Roles of Noncoding RNAs in Regulation of Mitochondrial Electron Transport Chain and Oxidative Phosphorylation
Abstract
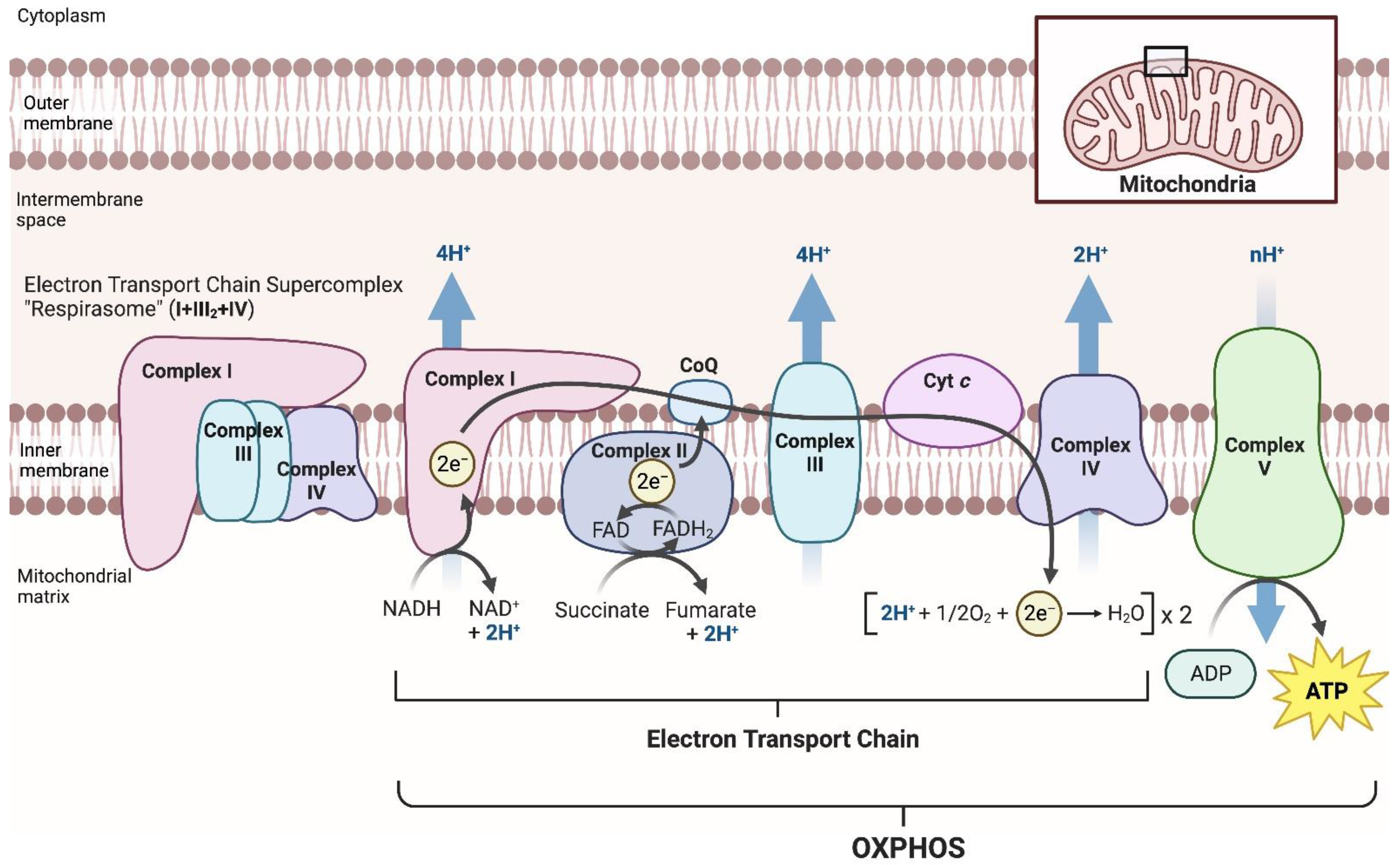
:1. Introduction
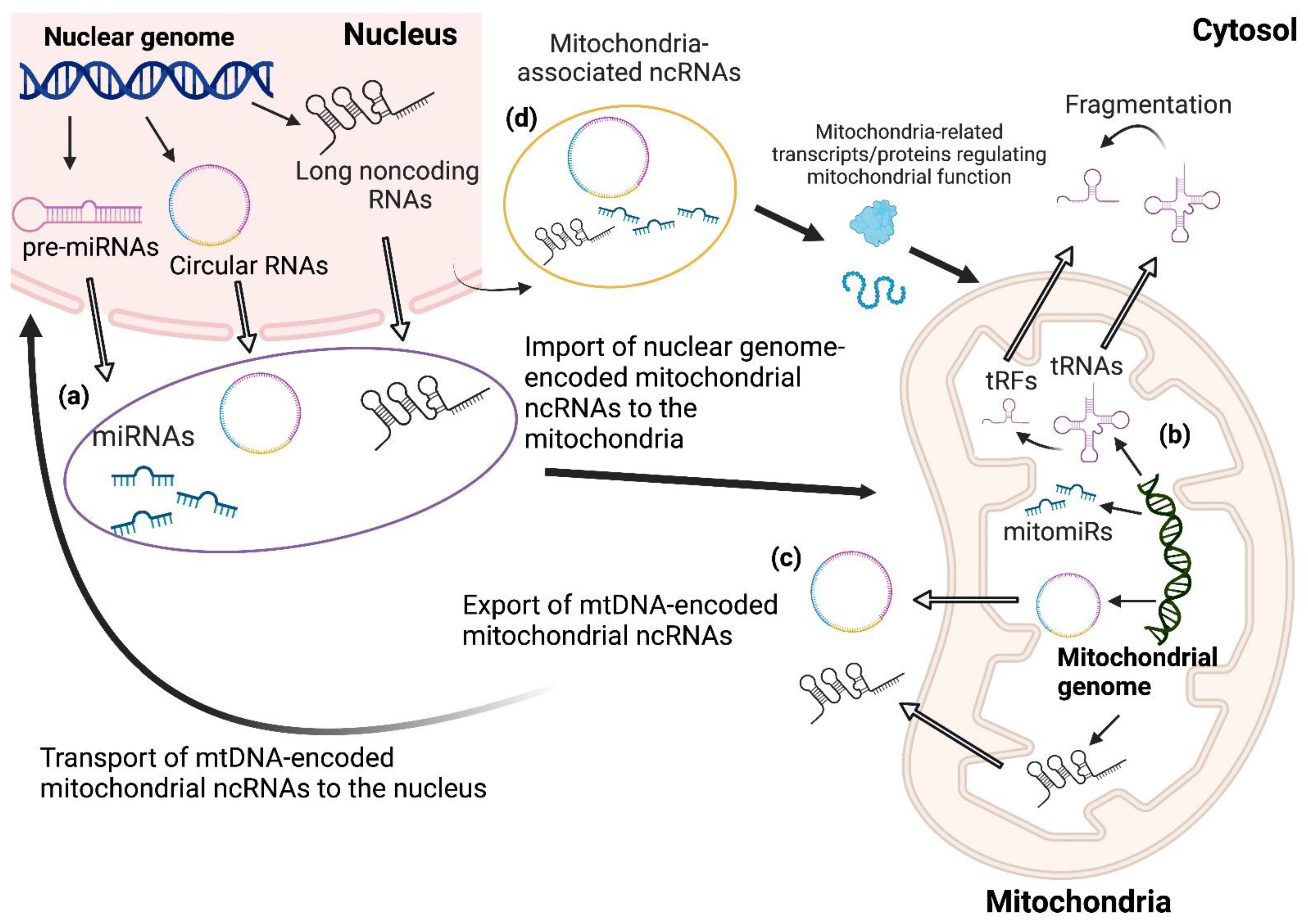
2. Species and Classification of ncRNAs Affecting Mitochondria–Nucleus Crosstalk
3. Roles of microRNAs in the Regulation of Mitochondrial ETC Complexes and OXPHOS
4. Roles of tRNA-Derived Small Fragments in the Regulation of Mitochondrial ETC Complexes
5. Roles of Long Noncoding RNAs in the Regulation of Mitochondrial ETC Complexes
6. Roles of Circular RNAs in the Regulation of Mitochondrial ETC Complexes
7. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Kobayashi, A.; Azuma, K.; Ikeda, K.; Inoue, S. Mechanisms Underlying the Regulation of Mitochondrial Respiratory Chain Complexes by Nuclear Steroid Receptors. Int. J. Mol. Sci. 2020, 21, 6683. [Google Scholar] [CrossRef]
- Schägger, H.; Pfeiffer, K. Supercomplexes in the respiratory chains of yeast and mammalian mitochondria. EMBO J. 2000, 19, 1777–1783. [Google Scholar] [CrossRef]
- Ikeda, K.; Shiba, S.; Horie-Inoue, K.; Shimokata, K.; Inoue, S. A stabilizing factor for mitochondrial respiratory supercomplex assembly regulates energy metabolism in muscle. Nat. Commun. 2013, 4, 2147. [Google Scholar] [CrossRef]
- Lapuente-Brun, E.; Moreno-Loshuertos, R.; Aciń-Pérez, R.; Latorre-Pellicer, A.; Colaś, C.; Balsa, E.; Perales-Clemente, E.; Quirós, P.M.; Calvo, E.; Rodríguez-Hernández, M.A.; et al. Supercomplex assembly determines electron flux in the mitochondrial electron transport chain. Science 2013, 340, 1567–1570. [Google Scholar] [CrossRef]
- Genova, M.L.; Lenaz, G. Functional role of mitochondrial respiratory supercomplexes. Biochim. Biophys. Acta-Bioenerg. 2014, 1837, 427–443. [Google Scholar] [CrossRef] [PubMed]
- Kobayashi, A.; Azuma, K.; Takeiwa, T.; Kitami, T.; Horie, K.; Ikeda, K.; Inoue, S. A FRET-based respirasome assembly screen identifies spleen tyrosine kinase as a target to improve muscle mitochondrial respiration and exercise performance in mice. Nat. Commun. 2023, 14, 312. [Google Scholar] [CrossRef]
- Maranzana, E.; Barbero, G.; Falasca, A.I.; Lenaz, G.; Genova, M.L. Mitochondrial respiratory supercomplex association limits production of reactive oxygen species from complex I. Antioxid. Redox Signal 2013, 19, 1469–1480. [Google Scholar] [CrossRef]
- Lopez-Fabuel, I.; Le Douce, J.; Logan, A.; James, A.M.; Bonvento, G.; Murphy, M.P.; Almeida, A.; Bolaños, J.P. Complex I assembly into supercomplexes determines differential mitochondrial ROS production in neurons and astrocytes. Proc. Natl. Acad. Sci. USA 2016, 113, 13063–13068. [Google Scholar] [CrossRef] [PubMed]
- Ikeda, K.; Horie-Inoue, K.; Suzuki, T.; Hobo, R.; Nakasato, N.; Takeda, S.; Inoue, S. Mitochondrial supercomplex assembly promotes breast and endometrial tumorigenesis by metabolic alterations and enhanced hypoxia tolerance. Nat. Commun. 2019, 10, 4108. [Google Scholar] [CrossRef] [PubMed]
- Schägger, H.; de Coo, R.; Bauer, M.F.; Hofmann, S.; Godinot, C.; Brandt, U. Significance of respirasomes for the assembly/stability of human respiratory chain complex I. J. Biol. Chem. 2004, 279, 36349–36353. [Google Scholar] [CrossRef]
- Boczonadi, V.; Ricci, G.; Horvath, R. Mitochondrial DNA transcription and translation: Clinical syndromes. Essays Biochem. 2018, 62, 321–340. [Google Scholar]
- Rötig, A. Human diseases with impaired mitochondrial protein synthesis. Biochim. Biophys. Acta 2011, 1807, 1198–1205. [Google Scholar] [CrossRef]
- DiMauro, S.; Schon, E.A.; Carelli, V.; Hirano, M. The clinical maze of mitochondrial neurology. Nat. Rev. Neurol. 2013, 9, 429–444. [Google Scholar] [CrossRef] [PubMed]
- Fernandez-Vizarra, E.; Zeviani, M. Mitochondrial disorders of the OXPHOS system. FEBS Lett. 2021, 595, 1062–1106. [Google Scholar] [CrossRef] [PubMed]
- Bhatti, J.S.; Bhatti, G.K.; Reddy, P.H. Mitochondrial dysfunction and oxidative stress in metabolic disorders—A step towards mitochondria based therapeutic strategies. Biochim. Biophys. Acta Mol. Basis Dis. 2017, 1863, 1066–1077. [Google Scholar] [CrossRef] [PubMed]
- Reinecke, F.; Smeitink, J.A.M.; van der Westhuizen, F.H. OXPHOS gene expression and control in mitochondrial disorders. Biochim. Biophys. Acta 2009, 1792, 1113–1121. [Google Scholar] [CrossRef]
- Das, S.; Ferlito, M.; Kent, O.A.; Fox-Talbot, K.; Wang, R.; Liu, D.; Raghavachari, N.; Yang, Y.; Wheelan, S.J.; Murphy, E.; et al. Nuclear miRNA regulates the mitochondrial genome in the heart. Circ. Res. 2012, 110, 1596–1603. [Google Scholar] [CrossRef]
- Zhao, Y.; Zhou, L.; Li, H.; Sun, T.; Wen, X.; Li, X.; Meng, Y.; Li, Y.; Liu, M.; Liu, S.; et al. Nuclear-Encoded lncRNA MALAT1 Epigenetically Controls Metabolic Reprogramming in HCC Cells through the Mitophagy Pathway. Mol. Ther. Nucleic Acids 2020, 23, 264–276. [Google Scholar] [CrossRef] [PubMed]
- Hyttinen, J.M.T.; Blasiak, J.; Kaarniranta, K. Non-Coding RNAs Regulating Mitochondrial Functions and the Oxidative Stress Response as Putative Targets against Age-Related Macular Degeneration (AMD). Int. J. Mol. Sci. 2023, 24, 2636. [Google Scholar] [CrossRef]
- Kamada, S.; Takeiwa, T.; Ikeda, K.; Horie-Inoue, K.; Inoue, S. Long Noncoding RNAs Involved in Metabolic Alterations in Breast and Prostate Cancers. Front. Oncol. 2020, 10, 593200. [Google Scholar] [CrossRef] [PubMed]
- Treiber, T.; Treiber, N.; Meister, G. Regulation of microRNA biogenesis and its crosstalk with other cellular pathways. Nat. Rev. Mol. Cell Biol. 2019, 20, 5–20. [Google Scholar] [CrossRef] [PubMed]
- Carthew, R.W.; Sontheimer, E.J. Origins and mechanisms of miRNAs and siRNAs. Cell 2009, 136, 642–655. [Google Scholar] [CrossRef] [PubMed]
- Chekulaeva, M.; Filipowicz, W. Mechanisms of miRNA-mediated post-transcriptional regulation in animal cells. Curr. Opin. Cell Biol. 2009, 21, 452–460. [Google Scholar] [CrossRef]
- Czech, B.; Hannon, G.J. Small RNA sorting: Matchmaking for Argonautes. Nat. Rev. Genet. 2011, 12, 19–31. [Google Scholar] [CrossRef]
- Iwakawa, H.O.; Tomari, Y. The Functions of MicroRNAs: MRNA Decay and Translational Repression. Trends Cell Biol. 2015, 25, 651–665. [Google Scholar] [CrossRef] [PubMed]
- Liu, H.; Lei, C.; He, Q.; Pan, Z.; Xiao, D.; Tao, Y. Nuclear functions of mammalian MicroRNAs in gene regulation, immunity and cancer. Mol. Cancer 2018, 17, 64. [Google Scholar] [CrossRef]
- Ni, W.J.; Leng, X.M. miRNA-Dependent Activation of mRNA Translation. MicroRNA 2016, 5, 83–86. [Google Scholar] [CrossRef]
- Barrey, E.; Saint-Auret, G.; Bonnamy, B.; Damas, D.; Boyer, O.; Gidrol, X. Pre-microRNA and Mature microRNA in Human Mitochondria. PLoS ONE 2011, 6, e20220. [Google Scholar] [CrossRef]
- Bandiera, S.; Matégot, R.; Girard, M.; Demongeot, J.; Henrion- Caude, A. MitomiRs delineating the intracellular localization of microRNAs at mitochondria. Free Radic. Biol. Med. 2013, 64, 12–19. [Google Scholar] [CrossRef]
- Thompson, D.M.; Lu, C.; Green, P.J.; Parker, R. tRNA cleavage is a conserved response to oxidative stress in eukaryotes. RNA 2008, 14, 2095–2103. [Google Scholar] [CrossRef]
- Thompson, D.M.; Parker, R. Stressing out over tRNA cleavage. Cell 2009, 138, 215–219. [Google Scholar] [CrossRef]
- Haussecker, D.; Huang, Y.; Lau, A.; Parameswaran, P.; Fire, A.Z.; Kay, M.A. Human tRNA-derived small RNAs in the global regulation of RNA silencing. RNA 2010, 16, 673–695. [Google Scholar] [CrossRef]
- Pederson, T. Regulatory RNAs derived from transfer RNA? RNA 2010, 16, 1865–1869. [Google Scholar] [CrossRef]
- Garcia-Silva, M.R.; Cabrera-Cabrera, F.; Güida, M.C.; Cayota, A. Hints of tRNA-Derived Small RNAs Role in RNA Silencing Mechanisms. Genes 2012, 3, 603–614. [Google Scholar] [CrossRef] [PubMed]
- Saikia, M.; Krokowski, D.; Guan, B.J.; Ivanov, P.; Parisien, M.; Hu, G.F.; Anderson, P.; Pan, T.; Hatzoglou, M. Genome-wide Identification and Quantitative Analysis of Cleaved tRNA Fragments Induced by Cellular Stress. J. Biol. Chem. 2012, 287, 42708–42725. [Google Scholar] [CrossRef]
- Gebetsberger, J.; Polacek, N. Slicing tRNAs to boost functional ncRNA diversity. RNA Biol. 2013, 10, 1798–1806. [Google Scholar] [CrossRef] [PubMed]
- Wang, Q.; Lee, I.; Ren, J.; Ajay, S.S.; Lee, Y.S.; Bao, X. Identification and functional characterization of tRNA-derived RNA fragments (tRFs) in respiratory syncytial virus infection. Mol. Ther. 2013, 21, 368–379. [Google Scholar] [CrossRef] [PubMed]
- Ro, S.; Ma, H.Y.; Park, C.; Ortogero, N.; Song, R.; Hennig, G.W.; Zheng, H.; Lin, Y.M.; Moro, L.; Hsieh, J.T.; et al. The mitochondrial genome encodes abundant small noncoding RNAs. Cell Res. 2013, 23, 759–774. [Google Scholar] [CrossRef]
- Telonis, A.G.; Loher, P.; Honda, S.; Jing, Y.; Palazzo, J.; Kirino, Y.; Rigoutsos, I. Dissecting tRNA-derived fragment complexities using personalized transcriptomes reveals novel fragment classes and unexpected dependencies. Oncotarget 2015, 6, 24797–24822. [Google Scholar] [CrossRef] [PubMed]
- Telonis, A.G.; Kirino, Y.; and Rigoutsos, I. Mitochondrial tRNA- look alikes in nuclear chromosomes: Could they be functional? RNA Biol. 2015, 12, 375–380. [Google Scholar] [CrossRef]
- Olvedy, M.; Scaravilli, M.; Hoogstrate, Y.; Visakorpi, T.; Jenster, G.; Martens- Uzunova, E.S. A comprehensive repertoire of tRNA-derived fragments in prostate cancer. Oncotarget 2016, 7, 24766–24777. [Google Scholar] [CrossRef] [PubMed]
- Pliatsika, V.; Loher, P.; Telonis, A.G.; Rigoutsos, I. MINTbase: A framework for the interactive exploration of mitochondrial and nuclear tRNA fragments. Bioinformatics 2016, 32, 2481–2489. [Google Scholar] [CrossRef] [PubMed]
- Pliatsika, V.; Loher, P.; Magee, R.; Telonis, A.G.; Londin, E.; Shigematsu, M.; Kirino, Y.; Rigoutsos, I. MINTbase v2. 0: A comprehensive database for tRNA- derived fragments that includes nuclear and mitochondrial fragments from all The Cancer Genome Atlas projects. Nucleic Acids Res. 2018, 46, D152–D159. [Google Scholar]
- Lant, J.T.; Berg, M.D.; Heinemann, I.U.; Brandl, C.J.; O’Donoghue, P. Pathways to disease from natural variations in human cytoplasmic tRNAs. J. Biol. Chem. 2019, 294, 5294–5308. [Google Scholar] [CrossRef]
- Salinas-Giegé, T.; Giegé, R.; Giegé, P. tRNA Biology in Mitochondria. Int. J. Mol. Sci. 2015, 16, 4518–4559. [Google Scholar] [CrossRef]
- Lei, H.T.; Wang, Z.H.; Li, B.; Sun, Y.; Mei, S.Q.; Yang, J.H.; Qu, L.H.; Zheng, L.L. tModBase: Deciphering the landscape of tRNA modifications and their dynamic changes from epitranscriptome data. Nucleic Acids Res. 2023, 51, D315–D327. [Google Scholar] [CrossRef] [PubMed]
- Pan, T. Modifications and functional genomics of human transfer RNA. Cell Res. 2018, 28, 395–404. [Google Scholar] [CrossRef]
- Lyons, S.M.; Fay, M.M.; Ivanov, P. The role of RNA modifications in the regulation of tRNA cleavage. FEBS Lett. 2018, 592, 2828–2844. [Google Scholar] [CrossRef]
- Tao, E.W.; Cheng, W.Y.; Li, W.L.; Yu, J.; Gao, Q.Y. tiRNAs: A novel class of small noncoding RNAs that helps cells respond to stressors and plays roles in cancer progression. J Cell Physiol. 2020, 235, 683–690. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.Y.; Deng, X.Y.; Li, Y.S.; Ma, X.C.; Feng, J.X.; Yu, B.; Chen, Y.; Luo, Y.L.; Wang, X.; Chen, M.L.; et al. Structure of Schlafen13 reveals a new class of tRNA/rRNA- targeting RNase engaged in translational control. Nat. Commun. 2018, 9, 1165. [Google Scholar] [CrossRef]
- Kumar, P.; Anaya, J.; Mudunuri, S.B.; Dutta, A. Meta-analysis of tRNA derived RNA fragments reveals that they are evolutionarily conserved and associate with AGO proteins to recognize specific RNA targets. BMC Biol. 2014, 12, 78. [Google Scholar] [CrossRef] [PubMed]
- Skeparnias, I.; Anastasakis, D.; Grafanaki, K.; Kyriakopoulos, G.; Alexopoulos, P.; Dougenis, D.; Scorilas, A.; Kontos, C.K.; Stathopoulos, C. Contribution of miRNAs, tRNAs and tRFs to Aberrant Signaling and Translation Deregulation in Lung Cancer. Cancers 2020, 12, 3056. [Google Scholar] [CrossRef] [PubMed]
- Kumar, P.; Mudunuri, S.B.; Anaya, J.; Dutta, A. tRFdb: A database for transfer RNA fragments. Nucleic Acids Res. 2015, 43, D141–D145. [Google Scholar] [CrossRef]
- Zheng, L.L.; Xu, W.L.; Liu, S.; Sun, W.J.; Li, J.H.; Wu, J.; Yang, J.H.; Qu, L.H. tRF2Cancer: A web server to detect tRNA-derived small RNA fragments (tRFs) and their expression in multiple cancers. Nucleic Acids Res. 2016, 44, W185–W193. [Google Scholar] [CrossRef]
- Looney, M.M.; Lu, Y.; Karakousis, P.C.; Halushka, M.K. Mycobacterium tuberculosis Infection Drives Mitochondria-Biased Dysregulation of Host Transfer RNA–Derived Fragments. J. Infect. Dis. 2021, 223, 1796–1805. [Google Scholar] [CrossRef]
- Loher, P.; Telonis, A.G.; Rigoutsos, I. MINTmap: Fast and exhaustive profiling of nuclear and mitochondrial tRNA fragments from short RNA-seq data. Sci. Rep. 2017, 7, 41184. [Google Scholar] [CrossRef]
- Meseguer, S.; Navarro-González, C.; Panadero, J.; Villarroya, M.; Boutoual, R.; Sánchez-Alcázar, J.A.; Armengod, M.E. The MELAS mutation m.3243A>G alters the expression of mitochondrial tRNA fragments. Biochim. Biophys. Acta Mol. Cell Res. 2019, 1866, 1433–1449.
- Slack, F.J.; Chinnaiyan, A.M. The Role of Non-coding RNAs in Oncology. Cell 2019, 179, 1033–1055. [Google Scholar] [CrossRef]
- Mitobe, Y.; Takayama, K.I.; Horie-Inoue, K.; Inoue, S. Prostate cancer-associated lncRNAs. Cancer Lett. 2018, 418, 159–166. [Google Scholar] [CrossRef] [PubMed]
- Takeiwa, T.; Ikeda, K.; Mitobe, Y.; Horie-Inoue, K.; Inoue, S. Long Noncoding RNAs Involved in the Endocrine Therapy Resistance of Breast Cancer. Cancers 2020, 12, 1424. [Google Scholar] [CrossRef]
- Takeiwa, T.; Ikeda, K.; Horie-Inoue, K.; Inoue, S. Mechanisms of Apoptosis-Related Long Noncoding RNAs in Ovarian Cancer. Front. Cell Dev. Biol. 2021, 9, 641963. [Google Scholar] [CrossRef]
- Noh, J.H.; Kim, K.M.; Abdelmohsen, K.; Yoon, J.H.; Panda, A.C.; Munk, R.; Kim, J.; Curtis, J.; Moad, C.A.; Wohler, C.M.; et al. HuR and GRSF1 modulate the nuclear export and mitochondrial localization of the lncRNA RMRP. Genes Dev. 2016, 30, 1224–1239. [Google Scholar] [CrossRef]
- Li, K.; Smagula, C.S.; Parsons, W.J.; Richardson, J.A.; Gonzalez, M.; Hagler, H.K.; Williams, R.S. Subcellular partitioning of MRP RNA assessed by ultrastructural and biochemical analysis. J. Cell Biol. 1994, 124, 871–882. [Google Scholar] [CrossRef]
- Jeandard, D.; Smirnova, A.; Tarassov, I.; Barrey, E.; Smirnov, A.; Entelis, N. Import of Non-Coding RNAs into Human Mitochondria: A Critical Review and Emerging Approaches. Cells 2019, 8, 286. [Google Scholar] [CrossRef]
- Gammage, P.A.; Frezza, C. Mitochondrial DNA: The overlooked oncogenome? BMC Biol. 2019, 17, 53. [Google Scholar] [CrossRef]
- Jeck, W.R.; Sorrentino, J.A.; Wang, K.; Slevin, M.K.; Burd, C.E.; Liu, J.; Marzluff, W.F.; Sharpless, N.E. Circular RNAs are abundant, conserved, and associated with ALU repeats. RNA 2013, 19, 141–157. [Google Scholar] [CrossRef]
- Salzman, J.; Chen, R.E.; Olsen, M.N.; Wang, P.L.; Brown, P.O. Cell-type specific features of circular RNA expression. PLoS Genet. 2013, 9, e1003777. [Google Scholar] [CrossRef]
- Chen, L.L. The biogenesis and emerging roles of circular RNAs. Nat. Rev. Mol. Cell Biol. 2016, 17, 205–211. [Google Scholar] [CrossRef] [PubMed]
- Kristensen, L.S.; Andersen, M.S.; Stagsted, L.V.W.; Ebbesen, K.K.; Hansen, T.B.; Kjems, J. The biogenesis, biology and characterization of circular RNAs. Nat. Rev. Genet. 2019, 20, 675–691. [Google Scholar] [CrossRef] [PubMed]
- Piwecka, M.; Glažar, P.; Hernandez-Miranda, L.R.; Memczak, S.; Wolf, S.A.; Rybak-Wolf, A.; Filipchyk, A.; Klironomos, F.; Cerda Jara, C.A.; Fenske, P.; et al. Loss of a mammalian circular RNA locus causes miRNA deregulation and affects brain function. Science 2017, 357, eaam8526. [Google Scholar] [CrossRef]
- Wilusz, J.E. A 360° view of circular RNAs: From biogenesis to functions. Wiley Interdiscip. Rev. RNA 2018, 9, e1478. [Google Scholar] [CrossRef] [PubMed]
- Jeck, W.R.; Sharpless, N.E. Detecting and characterizing circular RNAs. Nat. Biotechnol. 2014, 32, 453–461. [Google Scholar] [CrossRef]
- Chen, L.L. The expanding regulatory mechanisms and cellular functions of circular RNAs. Nat. Rev. Mol. Cell Biol. 2020, 21, 475–490. [Google Scholar] [CrossRef]
- Xiao, M.S.; Ai, Y.; Wilusz, J.E. Biogenesis and Functions of Circular RNAs Come into Focus. Trends Cell Biol. 2020, 30, 226–240. [Google Scholar] [CrossRef]
- Salzman, J.; Gawad, C.; Wang, P.L.; Lacayo, N.; Brown, P.O. Circular RNAs Are the Predominant Transcript Isoform from Hundreds of Human Genes in Diverse Cell Types. PLoS ONE 2012, 7, e30733. [Google Scholar] [CrossRef]
- Zhao, Q.; Liu, J.; Deng, H.; Ma, R.; Liao, J.Y.; Liang, H.; Hu, J.; Li, J.; Guo, J.; Cai, J.; et al. Targeting Mitochondria-Located circRNA SCAR Alleviates NASH via Reducing mROS Output. Cell 2020, 183, 76–93. [Google Scholar] [CrossRef] [PubMed]
- Wu, Z.; Sun, H.; Wang, C.; Liu, W.; Liu, M.; Zhu, Y.; Xu, W.; Jin, H.; Li, J. Mitochondrial Genome-Derived circRNA mc-COX2 Functions as an Oncogene in Chronic Lymphocytic Leukemia. Mol. Ther. Nucleic. Acids 2020, 20, 801–811. [Google Scholar] [CrossRef]
- Liu, X.; Wang, X.; Li, J.; Hu, S.; Deng, Y.; Yin, H.; Bao, X.; Zhang, Q.C.; Wang, G.; Wang, B.; et al. Identification of mecciRNAs and their roles in the mitochondrial entry of proteins. Sci. China Life Sci. 2020, 63, 1429–1449. [Google Scholar] [CrossRef] [PubMed]
- Liang, H.; Liu, J.; Su, S.; Zhao, Q. Mitochondrial noncoding RNAs: New wine in an old bottle. RNA Biol. 2021, 18, 2168–2182. [Google Scholar] [CrossRef] [PubMed]
- Cavalcante, G.C.; Magalhães, L.; Ribeiro-Dos-Santos, Â.; Vidal, A.F. Mitochondrial Epigenetics: Noncoding RNAs as a Novel Layer of Complexity. Int. J. Mol. Sci. 2020, 21, 1838. [Google Scholar] [CrossRef] [PubMed]
- Sharma, P.; Sharma, V.; Ahluwalia, T.S.; Dogra, N.; Kumar, S.; Singh, S. Let-7a induces metabolic reprogramming in breast cancer cells via targeting mitochondrial encoded ND4. Cancer Cell Int. 2021, 21, 629. [Google Scholar] [CrossRef]
- Yan, K.; An, T.; Zhai, M.; Huang, Y.; Wang, Q.; Wang, Y.; Zhang, R.; Wang, T.; Liu, J.; Zhang, Y.; et al. Mitochondrial miR-762 regulates apoptosis and myocardial infarction by impairing ND2. Cell Death Dis. 2019, 10, 500. [Google Scholar] [CrossRef] [PubMed]
- Jagannathan, R.; Thapa, D.; Nichols, C.E.; Shepherd, D.L.; Stricker, J.C.; Croston, T.L.; Baseler, W.A.; Lewis, S.E.; Martinez, I.; Hollander, J.M. Translational Regulation of the Mitochondrial Genome Following Redistribution of Mitochondrial MicroRNA in the Diabetic Heart. Circ. Cardiovasc. Genet. 2015, 8, 785–802. [Google Scholar] [CrossRef] [PubMed]
- Shepherd, D.L.; Hathaway, Q.A.; Pinti, M.V.; Nichols, C.E.; Durr, A.J.; Sreekumar, S.; Hughes, K.M.; Stine, S.M.; Martinez, I.; Hollander, J.M. Exploring the mitochondrial microRNA import pathway through Polynucleotide Phosphorylase (PNPase). J. Mol. Cell Cardiol. 2017, 110, 15–25. [Google Scholar] [CrossRef]
- Li, H.; Zhang, X.; Wang, F.; Zhou, L.; Yin, Z.; Fan, J.; Nie, X.; Wang, P.; Fu, X.D.; Chen, C.; et al. MicroRNA-21 Lowers Blood Pressure in Spontaneous Hypertensive Rats by Upregulating Mitochondrial Translation. Circulation 2016, 134, 734–751. [Google Scholar] [CrossRef]
- Zhang, X.; Zuo, X.; Yang, B.; Li, Z.; Xue, Y.; Zhou, Y.; Huang, J.; Zhao, X.; Zhou, J.; Yan, Y.; et al. MicroRNA Directly Enhances Mitochondrial Translation during Muscle Differentiation. Cell 2014, 158, 607–619. [Google Scholar] [CrossRef]
- Chen, W.; Wang, P.; Lu, Y.; Jin, T.; Lei, X.; Liu, M.; Zhuang, P.; Liao, J.; Lin, Z.; Li, B.; et al. Decreased expression of mitochondrial miR-5787 contributes to chemoresistance by reprogramming glucose metabolism and inhibiting MT-CO3 translation. Theranostics 2019, 9, 5739–5754. [Google Scholar] [CrossRef] [PubMed]
- Fan, S.; Tian, T.; Chen, W.; Lv, X.; Lei, X.; Zhang, H.; Sun, S.; Cai, L.; Pan, G.; He, L.; et al. Mitochondrial miRNA Determines Chemoresistance by Reprogramming Metabolism and Regulating Mitochondrial Transcription. Cancer Res. 2019, 79, 1069–1084. [Google Scholar] [CrossRef] [PubMed]
- Aschrafi, A.; Schwechter, A.D.; Mameza, M.G.; Natera-Naranjo, O.; Gioio, A.E.; Kaplan, B.B. MicroRNA-338 Regulates Local Cytochrome c Oxidase IV mRNA Levels and Oxidative Phosphorylation in the Axons of Sympathetic Neurons. J. Neurosci. 2008, 28, 12581–12590. [Google Scholar] [CrossRef] [PubMed]
- Chen, Z.; Li, Y.; Zhang, H.; Huang, P.; Luthra, R. Hypoxia-regulated microRNA-210 modulates mitochondrial function and decreases ISCU and COX10 expression. Oncogene 2010, 29, 4362–4368. [Google Scholar] [CrossRef] [PubMed]
- Zheng, S.Q.; Li, Y.X.; Zhang, Y.; Li, X.; Tang, H. MiR-101 regulates HSV-1 replication by targeting ATP5B. Antiviral Res. 2011, 89, 219–226. [Google Scholar] [CrossRef]
- Carden, T.; Singh, B.; Mooga, V.; Bajpai, P.; Singh, K.K. Epigenetic modification of miR-663 controls mitochondria-to-nucleus retrograde signaling and tumor progression. J. Biol. Chem. 2017, 292, 20694–20706. [Google Scholar] [CrossRef] [PubMed]
- Bayazit, M.B.; Jacovetti, C.; Cosentino, C.; Sobel, J.; Wu, K.; Brozzi, F.; Rodriguez-Trejo, A.; Stoll, L.; Guay, C.; Regazzi, R. Small RNAs derived from tRNA fragmentation regulate the functional maturation of neonatal β cells. Cell Rep. 2022, 40, 111069. [Google Scholar] [CrossRef] [PubMed]
- Leucci, E.; Vendramin, R.; Spinazzi, M.; Laurette, P.; Fiers, M.; Wouters, J.; Radaelli, E.; Eyckerman, S.; Leonelli, C.; Vanderheyden, K.; et al. Melanoma addiction to the long non-coding RNA SAMMSON. Nature 2016, 531, 518–522. [Google Scholar] [CrossRef] [PubMed]
- Dewaele, S.; Delhaye, L.; De Paepe, B.; de Bony, E.J.; De Wilde, J.; Vanderheyden, K.; Anckaert, J.; Yigit, N.; Nuytens, J.; Eynde, E.V.; et al. The long non-coding RNA SAMMSON is essential for uveal melanoma cell survival. Oncogene 2022, 41, 15–25. [Google Scholar] [CrossRef] [PubMed]
- Orre, C.; Dieu, X.; Guillon, J.; Gueguen, N.; Ahmadpour, S.T.; Dumas, J.F.; Khiati, S.; Reynier, P.; Lenaers, G.; Coqueret, O.; et al. The Long Non-Coding RNA SAMMSON Is a Regulator of Chemosensitivity and Metabolic Orientation in MCF-7 Doxorubicin-Resistant Breast Cancer Cells. Biology 2021, 10, 1156. [Google Scholar] [CrossRef]
- Durr, A.J.; Hathaway, Q.A.; Kunovac, A.; Taylor, A.D.; Pinti, M.V.; Rizwan, S.; Shepherd, D.L.; Cook, C.C.; Fink, G.K.; Hollander, J.M. Manipulation of the miR-378a/mt-ATP6 regulatory axis rescues ATP synthase in the diabetic heart and offers a novel role for lncRNA Kcnq1ot1. Am. J. Physiol. Cell Physiol. 2022, 322, C482–C495. [Google Scholar] [CrossRef]
- Sirey, T.M.; Roberts, K.; Haerty, W.; Bedoya-Reina, O.; Rogatti-Granados, S.; Tan, J.Y.; Li, N.; Heather, L.C.; Carter, R.N.; Cooper, S.; et al. The long non-coding RNA Cerox1 is a post transcriptional regulator of mitochondrial complex I catalytic activity. eLife 2019, 8, e45051. [Google Scholar] [CrossRef]
- Gong, W.; Xu, J.; Wang, Y.; Min, Q.; Chen, X.; Zhang, W.; Chen, J.; Zhan, Q. Nuclear genome-derived circular RNA circPUM1 localizes in mitochondria and regulates oxidative phosphorylation in esophageal squamous cell carcinoma. Signal Transduct. Target. Ther. 2022, 7, 40. [Google Scholar] [CrossRef]
- Chen, Z.; He, Q.; Lu, T.; Wu, J.; Shi, G.; He, L.; Zong, H.; Liu, B.; Zhu, P. mcPGK1-dependent mitochondrial import of PGK1 promotes metabolic reprogramming and self-renewal of liver TICs. Nat. Commun. 2023, 14, 1121. [Google Scholar] [CrossRef]
- Das, S.; Bedja, D.; Campbell, N.; Dunkerly, B.; Chenna, V.; Maitra, A.; Steenbergen, C. miR-181c Regulates the Mitochondrial Genome, Bioenergetics, and Propensity for Heart Failure In Vivo. PLoS ONE 2014, 9, e96820. [Google Scholar] [CrossRef]
- Gevaert, A.B.; Witvrouwen, I.; Van Craenenbroeck, A.H.; Van Laere, S.J.; Boen, J.R.A.; Van de Heyning, C.M.; Belyavskiy, E.; Mueller, S.; Winzer, E.; Duvinage, A.; et al. miR-181c level predicts response to exercise training in patients with heart failure and preserved ejection fraction: An analysis of the OptimEx-Clin trial. Eur. J. Prev. Cardiol. 2021, 28, 1722–1733. [Google Scholar] [CrossRef]
- Meseguer, S.; Martínez-Zamora, A.; García-Arumí, E.; Andreu, A.L.; Armengod, M.E. The ROS-sensitive microRNA-9/9∗ controls the expression of mitochondrial tRNA-modifying enzymes and is involved in the molecular mechanism of MELAS syndrome. Hum. Mol. Genet. 2015, 24, 167–184. [Google Scholar] [CrossRef] [PubMed]
- Meseguer, S.; Boix, O.; Navarro-González, C.; Villarroya, M.; Boutoual, R.; Emperador, S.; García-Arumí, E.; Montoya, J.; Armengod, M.E. microRNA-mediated differential expression of TRMU, GTPBP3 and MTO1 in cell models of mitochondrial-DNA diseases. Sci. Rep. 2017, 7, 6209. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.; Zhuang, Q.; Ji, K.; Wen, B.; Lin, P.; Zhao, Y.; Li, W.; Yan, C. Identification of miRNA, lncRNA and mRNA-associated ceRNA networks and potential biomarker for MELAS with mitochondrial DNA A3243G mutation. Sci. Rep. 2017, 7, 41639. [Google Scholar] [CrossRef]
- Meseguer, S.; Panadero, J.; Navarro-González, C.; Villarroya, M.; Boutoual, R.; Comi, G.P.; Armengod, M.E. The MELAS mutation m.3243A>G promotes reactivation of fetal cardiac genes and an epithelial-mesenchymal transition-like program via dysregulation of miRNAs. Biochim. Biophys. Acta Mol. Basis Dis. 2018, 1864, 3022–3037. [Google Scholar] [CrossRef]
- Anderson, S.; Bankier, A.T.; Barrell, B.G.; de Bruijn, M.H.; Coulson, A.R.; Drouin, J.; Eperon, I.C.; Nierlich, D.P.; Roe, B.A.; Sanger, F.; et al. Sequence and organization of the human mitochondrial genome. Nature 1981, 290, 457–465. [Google Scholar] [CrossRef]
- Gorman, G.S.; Chinnery, P.F.; DiMauro, S.; Hirano, M.; Koga, Y.; McFarland, R.; Suomalainen, A.; Thorburn, D.R.; Zeviani, M.; Turnbull, D.M. Mitochondrial diseases. Nat. Rev. Dis. Prim. 2016, 2, 16080. [Google Scholar] [CrossRef]
- Chinnery, P.F.; Johnson, M.A.; Wardell, T.M.; Singh-Kler, R.; Hayes, C.; Brown, D.T.; Taylor, R.W.; Bindoff, L.A.; Turnbull, D.M. The epidemiology of pathogenic mitochondrial DNA mutations. Ann. Neurol. 2000, 48, 188–193. [Google Scholar] [CrossRef] [PubMed]
- Majamaa, K.; Moilanen, J.S.; Uimonen, S.; Remes, A.M.; Salmela, P.I.; Kärppä, M.; Majamaa-Voltti, K.A.; Rusanen, H.; Sorri, M.; Peuhkurinen, K.J.; et al. Epidemiology of A3243G, the Mutation for Mitochondrial Encephalomyopathy, Lactic Acidosis, and Strokelike Episodes: Prevalence of the Mutation in an Adult Population. Am. J. Hum. Genet. 1998, 63, 447–454. [Google Scholar] [CrossRef]
- Uusimaa, J.; Moilanen, J.S.; Vainionpää, L.; Tapanainen, P.; Lindholm, P.; Nuutinen, M.; Löppönen, T.; Mäki-Torkko, E.; Rantala, H.; Majamaa, K. Prevalence, segregation, and phenotype of the mitochondrial DNA 3243A>G mutation in children. Ann. Neurol. 2007, 62, 278–287. [Google Scholar] [CrossRef] [PubMed]
- Fornuskova, D.; Brantova, O.; Tesarova, M.; Stiburek, L.; Honzik, T.; Wenchich, L.; Tietzeova, E.; Hansikova, H.; Zeman, J. The impact of mitochondrial tRNA mutations on the amount of ATP synthase differs in the brain compared to other tissues. Biochim. Biophys. Acta 2008, 1782, 317–325. [Google Scholar] [CrossRef] [PubMed]
- Dubeau, F.; De Stefano, N.; Zifkin, B.G.; Arnold, D.L.; Shoubridge, E.A. Oxidative phosphorylation defect in the brains of carriers of the tRNAleu(UUR) A3243G mutation in a MELAS pedigree. Ann. Neurol. 2000, 47, 179–185. [Google Scholar] [CrossRef] [PubMed]
- Börner, G.V.; Zeviani, M.; Tiranti, V.; Carrara, F.; Hoffmann, S.; Gerbitz, K.D.; Lochmüller, H.; Pongratz, D.; Klopstock, T.; Melberg, A.; et al. Decreased aminoacylation of mutant tRNAs in MELAS but not in MERRF patients. Hum. Mol. Genet. 2000, 9, 467–475. [Google Scholar]
- Chomyn, A.; Enriquez, J.A.; Micol, V.; Fernandez-Silva, P.; Attardi, G. The Mitochondrial Myopathy, Encephalopathy, Lactic Acidosis, and Stroke-like Episode Syndrome-associated Human Mitochondrial tRNALeu(UUR) Mutation Causes Aminoacylation Deficiency and Concomitant Reduced Association of mRNA with Ribosomes. J. Biol. Chem. 2000, 275, 19198–19209. [Google Scholar] [CrossRef] [PubMed]
- King, M.P.; Koga, Y.; Davidson, M.; Schon, E.A. Defects in Mitochondrial Protein Synthesis and Respiratory Chain Activity Segregate with the tRNALeu(UUR) Mutation Associated with Mitochondrial Myopathy, Encephalopathy, Lactic Acidosis, and Strokelike Episodes. Mol. Cell Biol. 1992, 12, 480–490. [Google Scholar] [CrossRef] [PubMed]
- El Meziane, A.; Lehtinen, S.K.; Hance, N.; Nijtmans, L.G.; Dunbar, D.; Holt, I.J.; Jacobs, H.T. A tRNA suppressor mutation in human mitochondria. Nat. Genet. 1998, 18, 350–353. [Google Scholar] [CrossRef]
- Park, H.; Davidson, E.; King, M.P. The Pathogenic A3243G Mutation in Human Mitochondrial tRNALeu(UUR) Decreases the Efficiency of Aminoacylation. Biochemistry 2003, 42, 958–964. [Google Scholar] [CrossRef] [PubMed]
- Schaefer, M.; Pollex, T.; Hanna, K.; Tuorto, F.; Meusburger, M.; Helm, M.; Lyko, F. RNA methylation by Dnmt2 protects transfer RNAs against stress- induced cleavage. Genes Dev. 2010, 24, 1590–1595. [Google Scholar] [CrossRef]
- Blanco, S.; Dietmann, S.; Flores, J.V.; Hussain, S.; Kutter, C.; Humphreys, P.; Lukk, M.; Lombard, P.; Treps, L.; Popis, M.; et al. Aberrant methylation of tRNAs links cellular stress to neuro-developmental disorders. EMBO J. 2014, 33, 2020–2039. [Google Scholar] [CrossRef]
- Zhang, Y.; Zhang, X.; Shi, J.; Tuorto, F.; Li, X.; Liu, Y.; Liebers, R.; Zhang, L.; Qu, Y.; Qian, J.; et al. Dnmt2 mediates intergenerational transmission of paternally acquired metabolic disorders through sperm small non-coding RNAs. Nat. Cell Biol. 2018, 20, 535–540. [Google Scholar] [CrossRef]
- Chen, Z.; Qi, M.; Shen, B.; Luo, G.; Wu, Y.; Li, J.; Lu, Z.; Zheng, Z.; Dai, Q.; Wang, H. Transfer RNA demethylase ALKBH3 promotes cancer progression via induction of tRNA-derived small RNAs. Nucleic Acids Res. 2019, 47, 2533–2545. [Google Scholar] [CrossRef] [PubMed]
- Rackham, O.; Shearwood, A.M.J.; Mercer, T.R.; Davies, S.M.K.; Mattick, J.S.; Filipovska, A. Long noncoding RNAs are generated from the mitochondrial genome and regulated by nuclear-encoded proteins. RNA 2011, 17, 2085–2093. [Google Scholar] [CrossRef] [PubMed]
- Fernandez-Vizarra, E.; Zeviani, M. Mitochondrial complex III Rieske Fe-S protein processing and assembly. Cell Cycle 2018, 17, 681–687. [Google Scholar] [CrossRef] [PubMed]

| Noncoding RNAs (ncRNAs) Related to Mitochondria | Definition | Genome | Location |
|---|---|---|---|
| miRNA (~22 nt) | |||
| mt-miRNAs | mtDNA-encoded miRNAs or miRNAs located in mitochondria | Nuclear | Mitochondria |
| Mitochondrial | Mitochondria or other compartments | ||
| MitomiRs | Mitochondria-located miRNAs | Nuclear or Mitochondrial | Mitochondria |
| Mitochondria-associated miRNAs | miRNAs that are neither located in mitochondria nor encoded by mtDNA but regulate mitochondrial functions | Nuclear | Not located in mitochondria |
| tRNA-derived fragments (tRFs) (16–35 nt) | |||
| mt-tRFs | mtDNA-encoded tRFs or tRFs located in mitochondria | Nuclear | Mitochondria |
| Mitochondrial | Mitochondria or other compartments | ||
| Mitochondria-associated tRFs | tRFs that are neither located in mitochondria nor encoded by mtDNA but regulate mitochondrial functions | Nuclear | Not located in mitochondria |
| Long noncoding RNAs (lncRNAs) (>200 nt) | |||
| mt-lncRNAs | mtDNA-encoded lncRNAs or lncRNAs located in mitochondria | Nuclear | Mitochondria |
| Mitochondrial | Mitochondria or other compartments | ||
| Mitochondria-associated lncRNAs | lncRNAs that are neither located in mitochondria nor encoded by mtDNA but regulate mitochondrial functions | Nuclear | Not located in mitochondria |
| Circular RNAs (circRNAs) | |||
| mt-circRNAs | mtDNA-encoded cicRNAs or circRNAs located in mitochondria | Nuclear | Mitochondria |
| Mitochondrial | Mitochondria or other compartments | ||
| Mitochondria-associated circRNAs | circRNAs that are neither located in mitochondria nor encoded by mtDNA but regulate mitochondrial functions | Nuclear | Not located in mitochondria |
| ncRNA | Species | Genome | Roles in Mitochondrial ETC and OXPHOS |
|---|---|---|---|
| miRNAs | |||
| mitomiRs | |||
| let-7a | Human | Nuclear | Decreases complex I activity by destabilizing ND4 mRNA in MCF-7 breast cancer cells [81] |
| miR-762 | Mouse | Nuclear | Decreases complex I activity through decreasing ND2 in murine cardiomyocytes upon anoxia/reoxygenation treatment [82] |
| miR-181c | Rat | Nuclear | Induces complex IV remodeling and increases ROS levels by suppressing COX1 translation in rat cardiomyocytes [17] |
| miR-378a | Mouse | Nuclear | Decreases a complex V subunit, ATP6, in IFM of a mouse model of type I diabetes mellitus [83] Downregulates ATP6 expression in SSM of a mouse model of type II diabetes mellitus (db/db mice) [84] |
| miR-21 | Rat | Nuclear | Promotes CYTB translation in H9c2 rat cardiomyocytes and in hearts of spontaneous hypertensive rats [85] |
| miR-1 | Mouse | Nuclear | Promotes the translation of COX1 and ND1 in mouse C2C12 myoblasts [86] |
| miR-5787 | Human | Nuclear | Promotes COX3 translation and is involved in metabolic reprogramming and cisplatin resistance in Cal27 and Scc25 TSCC cells [87] |
| miR-2392 | Human | Nuclear | Decreases the mtDNA-encoded ETC complex subunits by repressing polycistronic mtDNA transcription and enhances anaerobic respiration in Cal27 and Scc9 TSCC cells [88] |
| Mitochondria-associated miRNAs | |||
| miR-338 | Rat | Nuclear | Decreases the expression of COX4 to reduce mitochondrial oxygen consumption in superior cervical ganglia (SCG) neurons [89] |
| miR-210 | Human | Nuclear | Modulates mitochondrial function during hypoxia conditions by reducing expression levels of COX10 in HCT116 colon cancer cells [90] |
| miR-101 | Human | Nuclear | Inhibits the replication of herpes simplex virus-1 (HSV-1) by downregulating the expression of ATP5B [91] |
| miR-663 | Human | Nuclear | Increases the number of ETC complex (I, II, III, and IV) subunits and assembly factors, thereby stabilizing the ETC supercomplexes in MCF-7 breast cancer cells [92] |
| tRFs | |||
| mt-tRFs | |||
| mt-i-tRF Glu(UUC) | Human | Mitochondrial | Downregulates MPC1 and increases extracellular lactate levels in MELAS cybrid cells [57] |
| tiRNA-5His(GTG) | Rat | Nuclear | Decreases MPC1 and is involved in proliferation and insulin secretion of neonatal rat pancreatic β cells [93] |
| tiRNA-5Glu(CTC) | Rat | Nuclear | Decreases MPC1 and is involved in proliferation and insulin secretion of neonatal rat pancreatic β cells [93] |
| Mitochondria-associated tRFs | |||
| tiRNA-5Glu(TTC) | Rat | Nuclear | Decreases MPC1 levels and is involved in mitochondrial respiration, proliferation, and insulin secretion of neonatal rat pancreatic β cells [93] |
| lncRNA | |||
| mt-lncRNAs | |||
| MALAT1 | Human | Nuclear | Modulates the CpG methylation of mtDNA and the expression of mtDNA-encoded ETC-related genes in HepG2 HCC cells [18] |
| SAMMSON | Human | Nuclear | Modulates the expression of mtDNA-encoded ETC complex subunits by facilitating mitochondrial localization of p32 in melanoma cells [94,95] |
| SAMMSON silencing increases the expression of ETC complexes, cellular respiration, and mitochondrial replication in doxorubicin-resistant MCF-7 breast cancer cells [96] | |||
| Mitochondria-associated lncRNAs | |||
| Kcnq1ot1 | Mouse | Nuclear | Inhibits miR-378a to upregulate the ATP6 expression and complex V activity in murine cardiomyocytes [97] |
| Cerox1 | Mouse | Nuclear | Inhibits miR-488-3p to enhance the expression of multiple ETC complex I subunits and mitochondrial respiration in N2A neuroblastoma cells [98] |
| circRNAs | |||
| mt-circRNAs | |||
| circPUM1 | Human | Nuclear | Binds to UQCRC2 to modulate the formation of the UQCRC1-UQCRC2 dimer in mitochondrial complex III in KYSE30 and KYSE410 ESCC cells [99] |
| SCAR | Human | Mitochondrial | Binds to ATP5B to shut down mPTP, thus inhibiting mitochondrial ROS output in liver fibroblasts [76] |
| mcPGK1 | Human | Mitochondrial | Promotes mitochondrial import of PGK1 and is involved in metabolic reprogramming from OXPHOS to glycolysis in liver TICs [100] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kobayashi, A.; Takeiwa, T.; Ikeda, K.; Inoue, S. Roles of Noncoding RNAs in Regulation of Mitochondrial Electron Transport Chain and Oxidative Phosphorylation. Int. J. Mol. Sci. 2023, 24, 9414. https://doi.org/10.3390/ijms24119414
Kobayashi A, Takeiwa T, Ikeda K, Inoue S. Roles of Noncoding RNAs in Regulation of Mitochondrial Electron Transport Chain and Oxidative Phosphorylation. International Journal of Molecular Sciences. 2023; 24(11):9414. https://doi.org/10.3390/ijms24119414
Chicago/Turabian StyleKobayashi, Ami, Toshihiko Takeiwa, Kazuhiro Ikeda, and Satoshi Inoue. 2023. "Roles of Noncoding RNAs in Regulation of Mitochondrial Electron Transport Chain and Oxidative Phosphorylation" International Journal of Molecular Sciences 24, no. 11: 9414. https://doi.org/10.3390/ijms24119414
APA StyleKobayashi, A., Takeiwa, T., Ikeda, K., & Inoue, S. (2023). Roles of Noncoding RNAs in Regulation of Mitochondrial Electron Transport Chain and Oxidative Phosphorylation. International Journal of Molecular Sciences, 24(11), 9414. https://doi.org/10.3390/ijms24119414









