichment analysis of the 9 down-regulated biological pathways following reduction of neuroinflammation resulting from COX-2 inhibition.
Figure 9.
Venn diagrams of differentially expressed genes in each sex and shared genes in male and female neuroinflammation and relief. (a) Male and female differentially expressed neuroinflammatory genes compared between sexes for unique characteristics to males or females and those shared between sexes. Within each sex, neuroinflammatory gene sets originated from CCI vs. naïve transcriptome comparisons (female naïve n = 3, female CCI n = 2, male naïve n = 3, male CCI n = 3). (b) Neuroinflammatory relief gene expression profiles of males and females were compared for shared features and those unique to a sex. Within each sex, neuroinflammatory relief gene sets originated from 10X CXB-NE vs. DF-NE DRG transcriptome comparisons (female DF-NE n = 2, female 10X CXB-NE n = 2, male DF-NE n = 3, male 10X CXB-NE n = 3).
Figure 9.
Venn diagrams of differentially expressed genes in each sex and shared genes in male and female neuroinflammation and relief. (a) Male and female differentially expressed neuroinflammatory genes compared between sexes for unique characteristics to males or females and those shared between sexes. Within each sex, neuroinflammatory gene sets originated from CCI vs. naïve transcriptome comparisons (female naïve n = 3, female CCI n = 2, male naïve n = 3, male CCI n = 3). (b) Neuroinflammatory relief gene expression profiles of males and females were compared for shared features and those unique to a sex. Within each sex, neuroinflammatory relief gene sets originated from 10X CXB-NE vs. DF-NE DRG transcriptome comparisons (female DF-NE n = 2, female 10X CXB-NE n = 2, male DF-NE n = 3, male 10X CXB-NE n = 3).
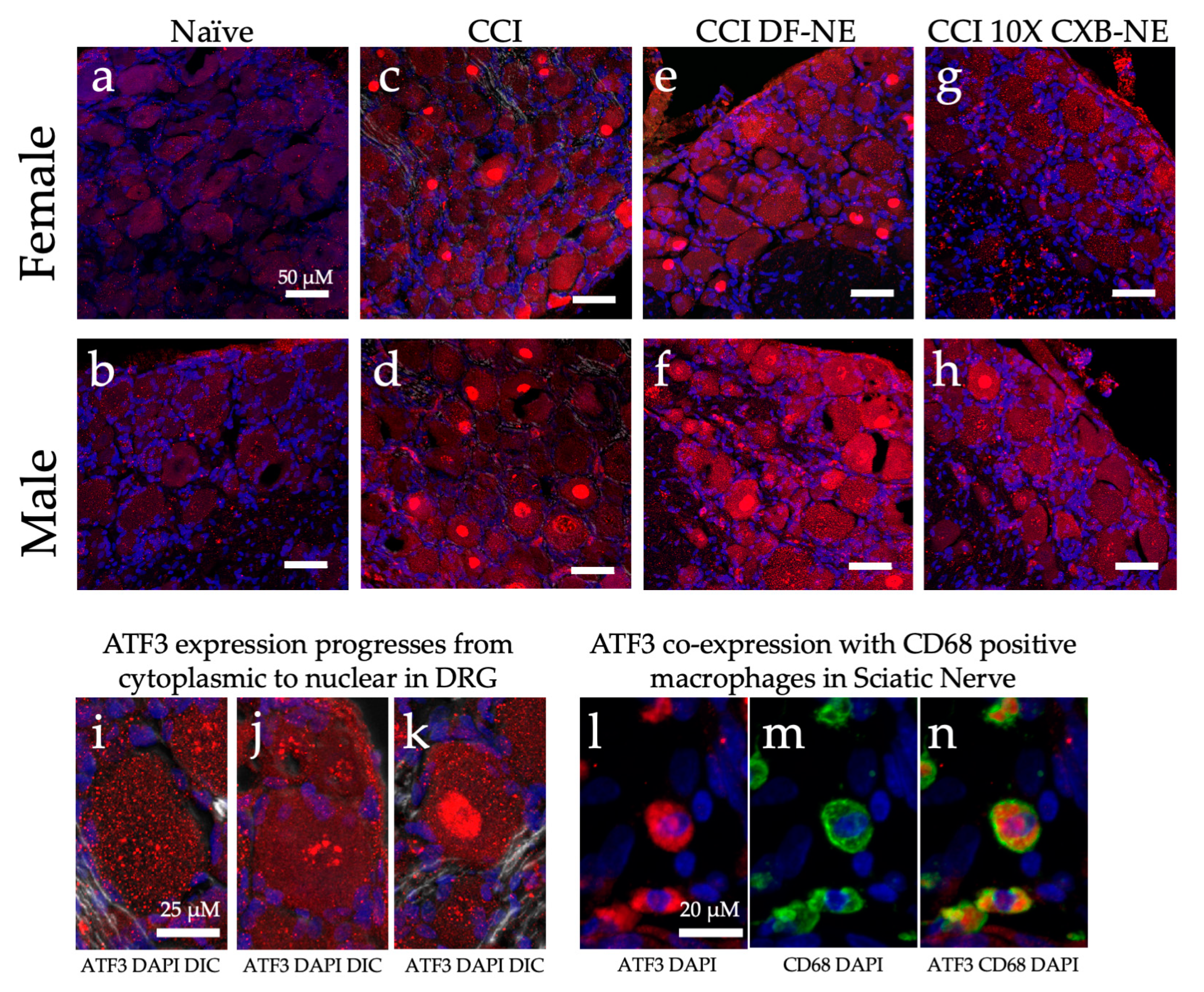
Figure 10.
ATF3 protein expression (red) changes in a sex specific manner in the ipsilateral DRG during hypersensitivity (CCI and CCI DF-NE) and with relief (CCI 10X CXB-NE). Tissue retrieved on day 12. Nuclei stained with DAPI (blue). Double stained merge images include DIC transmitted light. (a) and (b) Non-surgical naïve female and male DRG tissue with low level dispersed ATF3 located throughout the cytosol. (c) and (d) ATF3 increased expression and condensation in the nucleus following CCI neuroinflammatory hypersensitivity in both female and male ipsilateral DRG. (e) and (f) CCI treated with DF-NE continue to express high levels of ATF-3 in both cytoplasm and condensation in nuclei for both females and males. (g) Disappearance of nuclear condensation of ATF3 following pain-relieving theranostic nanoemulsion with 10X CXB-NE treatment in females. (h) Decreased nuclear condensation of ATF3 following 10X CXB-NE treatment in males. (i) Cytoplasmic ATF3 localization. (j) Perinuclear accumulation of ATF3. (k) Condensation of ATF3 in the nucleus. (l) Positive ATF3 (red) in macrophages of the sciatic nerve with (blue) DAPI nuclei. (m) The same field of view as (l), revealing positive staining of macrophages with the anti-CD68 (green). (n) Co-localization of ATF3 staining within CD68 positive macrophages within the sciatic nerve.
Figure 10.
ATF3 protein expression (red) changes in a sex specific manner in the ipsilateral DRG during hypersensitivity (CCI and CCI DF-NE) and with relief (CCI 10X CXB-NE). Tissue retrieved on day 12. Nuclei stained with DAPI (blue). Double stained merge images include DIC transmitted light. (a) and (b) Non-surgical naïve female and male DRG tissue with low level dispersed ATF3 located throughout the cytosol. (c) and (d) ATF3 increased expression and condensation in the nucleus following CCI neuroinflammatory hypersensitivity in both female and male ipsilateral DRG. (e) and (f) CCI treated with DF-NE continue to express high levels of ATF-3 in both cytoplasm and condensation in nuclei for both females and males. (g) Disappearance of nuclear condensation of ATF3 following pain-relieving theranostic nanoemulsion with 10X CXB-NE treatment in females. (h) Decreased nuclear condensation of ATF3 following 10X CXB-NE treatment in males. (i) Cytoplasmic ATF3 localization. (j) Perinuclear accumulation of ATF3. (k) Condensation of ATF3 in the nucleus. (l) Positive ATF3 (red) in macrophages of the sciatic nerve with (blue) DAPI nuclei. (m) The same field of view as (l), revealing positive staining of macrophages with the anti-CD68 (green). (n) Co-localization of ATF3 staining within CD68 positive macrophages within the sciatic nerve.
![Ijms 24 09163 g010]()
Figure 11.
COX-2 feedback loop transcriptionally regulated by ATF3. TLR4 is activated by cellular debris from neuronal injury as well as certain cytokines, which then activates parts of the MAPK signaling cascade as well as NF-κB. Production of PGE
2 provides a positive feedback loop to support continued production of prostaglandin, which further support increased immune cell infiltration and activation. PKA, which is activated through PGE
2 binding to its EP4 receptor, can phosphorylate TRPV1 inducing translocation of the receptor to the cell membrane; thereby, increasing neuronal hypersensitivity. Model influenced by our research and multiple citations [
33,
62,
65,
66]. Figure made with BioRender.
Figure 11.
COX-2 feedback loop transcriptionally regulated by ATF3. TLR4 is activated by cellular debris from neuronal injury as well as certain cytokines, which then activates parts of the MAPK signaling cascade as well as NF-κB. Production of PGE
2 provides a positive feedback loop to support continued production of prostaglandin, which further support increased immune cell infiltration and activation. PKA, which is activated through PGE
2 binding to its EP4 receptor, can phosphorylate TRPV1 inducing translocation of the receptor to the cell membrane; thereby, increasing neuronal hypersensitivity. Model influenced by our research and multiple citations [
33,
62,
65,
66]. Figure made with BioRender.
Table 1.
Top 25 significantly up-regulated genes in the female DRG following CCI injury of the sciatic nerve.
Table 1.
Top 25 significantly up-regulated genes in the female DRG following CCI injury of the sciatic nerve.
| Gene ID | FDR | Fold Change | ENSMBL Gene ID |
|---|
| Npy | 3.97 × 10−42 | 12.38 | ENSRNOG00000046449 |
| Atf3 | 2.17 × 10−23 | 7.83 | ENSRNOG00000003745 |
| Flrt3 | 2.46 × 10−23 | 2.51 | ENSRNOG00000004874 |
| Igfbp3 | 4.67 × 10−15 | 1.8 | ENSRNOG00000061910 |
| Gap43 | 8.54 × 10−15 | 1.58 | ENSRNOG00000001528 |
| Cacna2d1 | 7.36 × 10−13 | 1.91 | ENSRNOG00000033531 |
| Ktn1 | 1.04 × 10−12 | 1.53 | ENSRNOG00000012255 |
| Stac2 | 2.03 × 10−12 | 8.59 | ENSRNOG00000004805 |
| Gal | 5.39 × 10−12 | 2.92 | ENSRNOG00000015156 |
| Gadd45a | 7.04 × 10−12 | 2.25 | ENSRNOG00000005615 |
| Vip | 9.71 × 10−11 | 18.83 | ENSRNOG00000018808 |
| AC130970.1 | 1.41 × 10−10 | 3.77 | ENSRNOG00000042445 |
| Csrp3 | 3.07 × 10−10 | 43.49 | ENSRNOG00000014327 |
| Ccl2 | 1.04 × 10−9 | 2.76 | ENSRNOG00000007159 |
| Sema6a | 2.08 × 10−9 | 2.06 | ENSRNOG00000004033 |
| gene:ENSRNOG00000067276 | 2.18 × 10−9 | 1.81 | ENSRNOG00000067276 |
| Il7r | 2.44 × 10−9 | 1.98 | ENSRNOG00000065741 |
| Prkar2b | 3.04 × 10−9 | 1.46 | ENSRNOG00000009079 |
| Bdnf | 8.06 × 10−9 | 2.14 | ENSRNOG00000047466 |
| Vgf | 1.52 × 10−8 | 2.25 | ENSRNOG00000001416 |
| Stmn4 | 7.06 × 10−8 | 1.83 | ENSRNOG00000053334 |
| Sox11 | 1.17 × 10−7 | 2.53 | ENSRNOG00000030034 |
| Acsl4 | 1.23 × 10−7 | 1.43 | ENSRNOG00000019180 |
| Hs2st1 | 1.53 × 10−7 | 1.44 | ENSRNOG00000012549 |
| Il13ra1_2 | 1.69 × 10−7 | 1.76 | ENSRNOG00000013170 |
Table 2.
Top 25 significantly down-regulated genes in the female DRG following CCI injury of the sciatic nerve.
Table 2.
Top 25 significantly down-regulated genes in the female DRG following CCI injury of the sciatic nerve.
| Gene ID | FDR | Fold Change | ENSMBL Gene ID |
|---|
| Sema7a | 2.41 × 10−21 | −1.82 | ENSRNOG00000007687 |
| Igfbp6 | 7.15 × 10−15 | −2.12 | ENSRNOG00000010977 |
| RT1-CE16 | 8.18 × 10−15 | −3.45 | ENSRNOG00000065254 |
| Tagln | 4.07 × 10−11 | −2.27 | ENSRNOG00000017628 |
| Iscu | 9.71 × 10−11 | −1.51 | ENSRNOG00000000701 |
| Hapln4 | 7.04 × 10−10 | −1.9 | ENSRNOG00000049949 |
| Gsn | 2.18 × 10−9 | −1.46 | ENSRNOG00000018991 |
| Lgi3 | 2.08 × 10−8 | −1.44 | ENSRNOG00000011323 |
| Hcn2 | 1.02 × 10−7 | −1.46 | ENSRNOG00000008831 |
| gene:ENSRNOG00000070982 | 1.17 × 10−7 | −2.08 | ENSRNOG00000070982 |
| Capg | 1.17 × 10−7 | −3.01 | ENSRNOG00000013668 |
| gene:ENSRNOG00000006471 | 1.51 × 10−7 | −1.69 | ENSRNOG00000006471 |
| gene:ENSRNOG00000062930 | 1.97 × 10−7 | −1.95 | ENSRNOG00000062930 |
| Fxyd6 | 2.21 × 10−7 | −1.44 | ENSRNOG00000016412 |
| Islr | 3.48 × 10−7 | −1.76 | ENSRNOG00000048924 |
| Fxyd7 | 3.63 × 10−7 | −1.4 | ENSRNOG00000021067 |
| Abhd8 | 3.96 × 10−7 | −1.73 | ENSRNOG00000000054 |
| Tlcd3b | 4.91 × 10−7 | −1.4 | ENSRNOG00000019914 |
| Sptb | 4.94 × 10−7 | −1.39 | ENSRNOG00000006911 |
| Kcnh2 | 6.60 × 10−7 | −1.42 | ENSRNOG00000009872 |
| Kcnc3 | 7.18 × 10−7 | −1.41 | ENSRNOG00000019959 |
| Fbln2 | 7.35 × 10−7 | −1.37 | ENSRNOG00000007338 |
| Kcns1 | 7.67 × 10−7 | −1.67 | ENSRNOG00000013681 |
| Oaz1 | 1.08 × 10−6 | −1.4 | ENSRNOG00000019459 |
| Bphl | 1.08 × 10−6 | −1.92 | ENSRNOG00000017577 |
Table 3.
Top 25 significantly up-regulated genes in the male DRG following CCI injury of the sciatic nerve.
Table 3.
Top 25 significantly up-regulated genes in the male DRG following CCI injury of the sciatic nerve.
| Gene ID | FDR | Fold Change | ENSMBL Gene ID |
|---|
| Npy | 2.67 × 10−20 | 5.79 | ENSRNOG00000046449 |
| Thbs1 | 9.51 × 10−14 | 1.88 | ENSRNOG00000045829 |
| Cfh | 1.54 × 10−9 | 1.49 | ENSRNOG00000030715 |
| NEWGENE_1308171 | 1.77 × 10−7 | 1.58 | ENSRNOG00000029792 |
| RT1-Ba | 5.72 × 10−7 | 1.94 | ENSRNOG00000000451 |
| Atf3 | 1.26 × 10−6 | 4.12 | ENSRNOG00000003745 |
| gene:ENSRNOG00000067832 | 1.55 × 10−6 | 41.86 | ENSRNOG00000067832 |
| Lmo7 | 1.59 × 10−6 | 1.62 | ENSRNOG00000060775 |
| Slc13a4 | 2.38 × 10−6 | 1.63 | ENSRNOG00000011184 |
| H3f3c | 2.63 × 10−6 | 9.77 | ENSRNOG00000032401 |
| Phldb2 | 2.88 × 10−6 | 1.43 | ENSRNOG00000002171 |
| Fyb1 | 3.53 × 10−6 | 1.85 | ENSRNOG00000013886 |
| Aldh1a2 | 4.15 × 10−6 | 2.52 | ENSRNOG00000055049 |
| Cp | 4.44 × 10−6 | 1.37 | ENSRNOG00000011913 |
| Sema3c | 7.25 × 10−6 | 1.33 | ENSRNOG00000006526 |
| Fbln1 | 9.61 × 10−6 | 1.58 | ENSRNOG00000014137 |
| Emp1 | 1.65 × 10−5 | 1.55 | ENSRNOG00000008676 |
| Slc6a20 | 3.23 × 10−5 | 1.85 | ENSRNOG00000068642 |
| Thbd | 3.55 × 10−5 | 1.55 | ENSRNOG00000004687 |
| Cilp | 4.37 × 10−5 | 2.43 | ENSRNOG00000029911 |
| Crh | 5.12 × 10−5 | 33.5 | ENSRNOG00000012703 |
| Slc6a13 | 5.12 × 10−5 | 1.99 | ENSRNOG00000012876 |
| Lcp1 | 5.12 × 10−5 | 1.3 | ENSRNOG00000010319 |
| Cfb | 5.12 × 10−5 | 1.61 | ENSRNOG00000051235 |
| Fat4 | 6.83 × 10−5 | 1.32 | ENSRNOG00000028335 |
Table 4.
Top 25 significantly down-regulated genes in the male DRG following CCI injury of the sciatic nerve.
Table 4.
Top 25 significantly down-regulated genes in the male DRG following CCI injury of the sciatic nerve.
| Gene ID | FDR | Fold Change | ENSMBL Gene ID |
|---|
| Egr2 | 1.47 × 10−20 | −2.39 | ENSRNOG00000000640 |
| Zbtb16 | 1.27 × 10−12 | −3.05 | ENSRNOG00000029980 |
| Myh2 | 1.27 × 10−12 | −8.77 | ENSRNOG00000065740 |
| Sptbn5 | 1.39 × 10−10 | −1.45 | ENSRNOG00000059260 |
| Tnnc2 | 6.32 × 10−8 | −4.81 | ENSRNOG00000015155 |
| Dbp | 7.66 × 10−8 | −1.49 | ENSRNOG00000021027 |
| Ttn | 1.48 × 10−7 | −1.6 | ENSRNOG00000069271 |
| Tnnt3 | 1.77 × 10−7 | −3.66 | ENSRNOG00000020332 |
| Mertk | 1.90 × 10−7 | −1.44 | ENSRNOG00000017319 |
| Tsc22d3 | 4.98 × 10−7 | −1.38 | ENSRNOG00000056135 |
| Eno3 | 6.24 × 10−7 | −2.42 | ENSRNOG00000004078 |
| Gpd1 | 6.88 × 10−7 | −1.73 | ENSRNOG00000056457 |
| Sgk1 | 1.55 × 10−6 | −1.58 | ENSRNOG00000011815 |
| AABR07005775.1 | 1.55 × 10−6 | −3.71 | ENSRNOG00000047124 |
| Neb | 4.44 × 10−6 | −1.7 | ENSRNOG00000006783 |
| Hif3a | 7.25 × 10−6 | −5.79 | ENSRNOG00000017198 |
| Acta1 | 7.25 × 10−6 | −3.23 | ENSRNOG00000017786 |
| Per1 | 8.28 × 10−6 | −1.94 | ENSRNOG00000007387 |
| Aatk | 1.14 × 10−5 | −1.32 | ENSRNOG00000004392 |
| Fkbp5 | 1.19 × 10−5 | −1.56 | ENSRNOG00000022523 |
| Mb | 1.30 × 10−5 | −5.5 | ENSRNOG00000004583 |
| Prx | 1.34 × 10−5 | −1.3 | ENSRNOG00000018369 |
| Mylpf | 2.18 × 10−5 | −3.55 | ENSRNOG00000017645 |
| Ntng2 | 3.12 × 10−5 | −1.34 | ENSRNOG00000013694 |
| Atp1a3 | 3.38 × 10−5 | −1.28 | ENSRNOG00000020263 |
Table 5.
The significantly up-regulated genes in the female DRG following reduction of neuroinflammation with COX-2 inhibiting theranostic nanoemulsion.
Table 5.
The significantly up-regulated genes in the female DRG following reduction of neuroinflammation with COX-2 inhibiting theranostic nanoemulsion.
| Gene ID | FDR | Fold Change | ENSMBL Gene ID |
|---|
| Myom3 | 1.10 × 10−4 | 9.43 | ENSRNOG00000032994 |
| Grhl3 | 8.06 × 10−4 | 13.29 | ENSRNOG00000029427 |
| Txnip | 2.92 × 10−3 | 1.35 | ENSRNOG00000021201 |
| Hba-a3 | 4.00 × 10−2 | 2.51 | ENSRNOG00000047321 |
| Sptb | 0.02 | 1.31 | ENSRNOG00000006911 |
| Gas2l3 | 0.03 | 1.3 | ENSRNOG00000063755 |
Table 6.
The significantly down-regulated genes in the female DRG following reduction of neuroinflammation with COX-2 inhibiting theranostic nanoemulsion.
Table 6.
The significantly down-regulated genes in the female DRG following reduction of neuroinflammation with COX-2 inhibiting theranostic nanoemulsion.
| Gene ID | FDR | Fold Change | ENSMBL Gene ID |
|---|
| Ass1 | 3.44 × 10−4 | −2.4 | ENSRNOG00000008837 |
| Vgf | 4.22 × 10−4 | −1.63 | ENSRNOG00000001416 |
| Atf3 | 8.06 × 10−4 | −2.09 | ENSRNOG00000003745 |
| Sox11 | 1.35 × 10−3 | −1.66 | ENSRNOG00000030034 |
| Lce1f_2 | 8.47 × 10−3 | −5.85 | ENSRNOG00000066187 |
| Sema6a | 1.00 × 10−2 | −1.67 | ENSRNOG00000004033 |
| AABR07007068.1 | 2.00 × 10−2 | −1.3 | ENSRNOG00000017438 |
| Gadd45a | 3.00 × 10−2 | −1.49 | ENSRNOG00000005615 |
| Jun | 4.00 × 10−2 | −1.28 | ENSRNOG00000026293 |
| Rbp4 | 5.00 × 10−2 | −1.68 | ENSRNOG00000015518 |
Table 7.
Top 25 most significantly up-regulated genes in the male DRG following reduction of neuroinflammation with COX-2 inhibiting theranostic nanoemulsion.
Table 7.
Top 25 most significantly up-regulated genes in the male DRG following reduction of neuroinflammation with COX-2 inhibiting theranostic nanoemulsion.
| Gene ID | FDR | Fold Change | ENSMBL Gene ID |
|---|
| S100a9 | 9.37 × 10−30 | 5.68 | ENSRNOG00000011483 |
| S100a8 | 4.11 × 10−28 | 6.24 | ENSRNOG00000011557 |
| Mmp8 | 2.84 × 10−20 | 7.53 | ENSRNOG00000009907 |
| Ngp | 1.18 × 10−18 | 5.2 | ENSRNOG00000024330 |
| Ighm | 7.44 × 10−17 | 3.2 | ENSRNOG00000034190 |
| Ccl21 | 3.78 × 10−16 | 4.15 | ENSRNOG00000034290 |
| Slc4a1 | 2.75 × 10−13 | 3.69 | ENSRNOG00000020951 |
| Fcnb | 8.83 × 10−13 | 5.16 | ENSRNOG00000009342 |
| Ccn3 | 1.45 × 10−12 | 2.77 | ENSRNOG00000008697 |
| Mfap4 | 1.60 × 10−12 | 2.41 | ENSRNOG00000045683 |
| NEWGENE_1308171 | 3.27 × 10−11 | 1.45 | ENSRNOG00000029792 |
| Mpo | 2.04 × 10−10 | 5.12 | ENSRNOG00000008310 |
| Ptprc | 2.46 × 10−10 | 1.81 | ENSRNOG00000000655 |
| AABR07060872.1 | 4.76 × 10−10 | 4.5 | ENSRNOG00000049829 |
| Serpinb1b | 5.34 × 10−10 | 11.78 | ENSRNOG00000034229 |
| Mki67 | 5.34 × 10−10 | 2.22 | ENSRNOG00000028137 |
| Ccn2 | 1.13 × 10−9 | 2.03 | ENSRNOG00000015036 |
| Sell | 1.25 × 10−9 | 3.82 | ENSRNOG00000002776 |
| Lyz2 | 1.25 × 10−9 | 1.67 | ENSRNOG00000005825 |
| Prg2 | 3.12 × 10−9 | 4.31 | ENSRNOG00000008394 |
| RatNP-3b | 4.14 × 10−9 | 7.22 | ENSRNOG00000038135 |
| Hemgn | 1.29 × 10−8 | 3.6 | ENSRNOG00000009436 |
| Slfn4_1 | 1.39 × 10−8 | 2.24 | ENSRNOG00000057092 |
| Plac8 | 6.24 × 10−8 | 3.18 | ENSRNOG00000002217 |
| Acta2 | 1.26 × 10−7 | 1.77 | ENSRNOG00000058039 |
Table 8.
Top 25 most significantly down-regulated genes in the male DRG reduction of neuroinflammation with COX-2 inhibiting theranostic nanoemulsion.
Table 8.
Top 25 most significantly down-regulated genes in the male DRG reduction of neuroinflammation with COX-2 inhibiting theranostic nanoemulsion.
| Gene ID | FDR | Fold Change | ENSMBL Gene ID |
|---|
| Dbp | 8.42 × 10−15 | −1.72 | ENSRNOG00000021027 |
| Prim1 | 3.56 × 10−5 | −2.01 | ENSRNOG00000031993 |
| Hmgb1 | 6.42 × 10−5 | −2.03 | ENSRNOG00000058908 |
| Per1 | 9.79 × 10−5 | −1.53 | ENSRNOG00000007387 |
| Zbtb16 | 2.66 × 10−4 | −1.89 | ENSRNOG00000029980 |
| Gjc3 | 5.34 × 10−4 | −1.29 | ENSRNOG00000001329 |
| Sgk1 | 6.90 × 10−4 | −1.3 | ENSRNOG00000011815 |
| Clvs1 | 1.03 × 10−3 | −2.39 | ENSRNOG00000006919 |
| Chrna3 | 1.92 × 10−3 | −1.38 | ENSRNOG00000013829 |
| Acer2 | 1.95 × 10−3 | −1.52 | ENSRNOG00000007637 |
| Nptx1 | 2.13 × 10−3 | −1.23 | ENSRNOG00000003741 |
| Crh | 3.52 × 10−3 | −21.61 | ENSRNOG00000012703 |
| Trim45 | 4.35 × 10−3 | −2.29 | ENSRNOG00000015347 |
| AABR07034445.1 | 7.52 × 10−3 | −2.27 | ENSRNOG00000038610 |
| Ntng1 | 7.98 × 10−3 | −1.25 | ENSRNOG00000031136 |
| Klf9 | 8.75 × 10−3 | −1.23 | ENSRNOG00000014215 |
| Sema7a | 8.99 × 10−3 | −1.23 | ENSRNOG00000007687 |
| RGD1564899 | 0.01 | −2.2 | ENSRNOG00000049942 |
| Nr1d1 | 0.02 | −2.2 | ENSRNOG00000009329 |
| Gnai1 | 0.02 | −1.21 | ENSRNOG00000057096 |
| Kcng1 | 0.03 | −1.39 | ENSRNOG00000054314 |
| Chst2 | 0.03 | −1.19 | ENSRNOG00000047734 |
| Slitrk2 | 0.03 | −1.19 | ENSRNOG00000011562 |
| Svip | 0.03 | −1.19 | ENSRNOG00000037137 |
| Ncam2 | 0.03 | −1.19 | ENSRNOG00000002126 |
Table 9.
List of differentially expressed genes that show unique patterns of differential RNA expression across sex and or treatment. Table information adapted from output data provided by Metascape.com.
Table 9.
List of differentially expressed genes that show unique patterns of differential RNA expression across sex and or treatment. Table information adapted from output data provided by Metascape.com.
| Gene Name | Full Gene Name | Gene Ontology | Protein Function | Pattern of Expression |
|---|
| Atf3 | Activating transcription factor 3 | GO:0061394 regulation of transcription from RNA polymerase II promoter in response to arsenic-containing substance; GO:1903984 positive regulation of TRAIL-activated apoptotic signaling pathway; GO:1903121 regulation of TRAIL-activated apoptotic signaling pathway | Transcription factors: Basic domains; Predicted intracellular proteins | Up-regulated in both sexes following CCI, down-regulated with relief in females |
| Sema6a | Semaphorin 6A | GO:0106089 negative regulation of cell adhesion involved in sprouting angiogenesis; GO:0106088 regulation of cell adhesion involved in sprouting angiogenesis; GO:1900747 negative regulation of vascular endothelial growth factor signaling pathway | Predicted intracellular proteins | Up-regulated in both sexes following CCI, down-regulated with relief in females |
| Vgf | VGF nerve growth factor inducible | GO:0043084 penile erection; GO:0007620 copulation; GO:0002021 response to dietary excess | Predicted secreted proteins | Up-regulated in both sexes following CCI, down-regulated with relief in females |
| Flrt3 | Fibronectin leucine rich transmembrane protein 3 | GO:0003345 proepicardium cell migration involved in pericardium morphogenesis; GO:0003344 pericardium morphogenesis; GO:0060973 cell migration involved in heart development | Disease related genes; Human disease related genes: Endocrine and metabolic diseases: Hypothalamus and pituitary gland diseases | Up-regulated in both sexes following CCI. No change with relief. |
| Crh | Corticotropin releasing hormone | GO:0042322 negative regulation of circadian sleep/wake cycle, REM sleep; GO:0010841 positive regulation of circadian sleep/wake cycle, wakefulness; GO:0042321 negative regulation of circadian sleep/wake cycle, sleep | Predicted secreted proteins | Up-regulated in both sexes following CCI, down-regulated with relief in males |
| Prim1 | DNA primase subunit 1 | GO:0006269 DNA replication, synthesis of RNA primer; GO:0006270 DNA replication initiation; GO:0006261 DNA-templated DNA replication | Predicted intracellular proteins | Up-regulated in males following CCI, down-regulated with relief in males |
| Prx | Periaxin | GO:0032287 peripheral nervous system myelin maintenance; GO:0043217 myelin maintenance; GO:0022011 myelination in peripheral nervous system | Disease related genes; Human disease related genes: Nervous system diseases: Other nervous and sensory system diseases; Human disease related genes: Nervous system diseases: Neurodegenerative diseases; Predicted intracellular proteins | Down-regulated in both sexes following CCI, no significant change with treatment |
| Myh2 | Myosin Heavy Chain 2 | GO:0030049 muscle filament sliding; GO:0033275 actin-myosin filament sliding; GO:0070252 actin-mediated cell contraction | Disease related genes; Human disease related genes: Musculoskeletal diseases: Muscular diseases; Predicted intracellular proteins | Up-regulated in females following CCI, down-regulated in males following CCI |
| Tnnt3 | Troponin T3 | GO:1903612 positive regulation of calcium-dependent ATPase activity; GO:1903610 regulation of calcium-dependent ATPase activity; GO:0003009 skeletal muscle contraction | Disease related genes; Predicted intracellular proteins; Human disease related genes: Congenital malformations: Other congenital malformations | Up-regulated in females following CCI, down-regulated in males following CCI |
 DF-NE day 6 = 4.17 g, STD = 1.76,
DF-NE day 6 = 4.17 g, STD = 1.76,  DF-NE day 6 = 3.64, STD = 1.24,
DF-NE day 6 = 3.64, STD = 1.24,  DF-NE day 8 = 3.22 g, STD = 1.33,
DF-NE day 8 = 3.22 g, STD = 1.33,  DF-NE day 8 = 5.13, STD = 3.94). This indicates that both males and females are experiencing the same degree of provoked hypersensitivity resulting from the CCI surgery. Tail vein injections of either 10X CXB-NE or drug free nanoemulsion (DF-NE) were delivered on day 8 following von Frey behavior analysis. Receiving DF-NE did not decrease mechanical withdrawal threshold, nor did it produce any type of relief. On days 10 and 12, males (Figure 1b) and females (Figure 1c) both experienced significant relief when compared to their CCI counterparts (Day 10
DF-NE day 8 = 5.13, STD = 3.94). This indicates that both males and females are experiencing the same degree of provoked hypersensitivity resulting from the CCI surgery. Tail vein injections of either 10X CXB-NE or drug free nanoemulsion (DF-NE) were delivered on day 8 following von Frey behavior analysis. Receiving DF-NE did not decrease mechanical withdrawal threshold, nor did it produce any type of relief. On days 10 and 12, males (Figure 1b) and females (Figure 1c) both experienced significant relief when compared to their CCI counterparts (Day 10  p = 0.0338, Day 12
p = 0.0338, Day 12  p < 0.0001, Day 10
p < 0.0001, Day 10  p < 0.0001, Day 12
p < 0.0001, Day 12  p < 0.0001) and DF-NE counterparts (Day 10
p < 0.0001) and DF-NE counterparts (Day 10  p = 0.0404, Day 12
p = 0.0404, Day 12  p < 0.0001, Day 10
p < 0.0001, Day 10  p < 0.0001, Day 12
p < 0.0001, Day 12  p < 0.0001). We also find that on day 12, the 10X CXB-NE treated males and females exhibit no statistically significant difference in behavior as compared to their naïve baseline (2-way ANOVA with Tukey post hoc analysis, p > 0.9999). (
p < 0.0001). We also find that on day 12, the 10X CXB-NE treated males and females exhibit no statistically significant difference in behavior as compared to their naïve baseline (2-way ANOVA with Tukey post hoc analysis, p > 0.9999). ( 10X CXB-NE day 12 = 14.43 g, STD = 5.88,
10X CXB-NE day 12 = 14.43 g, STD = 5.88,  = 14.5 g, STD = 5.73). Furthermore, by assessing the female estrus cycle, we find that in this case, all the females were asynchronous, and their estrus cycle status did not correlate to any bias of their behavior (Supplementary Figure S1). Similar to what we have previously reported [11], the individual’s estrus cycle does not appear have an overt effect on the hypersensitivity behavior or the female response to CXB-NE. Dorsal root ganglia and sciatic nerves were recovered for subsequent analysis on day 12 because it is the day of maximum relief.
= 14.5 g, STD = 5.73). Furthermore, by assessing the female estrus cycle, we find that in this case, all the females were asynchronous, and their estrus cycle status did not correlate to any bias of their behavior (Supplementary Figure S1). Similar to what we have previously reported [11], the individual’s estrus cycle does not appear have an overt effect on the hypersensitivity behavior or the female response to CXB-NE. Dorsal root ganglia and sciatic nerves were recovered for subsequent analysis on day 12 because it is the day of maximum relief.  p = 0.0919,
p = 0.0919,  p = 0.3908) and day 12 (
p = 0.3908) and day 12 ( p = 0.8712,
p = 0.8712,  p = 0.8643) there is no longer a significant difference in behavior between drug treated animals of either sex and their baseline or naïve behavior. Recall that a different 1× CXB-NE formulation (0.24 mg/kg) presented to CCI male and female rats provided complete relief for males, but only partial relief for females [11] and that the 1× CXB-NE delivered to CFA mice also resulted in sexually dimorphic relief [27]. So, with the 10X CXB-NE, the higher level of celecoxib per nanodroplet could be influencing COX-1 activity in females. This model is consistent with other researcher’s observation that in a CFA inflammatory pain model using COX-1 and COX-2 knockout male and female mice, a difference in the involvement of these enzymes was detected between the sexes [28]. Chillingworth and colleagues [28] saw that male inflammatory relief was most effectively achieved by COX-2 inhibition while females appeared to have an additive effect based on the involvement of both COX-1 and COX-2 in the development of their inflammatory pain [28]. Given that celecoxib has a slight selectivity for COX-1 [29,30,31], this could mean that COX-1 and COX-2 are now effectively attenuated in the females and thus, complete relief similar to that seen in males can now be experienced. Given that males already have complete relief, more celecoxib can’t influence their behavior any further.
p = 0.8643) there is no longer a significant difference in behavior between drug treated animals of either sex and their baseline or naïve behavior. Recall that a different 1× CXB-NE formulation (0.24 mg/kg) presented to CCI male and female rats provided complete relief for males, but only partial relief for females [11] and that the 1× CXB-NE delivered to CFA mice also resulted in sexually dimorphic relief [27]. So, with the 10X CXB-NE, the higher level of celecoxib per nanodroplet could be influencing COX-1 activity in females. This model is consistent with other researcher’s observation that in a CFA inflammatory pain model using COX-1 and COX-2 knockout male and female mice, a difference in the involvement of these enzymes was detected between the sexes [28]. Chillingworth and colleagues [28] saw that male inflammatory relief was most effectively achieved by COX-2 inhibition while females appeared to have an additive effect based on the involvement of both COX-1 and COX-2 in the development of their inflammatory pain [28]. Given that celecoxib has a slight selectivity for COX-1 [29,30,31], this could mean that COX-1 and COX-2 are now effectively attenuated in the females and thus, complete relief similar to that seen in males can now be experienced. Given that males already have complete relief, more celecoxib can’t influence their behavior any further.