Broadly Applicable Control Approaches Improve Accuracy of ChIP-Seq Data
Abstract
1. Introduction
2. Results
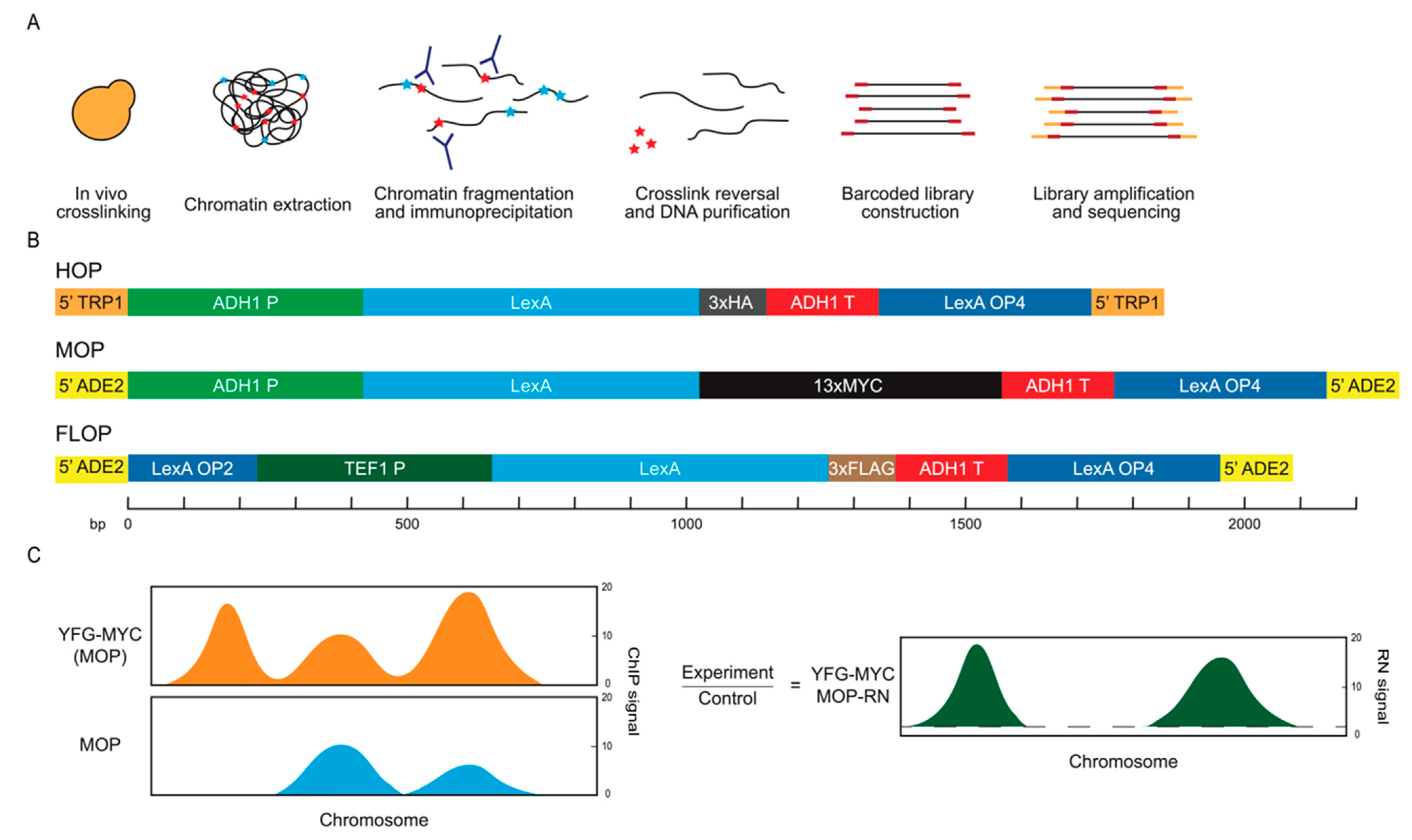
2.1. Expression of an Epitope-Tagged Protein as Normalization Control
2.2. Non-Specific Signals Are Pervasive in ChIP
2.3. Ratio Normalization by Epitope-Tagged Control Refines Data Quality for Target Proteins
2.4. Fkh1 Analysis in G1 Phase to Test Known Enrichment at Replication Origins
2.5. Analysis of Replication Origin Binding Proteins Validate Approach for HA
2.6. Controls Enhance Analysis of Potential Hyperchipable Loci
2.7. DNA-Binding Mutant May Be Ideal Control
3. Discussion
3.1. Expression of a Decoy Protein to Control for Non-Specific Sequence Enrichment
3.2. Multiple Factors Contribute to Non-Specific ChIP-Signal Enrichment
3.3. No Control, No Experiment
4. Materials and Methods
4.1. Plasmid Constructions
4.2. Yeast Strain Constructions
4.3. Other Methods
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Nakato, R.; Sakata, T. Methods for ChIP-seq analysis: A practical workflow and advanced applications. Methods 2021, 187, 44–53. [Google Scholar] [CrossRef] [PubMed]
- Nakato, R.; Shirahige, K. Recent advances in ChIP-seq analysis: From quality management to whole-genome annotation. Brief Bioinform. 2017, 18, 279–290. [Google Scholar] [CrossRef] [PubMed]
- Park, P.J. ChIP-seq: Advantages and challenges of a maturing technology. Nat. Rev. Genet. 2009, 10, 669–680. [Google Scholar] [CrossRef] [PubMed]
- Fan, X.; Struhl, K. Where Does Mediator Bind In Vivo? PLoS ONE 2009, 4, e5029. [Google Scholar] [CrossRef] [PubMed]
- Teytelman, L.; Thurtle, D.M.; Rine, J.; van Oudenaarden, A. Highly expressed loci are vulnerable to misleading ChIP localization of multiple unrelated proteins. Proc. Natl. Acad. Sci. USA 2013, 110, 18602–18607. [Google Scholar] [CrossRef]
- Wreczycka, K.; Franke, V.; Uyar, B.; Wurmus, R.; Bulut, S.; Tursun, B.; Akalin, A. HOT or not: Examining the basis of high-occupancy target regions. Nucleic Acids Res. 2019, 47, 5735–5745. [Google Scholar] [CrossRef]
- Jin, Y.; Liang, Z.; Lou, H. The Emerging Roles of Fox Family Transcription Factors in Chromosome Replication, Organization, and Genome Stability. Cells 2020, 9, 258. [Google Scholar] [CrossRef]
- Ostrow, A.Z.; Nellimoottil, T.; Knott, S.R.; Fox, C.A.; Tavare, S.; Aparicio, O.M. Fkh1 and Fkh2 bind multiple chromosomal elements in the S. cerevisiae genome with distinct specificities and cell cycle dynamics. PLoS ONE 2014, 9, e87647. [Google Scholar] [CrossRef]
- Knott, S.R.; Peace, J.M.; Ostrow, A.Z.; Gan, Y.; Rex, A.E.; Viggiani, C.J.; Tavare, S.; Aparicio, O.M. Forkhead transcription factors establish origin timing and long-range clustering in S. cerevisiae. Cell 2012, 148, 99–111. [Google Scholar] [CrossRef]
- Zhu, G.; Spellman, P.T.; Volpe, T.; Brown, P.O.; Botstein, D.; Davis, T.N.; Futcher, B. Two yeast forkhead genes regulate the cell cycle and pseudohyphal growth. Nature 2000, 406, 90–94. [Google Scholar] [CrossRef]
- Aparicio, O.M.; Weinstein, D.M.; Bell, S.P. Components and dynamics of DNA replication complexes in S. cerevisiae: Redistribution of MCM proteins and Cdc45p during S phase. Cell 1997, 91, 59–69. [Google Scholar] [CrossRef] [PubMed]
- Wyrick, J.J.; Aparicio, J.G.; Chen, T.; Barnett, J.D.; Jennings, E.G.; Young, R.A.; Bell, S.P.; Aparicio, O.M. Genome-wide distribution of ORC and MCM proteins in S. cerevisiae: High-resolution mapping of replication origins. Science 2001, 294, 2357–2360. [Google Scholar] [CrossRef] [PubMed]
- Nieduszynski, C.A.; Hiraga, S.-I.; Ak, P.; Benham, C.J.; Donaldson, A.D. OriDB: A DNA replication origin database. Nucleic Acids Res. 2006, 35, D40–D46. [Google Scholar] [CrossRef] [PubMed]
- Dukaj, L.; Rhind, N. The capacity of origins to load MCM establishes replication timing patterns. PLoS Genet. 2021, 17, e1009467. [Google Scholar] [CrossRef]
- Kueng, S.; Oppikofer, M.; Gasser, S.M. SIR Proteins and the Assembly of Silent Chromatin in Budding Yeast. Annu. Rev. Genet. 2013, 47, 275–306. [Google Scholar] [CrossRef]
- Huang, J.; Moazed, D. Association of the RENT complex with nontranscribed and coding regions of rDNA and a regional requirement for the replication fork block protein Fob1 in rDNA silencing. Genes Dev. 2003, 17, 2162–2176. [Google Scholar] [CrossRef]
- Clark, K.L.; Halay, E.D.; Lai, E.; Burley, S.K. Co-crystal structure of the HNF-3/fork head DNA-recognition motif resembles histone H5. Nature 1993, 364, 412–420. [Google Scholar] [CrossRef]
- Stroud, J.C.; Wu, Y.; Bates, D.L.; Han, A.; Nowick, K.; Paabo, S.; Tong, H.; Chen, L. Structure of the Forkhead Domain of FOXP2 Bound to DNA. Structure 2006, 14, 159–166. [Google Scholar] [CrossRef]
- Schnarr, M.; Oertel-Buchheit, P.; Kazmaier, M. DNA binding properties of the LexA repressor. Biochimie 1991, 73, 423–431. [Google Scholar] [CrossRef]
- Longtine, M.S.; McKenzie, A., 3rd; Demarini, D.J.; Shah, N.G.; Wach, A.; Brachat, A.; Philippsen, P.; Pringle, J.R. Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast 1998, 14, 953–961. [Google Scholar] [CrossRef]
- Li, J.; Coïc, E.; Lee, K.; Lee, C.-S.; Kim, J.-A.; Wu, Q.; Haber, J.E. Regulation of Budding Yeast Mating-Type Switching Donor Preference by the FHA Domain of Fkh1. PLoS Genet. 2012, 8, e1002630. [Google Scholar] [CrossRef]
- Ostrow, A.Z.; Kalhor, R.; Gan, Y.; Villwock, S.K.; Linke, C.; Barberis, M.; Chen, L.; Aparicio, O.M. Conserved forkhead dimerization motif controls DNA replication timing and spatial organization of chromosomes in S. cerevisiae. Proc. Natl. Acad. Sci. USA 2017, 114, E2411–E2419. [Google Scholar] [CrossRef] [PubMed]
- Gietz, R.D.; Schiestl, R.H. High-efficiency yeast transformation using the LiAc/SS carrier DNA/PEG method. Nat. Protoc. 2007, 2, 31–34. [Google Scholar] [CrossRef] [PubMed]
- Ostrow, A.Z.; Viggiani, C.J.; Aparicio, J.G.; Aparicio, O.M. ChIP-Seq to Analyze the Binding of Replication Proteins to Chromatin. Methods Mol. Biol. 2015, 1300, 155–168. [Google Scholar] [PubMed]
- McGuffee, S.R.; Smith, D.J.; Whitehouse, I. Quantitative, Genome-Wide Analysis of Eukaryotic Replication Initiation and Termination. Mol. Cell 2013, 50, 123–135. [Google Scholar] [CrossRef] [PubMed]
- Viggiani, C.J.; Aparicio, O.M. New vectors for simplified construction of BrdU-Incorporating strains of Saccharomyces cerevisiae. Yeast 2006, 23, 1045–1051. [Google Scholar] [CrossRef] [PubMed]
- He, Y.; Petrie, M.V.; Zhang, H.; Peace, J.M.; Aparicio, O.M. Rpd3 regulates single-copy origins independently of the rDNA array by opposing Fkh1-mediated origin stimulation. Proc. Natl. Acad. Sci. USA 2022, 119, e2212134119. [Google Scholar] [CrossRef]






Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Petrie, M.V.; He, Y.; Gan, Y.; Ostrow, A.Z.; Aparicio, O.M. Broadly Applicable Control Approaches Improve Accuracy of ChIP-Seq Data. Int. J. Mol. Sci. 2023, 24, 9271. https://doi.org/10.3390/ijms24119271
Petrie MV, He Y, Gan Y, Ostrow AZ, Aparicio OM. Broadly Applicable Control Approaches Improve Accuracy of ChIP-Seq Data. International Journal of Molecular Sciences. 2023; 24(11):9271. https://doi.org/10.3390/ijms24119271
Chicago/Turabian StylePetrie, Meghan V., Yiwei He, Yan Gan, Andrew Zachary Ostrow, and Oscar M. Aparicio. 2023. "Broadly Applicable Control Approaches Improve Accuracy of ChIP-Seq Data" International Journal of Molecular Sciences 24, no. 11: 9271. https://doi.org/10.3390/ijms24119271
APA StylePetrie, M. V., He, Y., Gan, Y., Ostrow, A. Z., & Aparicio, O. M. (2023). Broadly Applicable Control Approaches Improve Accuracy of ChIP-Seq Data. International Journal of Molecular Sciences, 24(11), 9271. https://doi.org/10.3390/ijms24119271





