Cracking the Floral Quartet Code: How Do Multimers of MIKCC-Type MADS-Domain Transcription Factors Recognize Their Target Genes?
Abstract
1. MADS-Domain Transcription Factors—A Primer
2. A Very Brief History of MADS-Box Genes
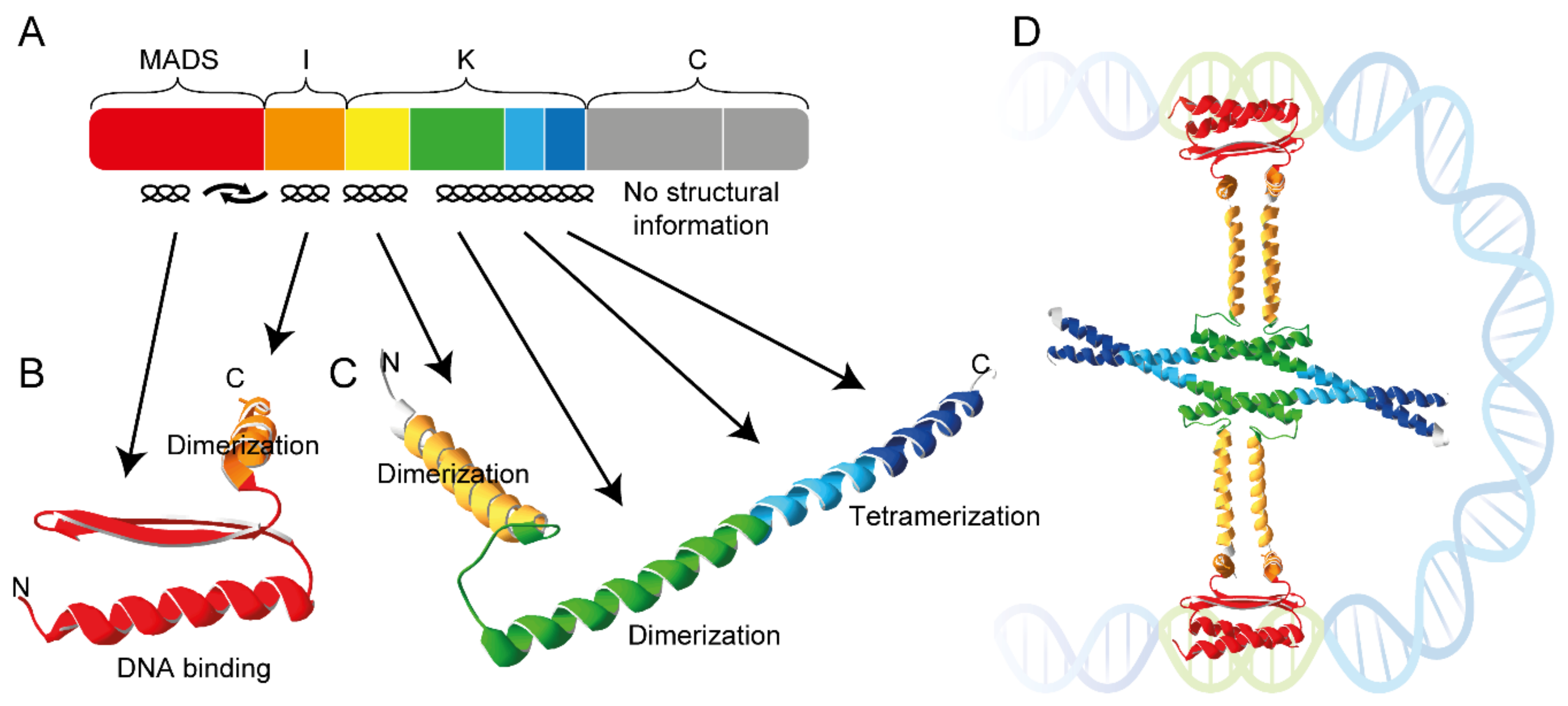
3. MIKC Blessing 2.0: A Prayer in C
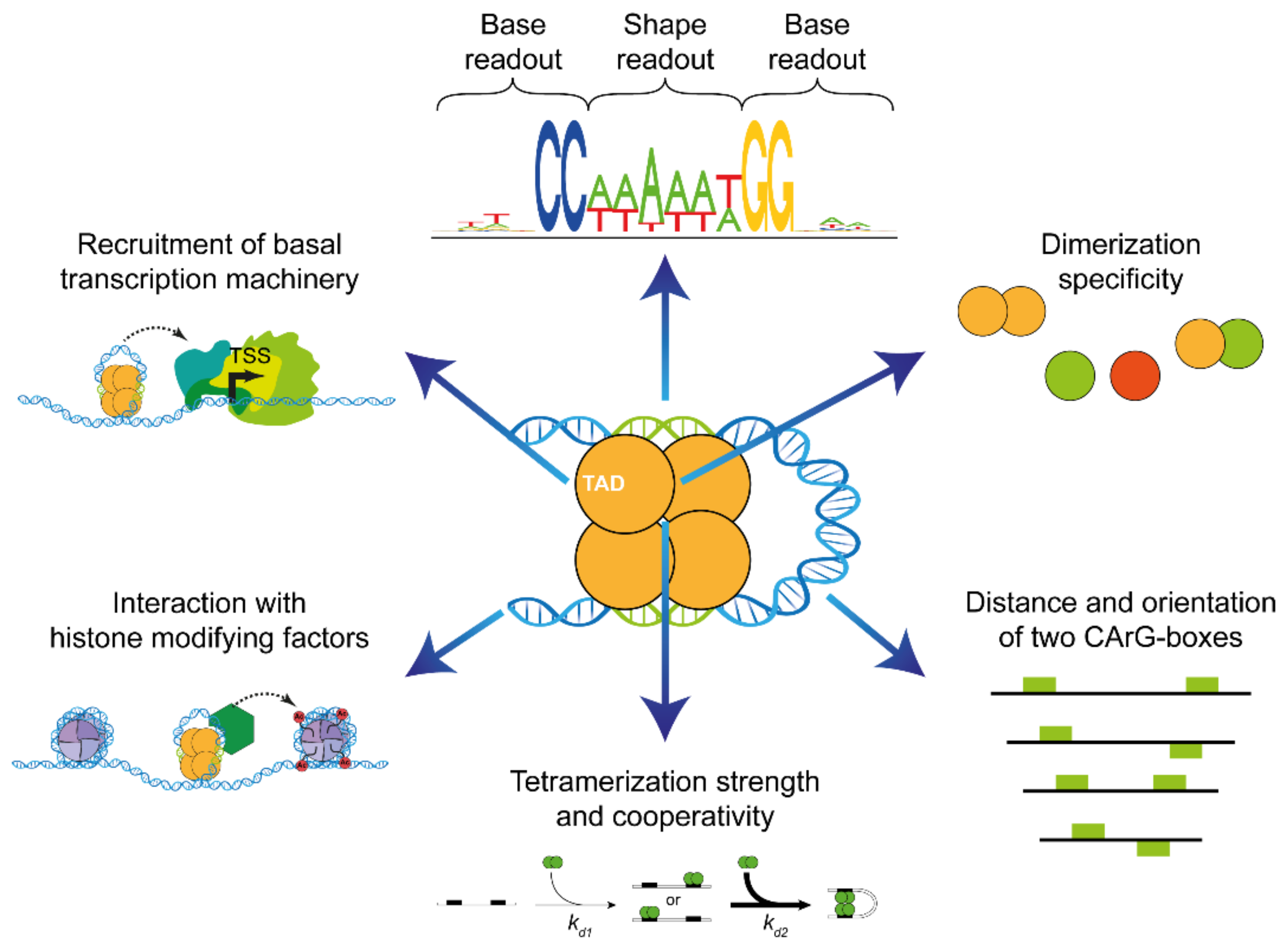
4. Recognition of DNA-Sequence Elements by MADS-Domain Proteins
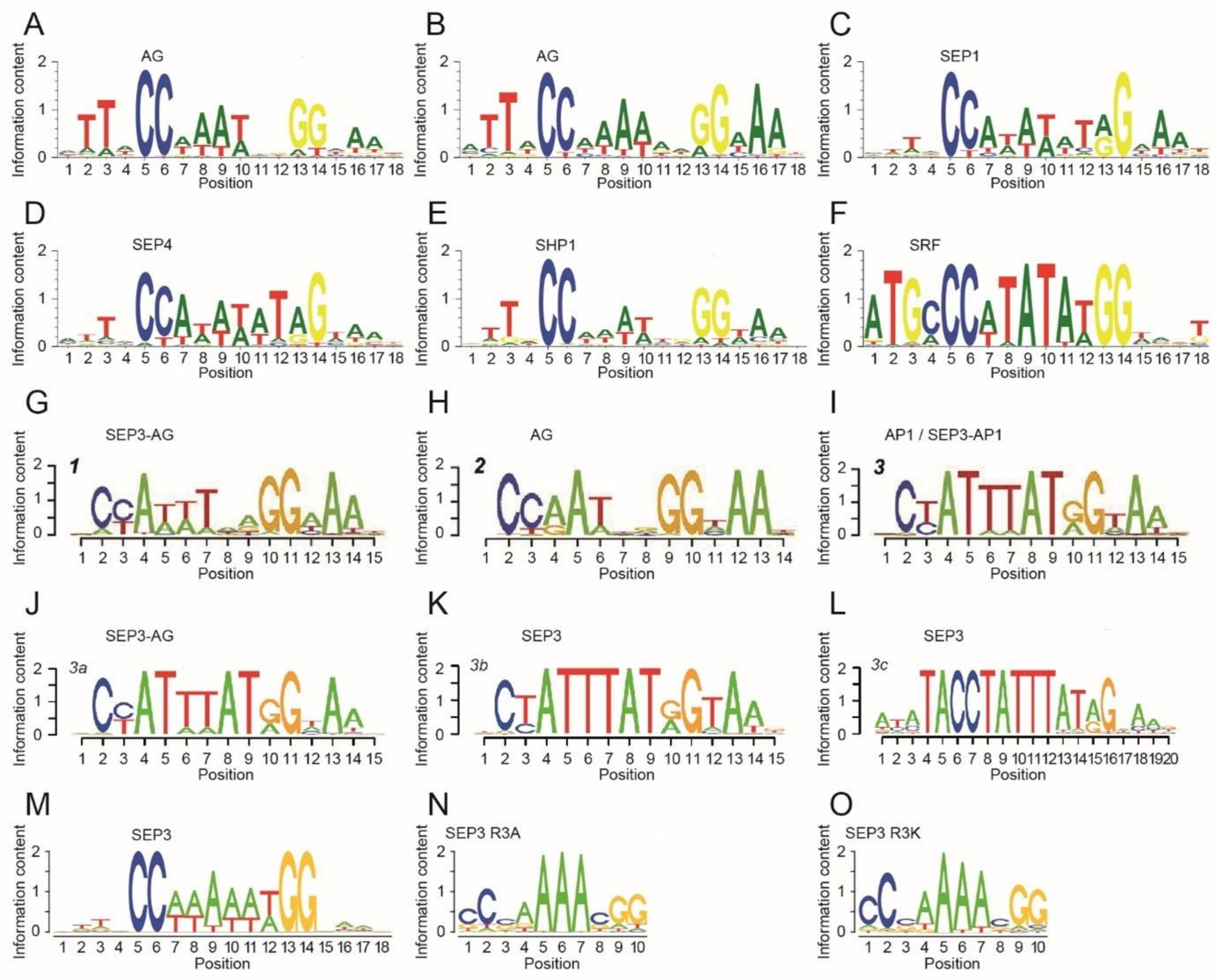
4.1. Base Readout
4.2. Shape Readout
4.3. Differences in DNA-Binding Specificity
4.4. Length of the DNA-Binding Motif
5. The Role of the Protein Structure
5.1. The General Contribution of MADS and I Domain to Target Gene Specificity
5.2. What Single Amino Acid Substitutions in the MADS and I Domain Tell Us
5.3. The Keratin-Like Domain—Mediator of Tetramerization
6. Origin and Evolution of FQCs
7. Why Quartets and FQCs?
8. Conclusions and Outlook
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Gramzow, L.; Ritz, M.S.; Theißen, G. On the origin of MADS-domain transcription factors. Trends Genet. 2010, 26, 149–153. [Google Scholar] [CrossRef]
- Messenguy, F.; Dubois, E. Role of MADS box proteins and their cofactors in combinatorial control of gene expression and cell development. Gene 2003, 316, 1–21. [Google Scholar] [CrossRef]
- Theißen, G.; Kim, J.T.; Saedler, H. Classification and phylogeny of the MADS-box multigene family suggest defined roles of MADS-box gene subfamilies in the morphological evolution of eukaryotes. J. Mol. Evol. 1996, 43, 484–516. [Google Scholar] [CrossRef] [PubMed]
- Becker, A.; Saedler, H.; Theißen, G. Distinct MADS-box gene expression patterns in the reproductive cones of the gymnosperm Gnetum gnemon. Dev. Genes Evol. 2003, 213, 567–572. [Google Scholar] [CrossRef]
- Schwarz-Sommer, Z.; Huijser, P.; Nacken, W.; Saedler, H.; Sommer, H. Genetic Control of Flower Development by Homeotic Genes in Antirrhinum majus. Science 1990, 250, 931–936. [Google Scholar] [CrossRef] [PubMed]
- Lai, X.; Vega-Léon, R.; Hugouvieux, V.; Blanc-Mathieu, R.; van der Wal, F.; Lucas, J.; Silva, C.S.; Jourdain, A.; Muino, J.M.; Nanao, M.H.; et al. The intervening domain is required for DNA-binding and functional identity of plant MADS transcription factors. Nat. Commun. 2021, 12, 1–13. [Google Scholar] [CrossRef]
- Puranik, S.; Acajjaoui, S.; Conn, S.; Costa, L.; Conn, V.; Vial, A.; Marcellin, R.; Melzer, R.; Brown, E.; Hart, D.; et al. Structural Basis for the Oligomerization of the MADS Domain Transcription Factor SEPALLATA3 in Arabidopsis. Plant Cell 2014, 26, 3603–3615. [Google Scholar] [CrossRef]
- Pellegrini, L.; Tan, S.; Richmond, T.J. Structure of serum response factor core bound to DNA. Nature 1995, 376, 490–498. [Google Scholar] [CrossRef]
- Santelli, E.; Richmond, T.J. Crystal structure of MEF2A core bound to DNA at 1.5 A resolution. J. Mol. Biol. 2000, 297, 437–449. [Google Scholar] [CrossRef]
- Huang, K.; Louis, J.M.; Donaldson, L.; Lim, F.L.; Sharrocks, A.D.; Clore, G.M. Solution structure of the MEF2A-DNA complex: Structural basis for the modulation of DNA bending and specificity by MADS-box transcription factors. EMBO J. 2000, 19, 2615–2628. [Google Scholar] [CrossRef] [PubMed]
- Theißen, G.; Melzer, R.; Rümpler, F. MADS-domain transcription factors and the floral quartet model of flower development: Linking plant development and evolution. Development 2016, 143, 3259–3271. [Google Scholar] [CrossRef]
- Goslin, K.; Finocchio, A.; Wellmer, F. Floral Homeotic Factors: A Question of Specificity. Plants 2023, 12, 1128. [Google Scholar] [CrossRef] [PubMed]
- Pollock, R.; Treisman, R. A sensitive method for the determination of protein-DNA binding specificities. Nucleic Acids Res. 1990, 18, 6197–6204. [Google Scholar] [CrossRef] [PubMed]
- De Folter, S.; Angenent, G.C. Trans meets cis in MADS science. Trends Plant Sci. 2006, 11, 224–231. [Google Scholar] [CrossRef]
- Wu, W.; Huang, X.; Cheng, J.; Li, Z.; de Folter, S.; Huang, Z.; Jiang, X.; Pang, H.; Tao, S. Conservation and Evolution in and among SRF- and MEF2-Type MADS Domains and Their Binding Sites. Mol. Biol. Evol. 2010, 28, 501–511. [Google Scholar] [CrossRef] [PubMed]
- Alvarez-Buylla, E.R.; Pelaz, S.; Liljegren, S.J.; Gold, S.E.; Burgeff, C.; Ditta, G.S.; de Pouplana, L.R.; Martínez-Castilla, L.; Yanofsky, M.F. An ancestral MADS-box gene duplication occurred before the divergence of plants and animals. Proc. Natl. Acad. Sci. USA 2000, 97, 5328–5333. [Google Scholar] [CrossRef] [PubMed]
- Gramzow, L.; Theißen, G. A hitchhiker’s guide to the MADS world of plants. Genome Biol. 2010, 11, 214. [Google Scholar] [CrossRef]
- De Bodt, S.; Raes, J.; Van de Peer, Y.; Theißen, G. And then there were many: MADS goes genomic. Trends Plant Sci. 2003, 8, 475–483. [Google Scholar] [CrossRef]
- Kaufmann, K.; Melzer, R.; Theißen, G. MIKC-type MADS-domain proteins: Structural modularity, protein interactions and network evolution in land plants. Gene 2005, 347, 183–198. [Google Scholar] [CrossRef]
- Nam, J.; Kim, J.; Lee, S.; An, G.; Ma, H.; Nei, M. Type I MADS-box genes have experienced faster birth-and-death evolution than type II MADS-box genes in angiosperms. Proc. Natl. Acad. Sci. USA 2004, 101, 1910–1915. [Google Scholar] [CrossRef]
- Gramzow, L.; Theißen, G. Phylogenomics of MADS-Box Genes in Plants—Two Opposing Life Styles in One Gene Family. Biology 2013, 2, 1150–1164. [Google Scholar] [CrossRef]
- Saedler, H.; Becker, A.; Winter, K.U.; Kirchner, C.; Theißen, G. MADS-box genes are involved in floral development and evolution. Acta Biochim. Pol. 2001, 48. [Google Scholar] [CrossRef]
- Becker, A.; Theißen, G. The major clades of MADS-box genes and their role in the development and evolution of flowering plants. Mol. Phylogenetics Evol. 2003, 29, 464–489. [Google Scholar] [CrossRef] [PubMed]
- Tanabe, Y.; Hasebe, M.; Sekimoto, H.; Nishiyama, T.; Kitani, M.; Henschel, K.; Münster, T.; Theißen, G.; Nozaki, H.; Ito, M. Characterization of MADS-box genes in charophycean green algae and its implication for the evolution of MADS-box genes. Proc. Natl. Acad. Sci. USA 2005, 102, 2436–2441. [Google Scholar] [CrossRef] [PubMed]
- Gramzow, L.; Tessari, C.; Rümpler, F.; Theißen, G. Deep evolution of MADS-box genes in Archaeplastida. bioRxiv 2023. [Google Scholar] [CrossRef]
- Feng, X.; Zheng, J.; Irisarri, I.; Yu, H.; Zheng, B.; Ali, Z.; de Vries, S.; Keller, J.; Fürst-Jansen, J.M.; Dadras, A.; et al. Chromosome-level genomes of multicellular algal sisters to land plants illuminate signaling network evolution. bioRxiv 2023. [Google Scholar] [CrossRef]
- Hugouvieux, V.; Silva, C.S.; Jourdain, A.; Stigliani, A.; Charras, Q.; Conn, V.; Conn, S.J.; Carles, C.C.; Parcy, F.; Zubieta, C. Tetramerization of MADS family transcription factors SEPALLATA3 and AGAMOUS is required for floral meristem determinacy in Arabidopsis. Nucleic Acids Res. 2018, 46, 4966–4977. [Google Scholar] [CrossRef]
- Smaczniak, C.; Immink, R.; Angenent, G.C.; Kaufmann, K. Developmental and evolutionary diversity of plant MADS-domain factors: Insights from recent studies. Development 2012, 139, 3081–3098. [Google Scholar] [CrossRef]
- Nishiyama, T.; Sakayama, H.; de Vries, J.; Buschmann, H.; Saint-Marcoux, D.; Ullrich, K.K.; Haas, F.B.; Vanderstraeten, L.; Becker, D.; Lang, D.; et al. The Chara Genome: Secondary Complexity and Implications for Plant Terrestrialization. Cell 2018, 174, 448–464.e24. [Google Scholar] [CrossRef]
- Henschel, K.; Kofuji, R.; Hasebe, M.; Saedler, H.; Münster, T.; Theißen, G. Two Ancient Classes of MIKC-type MADS-box Genes are Present in the Moss Physcomitrella patens. Mol. Biol. Evol. 2002, 19, 801–814. [Google Scholar] [CrossRef]
- Rümpler, F.; Tessari, C.; Gramzow, L.; Gafert, C.; Blohs, M.; Theißen, G. The origin of floral quartet formation—Ancient exon duplications shaped the evolution of MIKC-type MADS-domain transcription factor interactions. Mol. Biol. Evol. 2023, in press. [Google Scholar] [CrossRef] [PubMed]
- Theißen, G.; Gramzow, L. Structure and Evolution of Plant MADS-Domain Transcription Factors. In Plant Transcription Factors: Evolutionary, Structural and Functional Aspects; Gonzalez, D.H., Ed.; Elsevier: Philadelphia, PA, USA, 2016; pp. 127–138. [Google Scholar]
- Thangavel, G.; Nayar, S. A Survey of MIKC Type MADS-Box Genes in Non-seed Plants: Algae, Bryophytes, Lycophytes and Ferns. Front. Plant Sci. 2018, 9, 510. [Google Scholar] [CrossRef]
- Theißen, G.; Becker, A.; Di Rosa, A.; Kanno, A.; Kim, J.T.; Münster, T.; Winter, K.-U.; Saedler, H. A short history of MADS-box genes in plants. Plant Mol. Biol. 2000, 42, 115–149. [Google Scholar] [CrossRef]
- Sommer, H.; Beltrán, J.; Huijser, P.; Pape, H.; Lönnig, W.; Saedler, H.; Schwarz-Sommer, Z. Deficiens, a homeotic gene involved in the control of flower morphogenesis in Antirrhinum majus: The protein shows homology to transcription factors. EMBO J. 1990, 9, 605–613. [Google Scholar] [CrossRef] [PubMed]
- Yanofsky, M.F.; Ma, H.; Bowman, J.L.; Drews, G.N.; Feldmann, K.A.; Meyerowitz, E.M. The protein encoded by the Arabidopsis homeotic gene agamous resembles transcription factors. Nature 1990, 346, 35–39. [Google Scholar] [CrossRef]
- Gramzow, L.; Weilandt, L.; Theißen, G. MADS goes genomic in conifers: Towards determining the ancestral set of MADS-box genes in seed plants. Ann. Bot. 2014, 114, 1407–1429. [Google Scholar] [CrossRef] [PubMed]
- Theißen, G. Development of floral organ identity: Stories from the MADS house. Curr. Opin. Plant Biol. 2001, 4, 75–85. [Google Scholar] [CrossRef]
- Theißen, G.; Saedler, H. Plant biology. Floral quartets. Nature 2001, 409, 469–471. [Google Scholar] [CrossRef]
- Egea-Cortines, M.; Saedler, H.; Sommer, H. Ternary complex formation between the MADS-box proteins SQUAMOSA, DEFICIENS and GLOBOSA is involved in the control of floral architecture in Antirrhinum majus. EMBO J. 1999, 18, 5370–5379. [Google Scholar] [CrossRef]
- Oehler, S.; Eismann, E.R.; Krämer, H.; Müller-Hill, B. The three operators of the lac operon cooperate in repression. EMBO J. 1990, 9, 973–979. [Google Scholar] [CrossRef]
- Lewis, M. The lac repressor. C R Biol. 2005, 328, 521–548. [Google Scholar] [CrossRef] [PubMed]
- Hochschild, A.; Ptashne, M. Cooperative Binding of Lambda-Repressors to Sites Separated by Integral Turns of the DNA Helix. Cell 1986, 44, 681–687. [Google Scholar] [CrossRef]
- Shore, P.; Sharrocks, A.D. The MADS-Box Family of Transcription Factors. Eur. J. Biochem. 1995, 229, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Rohs, R.; Jin, X.; West, S.M.; Joshi, R.; Honig, B.; Mann, R.S. Origins of Specificity in Protein-DNA Recognition. Annu. Rev. Biochem. 2010, 79, 233–269. [Google Scholar] [CrossRef]
- Lai, X.; Daher, H.; Galien, A.; Hugouvieux, V.; Zubieta, C. Structural Basis for Plant MADS Transcription Factor Oligomerization. Comput. Struct. Biotechnol. J. 2019, 17, 946–953. [Google Scholar] [CrossRef]
- Tan, S.; Richmond, T.J. Crystal structure of the yeast MATα2/MCM1/DNA ternary complex. Nature 1998, 391, 660–666. [Google Scholar] [CrossRef] [PubMed]
- Han, A.; Pan, F.; Stroud, J.C.; Youn, H.-D.; Liu, J.O.; Chen, L. Sequence-specific recruitment of transcriptional co-repressor Cabin1 by myocyte enhancer factor-2. Nature 2003, 422, 730–734. [Google Scholar] [CrossRef]
- Parenicova, L.; de Folter, S.; Kieffer, M.; Horner, D.S.; Favalli, C.; Busscher, J.; Cook, H.E.; Ingram, R.M.; Kater, M.M.; Davies, B.; et al. Molecular and phylogenetic analyses of the complete MADS-box transcription factor family in Arabidopsis: New openings to the MADS world. Plant Cell 2003, 15, 1538–1551. [Google Scholar] [CrossRef]
- Käppel, S.; Melzer, R.; Rümpler, F.; Gafert, C.; Theißen, G. The floral homeotic protein SEPALLATA3 recognizes target DNA sequences by shape readout involving a conserved arginine residue in the MADS-domain. Plant J. 2018, 95, 341–357. [Google Scholar] [CrossRef]
- Machado, A.C.D.; Cooper, B.H.; Lei, X.; Di Felice, R.; Chen, L.; Rohs, R. Landscape of DNA binding signatures of myocyte enhancer factor-2B reveals a unique interplay of base and shape readout. Nucleic Acids Res. 2020, 48, 8529–8544. [Google Scholar] [CrossRef]
- Smaczniak, C.; Muiño, J.M.; Chen, D.; Angenent, G.C.; Kaufmann, K. Differences in DNA Binding Specificity of Floral Homeotic Protein Complexes Predict Organ-Specific Target Genes. Plant Cell 2017, 29, 1822–1835. [Google Scholar] [CrossRef] [PubMed]
- Muiño, J.M.; Smaczniak, C.; Angenent, G.C.; Kaufmann, K.; van Dijk, A.-J. Structural determinants of DNA recognition by plant MADS-domain transcription factors. Nucleic Acids Res. 2013, 42, 2138–2146. [Google Scholar] [CrossRef] [PubMed]
- Rohs, R.; West, S.M.; Sosinsky, A.; Liu, P.; Mann, R.S.; Honig, B. The role of DNA shape in protein–DNA recognition. Nature 2009, 461, 1248–1253. [Google Scholar] [CrossRef]
- Hud, N.V.; Plavec, J. A unified model for the origin of DNA sequence-directed curvature. Biopolymers 2003, 69, 144–158. [Google Scholar] [CrossRef]
- Koo, H.-S.; Wu, H.-M.; Crothers, D.M. DNA bending at adenine · thymine tracts. Nature 1986, 320, 501–506. [Google Scholar] [CrossRef] [PubMed]
- Käppel, S.; Eggeling, R.; Rümpler, F.; Groth, M.; Melzer, R.; Theißen, G. DNA-binding properties of the MADS-domain transcription factor SEPALLATA3 and mutant variants characterized by SELEX-seq. Plant Mol. Biol. 2021, 105, 543–557. [Google Scholar] [CrossRef]
- Aerts, N.; De Bruijn, S.; Van Mourik, H.; Angenent, G.C.; Van Dijk, A.D.J. Comparative analysis of binding patterns of MADS-domain proteins in Arabidopsis thaliana. BMC Plant Biol. 2018, 18, 131. [Google Scholar] [CrossRef] [PubMed]
- Mathelier, A.; Xin, B.; Chiu, T.-P.; Yang, L.; Rohs, R.; Wasserman, W.W. DNA Shape Features Improve Transcription Factor Binding Site Predictions In Vivo. Cell Syst. 2016, 3, 278–286.e4. [Google Scholar] [CrossRef] [PubMed]
- Tsukanov, A.V.; Mironova, V.V.; Levitsky, V.G. Motif models proposing independent and interdependent impacts of nucleotides are related to high and low affinity transcription factor binding sites in Arabidopsis. Front. Plant Sci. 2022, 13. [Google Scholar] [CrossRef]
- Lai, X.; Stigliani, A.; Lucas, J.; Hugouvieux, V.; Parcy, F.; Zubieta, C. Genome-wide binding of SEPALLATA3 and AGAMOUS complexes determined by sequential DNA-affinity purification sequencing. Nucleic Acids Res. 2020, 48, 9637–9648. [Google Scholar] [CrossRef]
- Yu, H.; Ito, T.; Wellmer, F.; Meyerowitz, E.M. Repression of AGAMOUS-LIKE 24 is a crucial step in promoting flower development. Nat. Genet. 2004, 36, 157–161. [Google Scholar] [CrossRef] [PubMed]
- Kaufmann, K.; Wellmer, F.; Muiño, J.M.; Ferrier, T.; Wuest, S.E.; Kumar, V.; Serrano-Mislata, A.; Madueño, F.; Krajewski, P.; Meyerowitz, E.M.; et al. Orchestration of Floral Initiation by APETALA1. Science 2010, 328, 85–89. [Google Scholar] [CrossRef] [PubMed]
- O’Maoileidigh, D.S.; E Wuest, S.; Rae, L.; Raganelli, A.; Ryan, P.T.; Kwasniewska, K.; Das, P.; Lohan, A.J.; Loftus, B.; Graciet, E.; et al. Control of reproductive floral organ identity specification in Arabidopsis by the C function regulator AGAMOUS. Plant Cell 2013, 25, 2482–2503. [Google Scholar] [CrossRef]
- Pajoro, A.; Madrigal, P.; Muiño, J.M.; Matus, J.T.; Jin, J.; Mecchia, M.A.; Debernardi, J.M.; Palatnik, J.F.; Balazadeh, S.; Arif, M.; et al. Dynamics of chromatin accessibility and gene regulation by MADS-domain transcription factors in flower development. Genome Biol. 2014, 15, R41. [Google Scholar] [CrossRef]
- Yan, W.; Chen, D.; Kaufmann, K. Molecular mechanisms of floral organ specification by MADS domain proteins. Curr. Opin. Plant Biol. 2016, 29, 154–162. [Google Scholar] [CrossRef]
- Wuest, S.E.; O’maoileidigh, D.S.; Rae, L.; Kwasniewska, K.; Raganelli, A.; Hanczaryk, K.; Lohan, A.J.; Loftus, B.; Graciet, E.; Wellmer, F. Molecular basis for the specification of floral organs by APETALA3 and PISTILLATA. Proc. Natl. Acad. Sci. USA 2012, 109, 13452–13457. [Google Scholar] [CrossRef] [PubMed]
- Huang, H.; Mizukami, Y.; Hu, Y.; Ma, H. Isolation and characterization of the binding sequences for the product of the Arabidopsis floral homeotic gene AGAMOUS. Nucleic Acids Res. 1993, 21, 4769–4776. [Google Scholar] [CrossRef]
- Huang, H.; Tudor, M.; Su, T.; Zhang, Y.; Hu, Y.; Ma, H. DNA binding properties of two Arabidopsis MADS domain proteins: Binding consensus and dimer formation. Plant Cell 1996, 8, 81–94. [Google Scholar]
- Huang, H.; Tudor, M.; Weiss, C.A.; Hu, Y.; Ma, H. The Arabidopsis MADS-box gene AGL3 is widely expressed and encodes a sequence-specific DNA-binding protein. Plant Mol. Biol. 1995, 28, 549–567. [Google Scholar] [CrossRef]
- Shiraishi, H.; Okada, K.; Shimura, Y. Nucleotide sequences recognized by the AGAMOUS MADS domain of Arabidopsis thaliana in vitro. Plant J. 1993, 4, 385–398. [Google Scholar] [CrossRef]
- Crooks, G.E.; Hon, G.; Chandonia, J.-M.; Brenner, S.E. WebLogo: A Sequence Logo Generator. Genome Res. 2004, 14, 1188–1190. [Google Scholar] [CrossRef]
- Schneider, T.; Stephens, R.M. Sequence logos: A new way to display consensus sequences. Nucleic Acids Res. 1990, 18, 6097–6100. [Google Scholar] [CrossRef] [PubMed]
- Mateos, J.L.; Madrigal, P.; Tsuda, K.; Rawat, V.; Richter, R.; Romera-Branchat, M.; Fornara, F.; Schneeberger, K.; Krajewski, P.; Coupland, G. Combinatorial activities of SHORT VEGETATIVE PHASE and FLOWERING LOCUS C define distinct modes of flowering regulation in Arabidopsis. Genome Biol. 2015, 16, 1–23. [Google Scholar] [CrossRef] [PubMed]
- Deng, W.; Ying, H.; Helliwell, C.A.; Taylor, J.M.; Peacock, W.J.; Dennis, E.S. FLOWERING LOCUS C (FLC) regulates development pathways throughout the life cycle of Arabidopsis. Proc. Natl. Acad. Sci. USA 2011, 108, 6680–6685. [Google Scholar] [CrossRef]
- Posé, D.; Verhage, L.; Ott, F.; Yant, L.; Mathieu, J.; Angenent, G.C.; Immink, R.G.H.; Schmid, M. Temperature-dependent regulation of flowering by antagonistic FLM variants. Nature 2013, 503, 414–417. [Google Scholar] [CrossRef] [PubMed]
- Kaufmann, K.; Muiño, J.M.; Jauregui, R.; Airoldi, C.A.; Smaczniak, C.; Krajewski, P.; Angenent, G.C. Target Genes of the MADS Transcription Factor SEPALLATA3: Integration of Developmental and Hormonal Pathways in the Arabidopsis Flower. PLoS Biol. 2009, 7, e1000090. [Google Scholar] [CrossRef]
- Immink, R.G.; Posé, D.; Ferrario, S.; Ott, F.; Kaufmann, K.; Valentim, F.L.; de Folter, S.; van der Wal, F.; van Dijk, A.D.; Schmid, M.; et al. Characterization of SOC1’s Central Role in Flowering by the Identification of Its Upstream and Downstream Regulators. Plant Physiol. 2012, 160, 433–449. [Google Scholar] [CrossRef]
- Gregis, V.; Andrés, F.; Sessa, A.; Guerra, R.F.; Simonini, S.; Mateos, J.L.; Torti, S.; Zambelli, F.; Prazzoli, G.M.; Bjerkan, K.N.; et al. Identification of pathways directly regulated by SHORT VEGETATIVE PHASE during vegetative and reproductive development in Arabidopsis. Genome Biol. 2013, 14, 1–26. [Google Scholar] [CrossRef]
- Melzer, R.; Kaufmann, K.; Theißen, G. Missing Links: DNA-Binding and Target Gene Specificity of Floral Homeotic Proteins. Adv. Bot. Res. 2006, 44, 209–236. [Google Scholar] [CrossRef]
- Riechmann, J.L.; Meyerowitz, E.M. Determination of floral organ identity by Arabidopsis MADS domain homeotic proteins AP1, AP3, PI, and AG is independent of their DNA-binding specificity. Mol. Biol. Cell 1997, 8, 1243–1259. [Google Scholar] [CrossRef]
- Krizek, B.A.; Meyerowitz, E.M. Mapping the protein regions responsible for the functional specificities of the Arabidopsis MADS domain organ-identity proteins. Proc. Natl. Acad. Sci. USA 1996, 93, 4063–4070. [Google Scholar] [CrossRef] [PubMed]
- Pon, J.R.; Wong, J.; Saberi, S.; Alder, O.; Moksa, M.; Cheng, S.W.G.; Morin, G.B.; Hoodless, P.A.; Hirst, M.; Marra, M.A. MEF2B mutations in non-Hodgkin lymphoma dysregulate cell migration by decreasing MEF2B target gene activation. Nat. Commun. 2015, 6, 7953. [Google Scholar] [CrossRef]
- Lei, X.; Kou, Y.; Fu, Y.; Rajashekar, N.; Shi, H.; Wu, F.; Xu, J.; Luo, Y.; Chen, L. The Cancer Mutation D83V Induces an alpha-Helix to beta-Strand Conformation Switch in MEF2B. J. Mol. Biol. 2018, 430, 1157–1172. [Google Scholar] [CrossRef] [PubMed]
- Ma, H.; Yanofsky, M.F.; Meyerowitz, E.M. AGL1-AGL6, an Arabidopsis gene family with similarity to floral homeotic and transcription factor genes. Genes Dev. 1991, 5, 484–495. [Google Scholar] [CrossRef]
- Yang, Y.; Fanning, L.; Jack, T. The K domain mediates heterodimerization of the Arabidopsis floral organ identity proteins, APETALA3 and PISTILLATA. Plant J. 2003, 33, 47–59. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.; Jack, T. Defining subdomains of the K domain important for protein-protein interactions of plant MADS proteins. Plant Mol. Biol. 2004, 55, 45–59. [Google Scholar] [CrossRef]
- Rümpler, F.; Theißen, G.; Melzer, R. A conserved leucine zipper-like motif accounts for strong tetramerization capabilities of SEPALLATA-like MADS-domain transcription factors. J. Exp. Bot. 2018, 69, 1943–1954. [Google Scholar] [CrossRef]
- Mason, J.; Arndt, K.M. Coiled Coil Domains: Stability, Specificity, and Biological Implications. Chembiochem 2004, 5, 170–176. [Google Scholar] [CrossRef]
- Mason, J.M.; Hagemann, U.B.; Arndt, K.M. Role of Hydrophobic and Electrostatic Interactions in Coiled Coil Stability and Specificity. Biochemistry 2009, 48, 10380–10388. [Google Scholar] [CrossRef]
- Alber, T. Structure of the leucine zipper. Curr. Opin. Genet. Dev. 1992, 2, 205–210. [Google Scholar] [CrossRef]
- Hu, J.C.; Sauer, R.T. The Basic-Region Leucine-Zipper Family of DNA Binding Proteins. In Nucleic Acids and Molecular Biology; Springer: Berlin/Heidelberg, Germany, 1992; pp. 82–101. [Google Scholar] [CrossRef]
- Lupas, A.N.; Bassler, J. Coiled Coils—A Model System for the 21st Century. Trends Biochem. Sci. 2016, 42, 130–140. [Google Scholar] [CrossRef]
- Hugouvieux, V.; Zubieta, C. MADS transcription factors cooperate: Complexities of complex formation. J. Exp. Bot. 2018, 69, 1821–1823. [Google Scholar] [CrossRef]
- Azuma, Y.; Kükenshöner, T.; Ma, G.; Yasunaga, J.-I.; Imanishi, M.; Tanaka, G.; Nakase, I.; Maruno, T.; Kobayashi, Y.; Arndt, K.M.; et al. Controlling leucine-zipper partner recognition in cells through modification of a–g interactions. Chem. Commun. 2014, 50, 6364–6367. [Google Scholar] [CrossRef]
- Kükenshöner, T.; Wohlwend, D.; Niemöller, C.; Dondapati, P.; Speck, J.; Adeniran, A.V.; Nieth, A.; Gerhardt, S.; Einsle, O.; Müller, K.M.; et al. Improving coiled coil stability while maintaining specificity by a bacterial hitchhiker selection system. J. Struct. Biol. 2014, 186, 335–348. [Google Scholar] [CrossRef]
- Yang, Y.; Xiang, H.; Jack, T. pistillata-5, an Arabidopsis B class mutant with strong defects in petal but not in stamen development. Plant J. 2003, 33, 177–188. [Google Scholar] [CrossRef]
- Rümpler, F. Evolution of the Interaction of Floral Homeotic Proteins. Ph.D. Thesis, Friedrich Schiller University, Jena, Germany, 2017. [Google Scholar]
- De Folter, S.; Immink, R.G.; Kieffer, M.; Parenicova, L.; Henz, S.R.; Weigel, D.; Busscher, M.; Kooiker, M.; Colombo, L.; Kater, M.M.; et al. Comprehensive interaction map of the Arabidopsis MADS Box transcription factors. Plant Cell 2005, 17, 1424–1433. [Google Scholar] [CrossRef]
- Immink, R.G.H.; Tonaco, I.A.N.; de Folter, S.; Shchennikova, A.; Van Dijk, A.D.J.; Busscher-Lange, J.; Borst, J.W.; Angenent, G.C. SEPALLATA3: The ’glue’ for MADS box transcription factor complex formation. Genome Biol. 2009, 10, R24. [Google Scholar] [CrossRef]
- Alhindi, T.; Zhang, Z.; Ruelens, P.; Coenen, H.; Degroote, H.; Iraci, N.; Geuten, K. Protein interaction evolution from promiscuity to specificity with reduced flexibility in an increasingly complex network. Sci. Rep. 2017, 7, 44948. [Google Scholar] [CrossRef]
- Causier, B.; Cook, H.; Davies, B. An Antirrhinum ternary complex factor specifically interacts with C-function and SEPALLATA-like MADS-box factors. Plant Mol. Biol. 2003, 52, 1051–1062. [Google Scholar] [CrossRef]
- Immink, R.G.H.; Ferrario, S.; Busscher-Lange, J.; Kooiker, M.; Busscher, M.; Angenent, G.C. Analysis of the petunia MADS-box transcription factor family. Mol. Genet. Genom. 2003, 268, 598–606. [Google Scholar] [CrossRef] [PubMed]
- Leseberg, C.H.; Eissler, C.L.; Wang, X.; Johns, M.A.; Duvall, M.R.; Mao, L. Interaction study of MADS-domain proteins in tomato. J. Exp. Bot. 2008, 59, 2253–2265. [Google Scholar] [CrossRef]
- Ruokolainen, S.; Ng, Y.P.; Albert, V.A.; Elomaa, P.; Teeri, T.H. Large scale interaction analysis predicts that the Gerbera hybrida floral E function is provided both by general and specialized proteins. BMC Plant Biol. 2010, 10, 129. [Google Scholar] [CrossRef] [PubMed]
- Liu, C.; Zhang, J.; Zhang, N.; Shan, H.; Su, K.; Meng, Z.; Kong, H.; Chen, Z. Interactions among Proteins of Floral MADS-Box Genes in Basal Eudicots: Implications for Evolution of the Regulatory Network for Flower Development. Mol. Biol. Evol. 2010, 27, 1598–1611. [Google Scholar] [CrossRef] [PubMed]
- Cooper, B.; Clarke, J.D.; Budworth, P.; Kreps, J.; Hutchison, D.; Park, S.; Guimil, S.; Dunn, M.; Luginbühl, P.; Ellero, C.; et al. A network of rice genes associated with stress response and seed development. Proc. Natl. Acad. Sci. USA 2003, 100, 4945–4950. [Google Scholar] [CrossRef]
- Whipple, C.J.; Ciceri, P.; Padilla, C.M.; Ambrose, B.A.; Bandong, S.L.; Schmidt, R.J. Conservation of B-class floral homeotic gene function between maize and Arabidopsis. Development 2004, 131, 6083–6091. [Google Scholar] [CrossRef]
- Abraham-Juárez, M.J.; Schrager-Lavelle, A.; Man, J.; Whipple, C.; Handakumbura, P.; Babbitt, C.; Bartlett, M. Evolutionary Variation in MADS Box Dimerization Affects Floral Development and Protein Abundance in Maize. Plant Cell 2020, 32, 3408–3424. [Google Scholar] [CrossRef]
- Li, L.; Yu, X.-X.; Guo, C.-C.; Duan, X.-S.; Shan, H.-Y.; Zhang, R.; Xu, G.-X.; Kong, H.-Z. Interactions among proteins of floral MADS-box genes in Nuphar pumila (Nymphaeaceae) and the most recent common ancestor of extant angiosperms help understand the underlying mechanisms of the origin of the flower. J. Syst. Evol. 2015, 53, 285–296. [Google Scholar] [CrossRef]
- Melzer, R.; Härter, A.; Rümpler, F.; Kim, S.; Soltis, P.S.; Soltis, D.E.; Theißen, G. DEF- and GLO-like proteins may have lost most of their interaction partners during angiosperm evolution. Ann. Bot. 2014, 114, 1431–1443. [Google Scholar] [CrossRef]
- Peréz-Mesa, P.; Suárez-Baron, H.; Ambrose, B.A.; González, F.; Pabón-Mora, N. Floral MADS-box protein interactions in the early diverging angiosperm Aristolochia fimbriata Cham. (Aristolochiaceae: Piperales). Evol. Dev. 2019, 21, 96–110. [Google Scholar] [CrossRef]
- Wang, Y.-Q.; Melzer, R.; Theißen, G. Molecular interactions of orthologues of floral homeotic proteins from the gymnosperm Gnetum gnemon provide a clue to the evolutionary origin of ‘floral quartets’. Plant J. 2010, 64, 177–190. [Google Scholar] [CrossRef]
- Winter, K.-U.; Weiser, C.; Kaufmann, K.; Bohne, A.; Kirchner, C.; Kanno, A.; Saedler, H.; Theißen, G. Evolution of Class B Floral Homeotic Proteins: Obligate Heterodimerization Originated from Homodimerization. Mol. Biol. Evol. 2002, 19, 587–596. [Google Scholar] [CrossRef]
- Ruelens, P.; Zhang, Z.; van Mourik, H.; Maere, S.; Kaufmann, K.; Geuten, K. The Origin of Floral Organ Identity Quartets. Plant Cell 2017, 29, 229–242. [Google Scholar] [CrossRef] [PubMed]
- Melzer, R.; Theißen, G. Reconstitution of ‘floral quartets’ in vitro involving class B and class E floral homeotic proteins. Nucleic Acids Res. 2009, 37, 2723–2736. [Google Scholar] [CrossRef]
- Smaczniak, C.; Immink, R.G.H.; Muiño, J.M.; Blanvillain, R.; Busscher, M.; Busscher-Lange, J.; Dinh, Q.D.; Liu, S.; Westphal, A.H.; Boeren, S.; et al. Characterization of MADS-domain transcription factor complexes in Arabidopsis flower development. Proc. Natl. Acad. Sci. USA 2012, 109, 1560–1565. [Google Scholar] [CrossRef] [PubMed]
- Espinosa-Soto, C.; Immink, R.G.; Angenent, G.C.; Alvarez-Buylla, E.R.; de Folter, S. Tetramer formation in Arabidopsis MADS domain proteins: Analysis of a protein-protein interaction network. BMC Syst. Biol. 2014, 8, 9. [Google Scholar] [CrossRef]
- Jetha, K.; Theißen, G.; Melzer, R. Arabidopsis SEPALLATA proteins differ in cooperative DNA-binding during the formation of floral quartet-like complexes. Nucleic Acids Res. 2014, 42, 10927–10942. [Google Scholar] [CrossRef]
- Kaufmann, K.; Pajoro, A.; Angenent, G.C. Regulation of transcription in plants: Mechanisms controlling developmental switches. Nat. Rev. Genet. 2010, 11, 830–842. [Google Scholar] [CrossRef]
- Theißen, G.; Melzer, R. Molecular mechanisms underlying origin and diversification of the angiosperm flower. Ann. Bot. 2007, 100, 603–619. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Käppel, S.; Rümpler, F.; Theißen, G. Cracking the Floral Quartet Code: How Do Multimers of MIKCC-Type MADS-Domain Transcription Factors Recognize Their Target Genes? Int. J. Mol. Sci. 2023, 24, 8253. https://doi.org/10.3390/ijms24098253
Käppel S, Rümpler F, Theißen G. Cracking the Floral Quartet Code: How Do Multimers of MIKCC-Type MADS-Domain Transcription Factors Recognize Their Target Genes? International Journal of Molecular Sciences. 2023; 24(9):8253. https://doi.org/10.3390/ijms24098253
Chicago/Turabian StyleKäppel, Sandra, Florian Rümpler, and Günter Theißen. 2023. "Cracking the Floral Quartet Code: How Do Multimers of MIKCC-Type MADS-Domain Transcription Factors Recognize Their Target Genes?" International Journal of Molecular Sciences 24, no. 9: 8253. https://doi.org/10.3390/ijms24098253
APA StyleKäppel, S., Rümpler, F., & Theißen, G. (2023). Cracking the Floral Quartet Code: How Do Multimers of MIKCC-Type MADS-Domain Transcription Factors Recognize Their Target Genes? International Journal of Molecular Sciences, 24(9), 8253. https://doi.org/10.3390/ijms24098253





