Network Analysis of Biomarkers Associated with Occupational Exposure to Benzene and Malathion
Abstract
:1. Introduction
2. Results
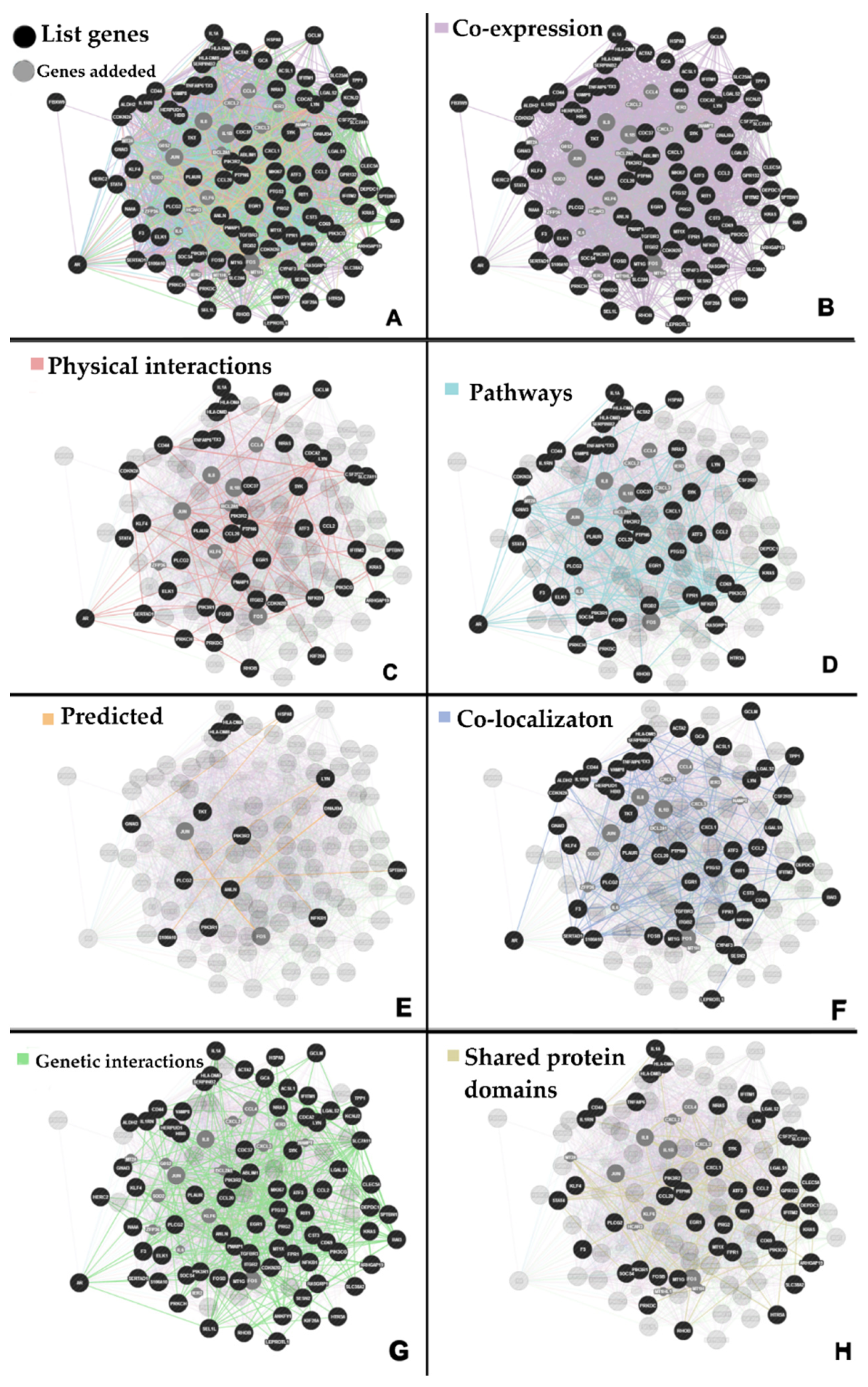
2.1. Interaction Network
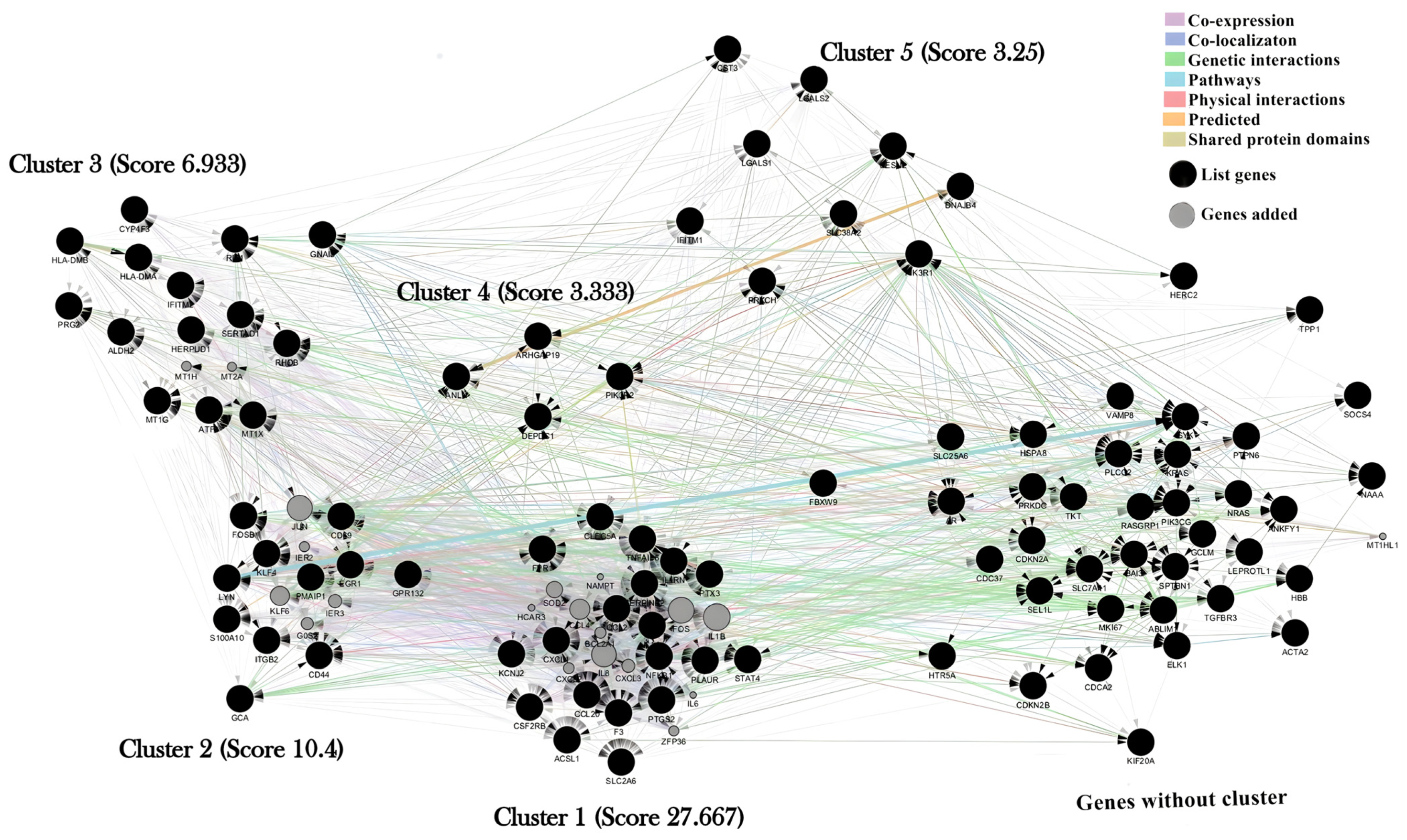
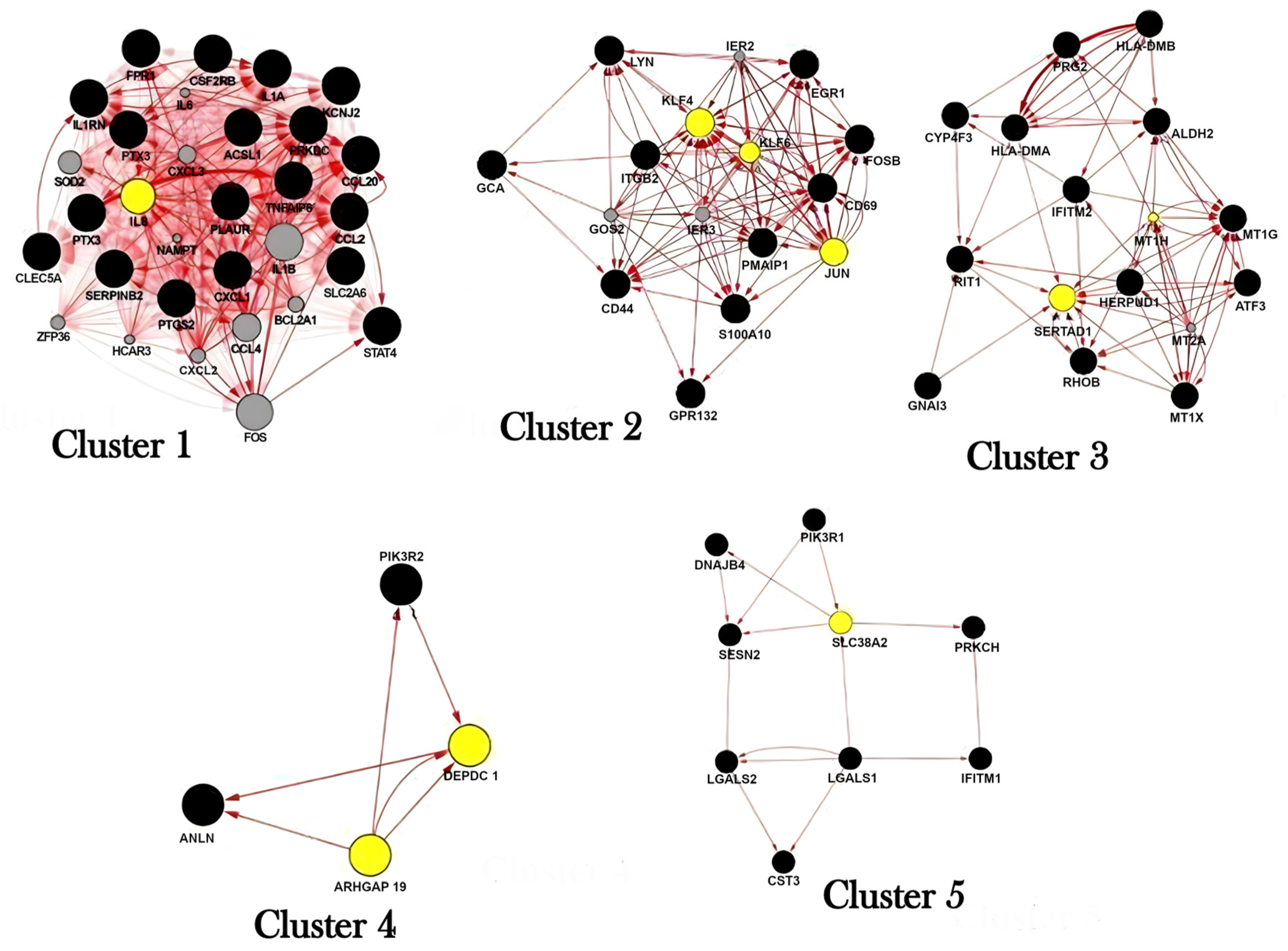
2.2. Cluster Analysis
2.3. Centiscape Analysis
2.4. GO Overrepresentation Analysis (BiNGO)
3. Discussion
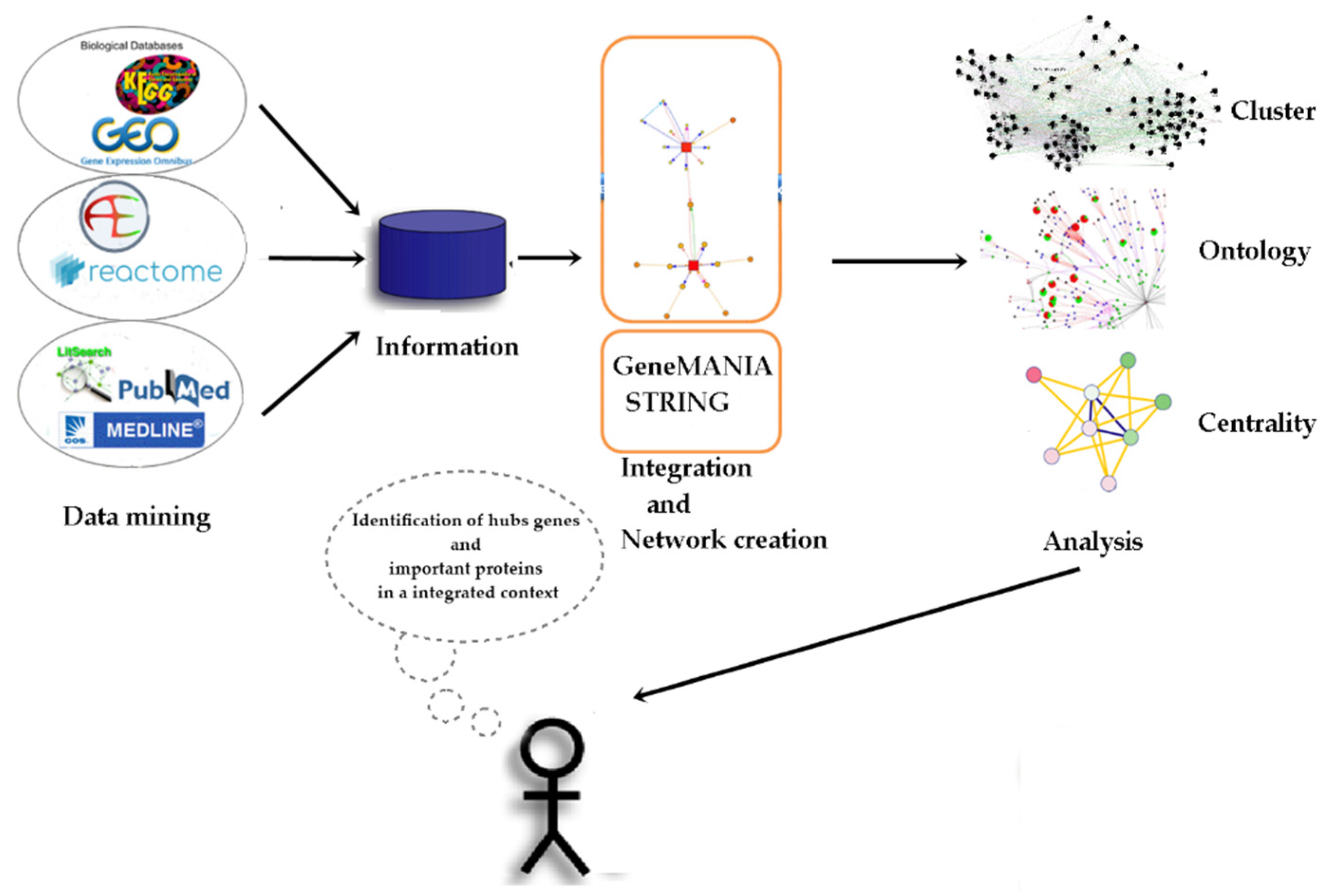
4. Materials and Methods
4.1. Data Collection
4.2. Construction of the Protein-Protein Interaction (PPI) Network
4.3. Extended Interaction Network Analysis
4.4. Identification of Molecular Complexes
4.5. GO Category Representation
4.6. Centrality Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Zhang, Q.; Li, J.; Xue, H.; Kong, L.; Wang, Y. Network-based methods for identifying critical pathways of complex diseases: A survey. Mol. Biosyst. 2016, 12, 1082–1089. [Google Scholar] [CrossRef] [PubMed]
- Wu, X.Y.; Liu, W.T.; Wu, Z.F.; Chen, C.; Liu, J.Y.; Wu, G.N.; Yao, X.Q.; Liu, F.K.; Li, G. Identification of HRAS as cancer-promoting gene in gastric carcinoma cell aggressiveness. Am. J. Cancer Res. 2016, 6, 1935–1948. [Google Scholar] [PubMed]
- Chen, L.; Wang, R.-S.; Zhang, X. Biomolecular Networks: Methods and Applications in Systems Biology, 1st ed.; Wiley: Hoboken, NJ, USA, 2009; p. 387. [Google Scholar]
- Barabási, A.L.; Gulbahce, N.; Loscalzo, J. Network medicine: A network-based approach to human disease. Nat. Rev. Genet. 2011, 12, 56–68. [Google Scholar] [CrossRef] [PubMed]
- Cowen, L.; Ideker, T.; Raphael, B.J.; Sharan, R. Network propagation: A universal amplifier of genetic associations. Nat. Rev. Genet. 2017, 18, 551–562. [Google Scholar] [CrossRef]
- Stuart, J.M. A Gene-Coexpression Network for Global Discovery of Conserved Genetic Modules. Science 2003, 302, 249–255. [Google Scholar] [CrossRef]
- Zhang, L.V.; King, O.D.; Wong, S.L.; Goldberg, D.S.; Tong, A.H.Y.; Lesage, G.; Andrews, B.; Bussey, H.; Boone, C.; Roth, F.P. Motifs, themes and thematic maps of an integrated Saccharomyces cerevisiae interaction network. J. Biol. 2005, 4, 6. [Google Scholar] [CrossRef]
- Giaever, G.; Shoemaker, D.D.; Jones, T.W.; Liang, H.; Winzeler, E.A.; Astromoff, A.; Davis, R.W. Genomic profiling of drug sensitivities via induced haploinsufficiency. Nat. Genet. 1999, 1, 278–283. [Google Scholar] [CrossRef]
- Uetz, P.; Giot, L.; Cagney, G.; Mansfield, T.A.; Judson, R.S.; Knight, J.R.; Lockshon, D.; Narayan, V.; Srinivasan, M.; Pochart, P.; et al. A comprehensive analysis of protein-protein interactions in Saccharomyces cerevisiae. Nature 2000, 403, 623–627. [Google Scholar] [CrossRef]
- Mering, C.V.; Krause, R.; Snel, B.; Cornell, M.; Oliver, S.G.; Fields, S.; Bork, P. Comparative assessment of large-scale data sets of protein-protein interactions. Nature 2002, 417, 399–403. [Google Scholar] [CrossRef]
- Polacco, B.J.; Babbitt, P.C. Automated discovery of 3D motifs for protein function annotation. Bioinformatics 2006, 22, 723–730. [Google Scholar] [CrossRef]
- Pellegrini, M.; Marcotte, E.M.; Thompson, M.J.; Eisenberg, D.; Yeates, T.O. Assigning protein functions by comparative genome analysis: Protein phylogenetic profiles. Proc. Natl. Acad. Sci. USA 1999, 96, 4285–4288. [Google Scholar] [CrossRef]
- Hegyi, H.; Gerstein, M. The relationship between protein structure and function: A comprehensive survey with application to the yeast genome. J. Mol. Biol. 1999, 288, 147–164. [Google Scholar] [CrossRef]
- Akavia, U.D.; Litvin, O.; Kim, J.; Sanchez-Garcia, F.; Kotliar, D.; Causton, H.C.; Pochanard, P.; Mozes, E.; Garraway, L.A.; Pe’er, D. An Integrated Approach to Uncover Drivers of Cancer. Cell 2010, 143, 1005–1017. [Google Scholar] [CrossRef]
- Zhang, L.; McHale, C.M.; Rothman, N.; Li, G.; Ji, Z.; Vermeulen, R.; Hubbard, A.E.; Ren, X.; Shen, M.; Rappaport, S.M.; et al. Systems biology of human benzene exposure. Chem. Biol. Interact. 2010, 184, 86–93. [Google Scholar] [CrossRef]
- International Agency for Research on Cancer (IARC). Chemical Agents and Related Occupations; IARC: Lyon, France, 2012; Volume 100F.
- Irons, R.D.; Chen, Y.; Wang, X.; Ryder, J.; Kerzic, P.J. Acute myeloid leukemia following exposure to benzene more closely resembles de novo than therapy related-disease. Genes Chromosomes Cancer 2013, 52, 887–894. [Google Scholar] [CrossRef]
- Lagorio, S.; Ferrante, D.; Ranucci, A.; Negri, S.; Sacco, P.; Rondelli, R.; Cannizzaro, S.; Torregrossa, M.V.; Cocco, P.; Forastiere, F.; et al. Exposure to benzene and childhood leukaemia: A pilot case-control study. BMJ Open 2013, 3, e002275. [Google Scholar] [CrossRef]
- Li, G.; Yin, S. Progress of epidemiological and molecular epidemiological studies on benzene in China. Ann. N. Y. Acad. Sci. 2006, 1076, 800–809. [Google Scholar] [CrossRef]
- Snyder, R. Leukemia and Benzene. Int. J. Environ. Res. Public Health 2012, 9, 2875–2893. [Google Scholar] [CrossRef]
- Fonseca, A.S.A.; Costa, D.F.; Dapper, V.; Machado, J.M.H.; Valente, D.; Carvalho, L.V.B.; Costa-Amaral, I.S.; Alves, S.R.; Sarcinelli, P.N.; Menezes, M.A.C.; et al. Classificação clínico-laboratorial para manejo clínico de trabalhadores expostos ao benzeno em postos de revenda de combustíveis. Rev. Bras. Saúde Ocup. 2017, 42, e5s. [Google Scholar] [CrossRef]
- Glass, D.C.; Gray, C.N.; Jolley, D.J.; Gibbons, C.; Sim, M.R.; Fritschi, L.; Adams, G.G.; Bisby, J.A.; Manuell, R. Leukemia risk associated with low-level benzene exposure. Epidemiol. Camb. Mass. 2003, 14, 569–577. [Google Scholar] [CrossRef]
- Lan, Q.; Zhang, L.; Li, G.; Vermeulen, R.; Weinberg, R.S.; Dosemeci, M.; Rappaport, S.M.; Shen, M.; Alter, B.P.; Wu, Y.; et al. Hematotoxicity in workers exposed to low levels of benzene. Science 2004, 306, 1774–1776. [Google Scholar] [CrossRef] [PubMed]
- Pesatori, A.C.; Garte, S.; Popov, T.; Georgieva, T.; Panev, T.; Bonzini, M.; Consonni, D.; Carugno, M.; Goldstein, B.D.; Taioli, E.; et al. Early effects of low benzene exposure on blood cell counts in Bulgarian petrochemical workers. Med. Lav. 2009, 100, 83–90. [Google Scholar] [PubMed]
- Swaen, G.M.H.; Amelsvoort, L.V.; Twisk, J.J.; Verstraeten, E.; Slootweg, R.; Collins, J.J.; Burns, C.J. Low level occupational benzene exposure and hematological parameters. Chem. Biol. Interact. 2010, 184, 94–100. [Google Scholar] [CrossRef] [PubMed]
- Smith, M.T. Advances in understanding benzene health effects and susceptibility. Annu. Rev. Public Health 2010, 31, 133–148. [Google Scholar] [CrossRef]
- Costa-Amaral, I.C.; Carvalho, L.V.B.; Pimentel, J.N.S.; Pereira, A.C.; Vieira, J.A.; Castro, V.S.; Borges, R.M.; Alvez, S.R.; Nogueira, S.M.; Tabalipa, M.d.M.; et al. Avaliação ambiental de BTEX (benzeno, tolueno, etilbenzeno, xilenos) e biomarcadores de genotoxicidade em trabalhadores de postos de combustíveis. Rev. Bras. Saúde Ocup. 2017, 42, e8s. [Google Scholar] [CrossRef]
- Valente, D.; Costa-Amaral, I.C.; Carvalho, L.V.B.; Santos, M.V.C.; Castro, V.S.; Rodrigues, D.R.F.; Falco, A.; Silva, C.B.; Nogueira, S.M.; Golçaolves, E.S.; et al. Utilização de biomarcadores de genotoxicidade e expressão gênica na avaliação de trabalhadores de postos de combustíveis expostos a vapores de gasolina. Rev. Bras. Saúde Ocup. 2017, 42, e2s. [Google Scholar] [CrossRef]
- McHale, C.M.; Zhang, L.; Smith, M.T. Current understanding of the mechanism of benzene-induced leukemia in humans: Implications for risk assessment. Carcinogenesis 2012, 33, 240–252. [Google Scholar] [CrossRef]
- Moore, P.D.; Yedjou, C.G.; Tchounwou, P.B. Malathion-Induced Oxidative Stress, Cytotoxicity, and Genotoxicity in Human Liver Carcinoma (HepG2) Cells. Environ. Toxicol. 2010, 25, 221–226. [Google Scholar] [CrossRef]
- Navarrete-Meneses, M.P.; Pedraza-Meléndez, A.I.; Salas-Labadía, C.; Moreno-Lorenzana, D.; Pérez-Vera, P. Low concentrations of permethrin and malathion induce numerical and structural abnormalities in KMT2A and IGH genes in vitro: Permethrin and malathion induce abnormalities in KMT2A and IGH genes. J. Appl. Toxicol. 2018, 38, 1262–1270. [Google Scholar] [CrossRef]
- Cabello, G.; Valenzuela, M.; Vilaxa, A.; Durán, V.; Rudolph, I.; Hrepic, N.; Calaf, G. A Rat Mammary Tumor Model Induced by the Organophosphorous Pesticides Parathion and Malathion, Possibly through Acetylcholinesterase Inhibition. Environ. Health Perspect. 2001, 109, 471–479. [Google Scholar] [CrossRef]
- Bonner, M.R.; Coble, J.; Blair, A.; Freeman, L.E.B.; Hoppin, J.A.; Sandler, D.P.; Alavanja, M.C.R. Malathion exposure and the incidence of cancer in the Agricultural Health Study. Am. J. Epidemiol. 2007, 166, 1023–1103. [Google Scholar] [CrossRef]
- Lerro, C.C.; Koutros, S.; Andreotti, G.; Friesen, M.C.; Alavanja, M.C.; Blair, A.; Hoppin, J.A.; Sandler, D.P.; Lubin, J.H.; Ma, X.; et al. Organophosphate insecticide use and cancer incidence among spouses of pesticide applicators in the Agricultural Health Study. Occup. Environ. Med. 2015, 72, 736–744. [Google Scholar] [CrossRef]
- Reuber, M.D. Carcinogenicity and toxicity of malathion and malaoxon. Environ. Res. 1985, 37, 119–153. [Google Scholar] [CrossRef]
- Smith, M.T.; Zhang, L.; McHale, C.M.; Skibola, C.F.; Rappaport, S.M. Benzene, the exposome and future investigations of leukemia etiology. Chem. Biol. Interact. 2011, 192, 155–159. [Google Scholar] [CrossRef]
- Roy, S.; Bhattacharyya, D.K.; Kalita, J.K. Reconstruction of gene co-expression network from microarray data using local expression patterns. BMC Bioinform. 2014, 15, S10. [Google Scholar] [CrossRef]
- Luo, J.; Qi, Y. Identification of essential proteins based on a new combination of local interaction density and protein complexes. PLoS ONE 2015, 10, e0131418. [Google Scholar] [CrossRef]
- Li, M.; Lu, Y.; Niu, Z.; Wu, F.X.; Pan, Y. Identification of essential proteins by using complexes and interaction network. In Bioinformatics Research and Applications; Basu, M., Pan, Y., Wang, J., Eds.; Springer International Publishing: Cham, Switzerland, 2014; pp. 255–265. [Google Scholar]
- Hu, P.; Mei, T. Ranking influential nodes in complex networks with structural holes. Phys. A Stat. Mech. Appl. 2018, 490, 624–631. [Google Scholar] [CrossRef]
- Cinaglia, P.; Cannataro, M. Network alignment and motif discovery in dynamic networks. Netw. Model. Anal. Health Inform. Bioinform. 2022, 11, 38. [Google Scholar] [CrossRef]
- Dai, C.; He, J.; Hu, K.; Ding, Y. Identifying essential proteins in dynamic protein networks based on an improved h-index algorithm. BMC Med. Inform. Decis. Mak. 2020, 20, 110. [Google Scholar] [CrossRef]
- Zhang, W.; Xu, J.; Li, Y.; Zou, X. Detecting Essential Proteins Based on Network Topology, Gene Expression Data, and Gene Ontology Information. IEEE/ACM Trans. Comput. Biol. Bioinform. 2018, 15, 109–116. [Google Scholar] [CrossRef]
- Lei, X.; Wang, S.; Wu, F.X. Identification of essential proteins based on improved HITS algorithm. IEEE/ACM Trans. Comput. Biol. Bioinform. Genes 2019, 10, 177. [Google Scholar] [CrossRef] [PubMed]
- Mistry, D.; Wise, R.; Dickerson, J. DiffSLC: A graph centrality method to detect essential proteins of a protein-protein interaction network. PLoS ONE 2017, 12, e0187091. [Google Scholar] [CrossRef] [PubMed]
- Zaki, N.; Berengueres, J.; Efimov, D. Detection of protein complexes using a protein ranking algorithm. Proteins Struct. Funct. Bioinform. 2012, 80, 2459–2468. [Google Scholar] [CrossRef] [PubMed]
- McHale, C.M.; Zhang, L.; Lan, Q.; Vermeulen, R.; Li, G.; Hubbard, A.E.; Porter, K.E.; Thomas, R.; Portier, C.J.; Shenet, M.; et al. Global Gene Expression Profiling of a Population Exposed to a Range of Benzene Levels. Environ. Health Perspect. 2010, 119, 628–634. [Google Scholar] [CrossRef] [PubMed]
- Bi, Y.; Li, Y.; Kong, M.; Xiao, X.; Zhao, Z.; He, X.; Ma, Q. Gene expression in benzene-exposed workers by microarray analysis of peripheral mononuclear blood cells: Induction and silencing of CYP4F3A and regulation of DNA-dependent protein kinase catalytic subunit in DNA double strand break repair. Chem. Biol. Interact. 2010, 184, 207–211. [Google Scholar] [CrossRef]
- Xing, C.; Wang, Q.; Li, B.; Tian, H.; Ni, Y.; Yin, S.; Li, G. Methylation and expression analysis of tumor suppressor genes p15 and p16 in benzene poisoning. Chem. Biol. Interact. 2010, 184, 306–309. [Google Scholar] [CrossRef]
- Sarma, S.N.; Kim, Y.-J.; Ryu, J.-C. Differential gene expression profiles of human leukemia cell lines exposed to benzene and its metabolites. Environ. Toxicol. Pharmacol. 2011, 32, 285–295. [Google Scholar] [CrossRef]
- Gao, A.; Yang, J.; Yang, G.; Niu, P.; Tian, L. Differential gene expression profiling analysis in workers occupationally exposed to benzene. Sci. Total Environ. 2014, 472, 872–879. [Google Scholar] [CrossRef]
- Anjitha, R.; Antony, A.; Shilpa, O.; Anupama, K.P.; Mallikarjunaiah, S.; Gurushankara, H.P. Malathion induced cancer-linked gene expression in human lymphocytes. Environ. Res. 2020, 182, 109–131. [Google Scholar] [CrossRef]
- Chow, K.; Sarkar, A.; Elhesha, R.; Cinaglia, P.; Ay, A.; Kahveci, T. ANCA: Alignment-Based Network Construction Algorithm. IEEE/ACM Trans. Comput. Biol. Bioinf. 2021, 18, 512–524. [Google Scholar] [CrossRef]
- Waugh, D.J.J.; Wilson, C. The Interleukin-8 Pathway in Cancer. Clin. Cancer Res. 2008, 14, 6735–6741. [Google Scholar] [CrossRef]
- Moro, A.M.; Charão, M.F.; Brucker, N.; Durgante, J.; Baierle, M.; Bubols, G.; Goethel, G.; Fracasso, R.; Nascimento, S.; Bulcão, R.; et al. Genotoxicity and oxidative stress in gasoline station attendants. Mutat. Res. Toxicol. Environ. Mutagen. 2013, 754, 63–70. [Google Scholar] [CrossRef]
- Lan, Q. Polymorphisms in Cytokine and Cellular Adhesion Molecule Genes and Susceptibility to Hematotoxicity among Workers Exposed to Benzene. Cancer Res. 2005, 65, 9574–9581. [Google Scholar] [CrossRef]
- Bader, G.D.; Hogue, C.W.V. An automated method for finding molecular complexes in large protein interaction networks. BMC Bioinform. 2003, 4, 2. [Google Scholar] [CrossRef]
- Mostafavi, S.; Ray, D.; Warde-Farley, D.; Grouios, C.; Morris, Q. GeneMANIA: A real-time multiple association network integration algorithm for predicting gene function. Genome Biol. 2008, 9, S4. [Google Scholar] [CrossRef]
- Bironaite, D.; Siegel, D.; Moran, J.L.; Weksler, B.B.; Ross, D. Stimulation of endothelial IL-8 (eIL-8) production and apoptosis by phenolic metabolites of benzene in HL-60 cells and human bone marrow endothelial cells. Chem. Biol. Interact. 2004, 149, 177–188. [Google Scholar] [CrossRef]
- Gillis, B.; Gavin, I.M.; Arbieva, Z.; King, S.T.; Jayaraman, S.; Prabhakar, B.S. Identification of human cell responses to benzene and benzene metabolites. Genomics 2007, 90, 324–333. [Google Scholar] [CrossRef]
- Lord, K.A.; Abdollahi, A.; Hoffman-Liebermann, B.; Liebermann, D.A. Proto-oncogenes of the fos/jun family of transcription factors are positive regulators of myeloid differentiation. Mol. Cell. Biol. 1993, 13, 841–851. [Google Scholar]
- Narla, G. KLF6, a Candidate Tumor Suppressor Gene Mutated in Prostate Cancer. Science 2001, 294, 2563–2566. [Google Scholar] [CrossRef]
- Mukai, S. Involvement of Kr?ppel-like factor 6 (KLF6) mutation in the development of nonpolypoid colorectal carcinoma. World J. Gastroenterol. 2007, 13, 3932. [Google Scholar] [CrossRef]
- Ito, G.; Uchiyama, M.; Kondo, M.; Mori, S.; Usami, N.; Maeda, O.; Kawabe, T.; Hasegawa, Y.; Shimokata, K.; Sekido, Y. Kruppel-Like Factor 6 Is Frequently Down-Regulated and Induces Apoptosis in Non-Small Cell Lung Cancer Cells. Cancer Res. 2004, 64, 3838–3843. [Google Scholar] [CrossRef] [PubMed]
- DiFeo, A. Roles of KLF6 and KLF6-SV1 in Ovarian Cancer Progression and Intraperitoneal Dissemination. Clin. Cancer Res. 2006, 12, 3730–3739. [Google Scholar] [CrossRef] [PubMed]
- Camacho-Vanegas, O.; Narla, G.; Teixeira, M.S.; DiFeo, A.; Misra, A.; Singh, G.; Chan, A.M.; Friedman, S.L.; Feuerstein, B.G.; Martignetti, J.A. Functional inactivation of the KLF6 tumor suppressor gene by loss of heterozygosity and increased alternative splicing in glioblastoma. Int. J. Cancer 2007, 121, 1390–1395. [Google Scholar] [CrossRef]
- Teixeira, M.S.; Camacho-Vanegas, O.; Fernandez, Y.; Narla, G.; DiFeo, A.; Lee, B.; Kalir, T.; Friedman, S.L.; Schlecht, N.F.; Genden, E.M.; et al. KLF6 allelic loss is associated with tumor recurrence and markedly decreased survival in head and neck squamous cell carcinoma. Int. J. Cancer 2007, 121, 1976–1983. [Google Scholar] [CrossRef] [PubMed]
- Song, J.; Kim, C.J.; Cho, Y.G.; Kim, S.Y.; Nam, S.W.; Lee, S.H.; Yoo, N.J.; Lee, J.Y.; Park, W.S. Genetic and epigenetic alterations of the KLF6 gene in hepatocellular carcinoma. J. Gastroenterol. Hepatol. 2006, 21, 1286–1289. [Google Scholar] [CrossRef]
- Slavin, D.A.; Koritschoner, N.P.; Prieto, C.C.; López-Díaz, F.J.; Chatton, B.; Bocco, J.L. A new role for the Krüppel-like transcription factor KLF6 as an inhibitor of c-Jun proto-oncoprotein function. Oncogene 2004, 23, 8196–8205. [Google Scholar] [CrossRef]
- Qiu, S.; Liu, S.; Yu, T.; Yu, J.; Wang, M.; Rao, Q.; Xing, H.; Tang, K.; Mi, Y.; Wang, J. Sertad1 antagonizes iASPP function by hindering its entrance into nuclei to interact with P53 in leukemic cells. BMC Cancer 2017, 17, 795. [Google Scholar] [CrossRef]
- Kompier, L.C.; Lurkin, I.; van-der-Aa, M.N.M.; van-Rhijn, B.W.G.; van-der-Kwast, T.H.; Zwarthoff, E.C. FGFR3, HRAS, KRAS, NRAS and PIK3CA mutations in bladder cancer and their potential as biomarkers for surveillance and therapy. PLoS ONE 2010, 5, e13821. [Google Scholar] [CrossRef]
- Raz, R.; Durbin, J.E.; Levy, D.E. Acute phase response factor and additional members of the interferon-stimulated gene factor 3 family integrate diverse signals from cytokines, interferons, and growth factors. J. Biol. Chem. 1994, 269, 24391–24395. [Google Scholar] [CrossRef]
- van-Engen-van Grunsven, A.C.H.; van-Dijk, M.C.R.F.; Ruiter, D.J.; Klaasen, A.; Mooi, W.J.; Blokx, W.A.M. HRAS-mutated Spitz Tumors: A Subtype of Spitz Tumors With Distinct Features. Am. J. Surg. Pathol. 2010, 34, 1436–1441. [Google Scholar] [CrossRef]
- Calaf, G.; Roy, D. Cancer genes induced by malathion and parathion in the presence of estrogen in breast cells. Int. J. Mol. Med. 2008, 2, 261–268. [Google Scholar] [CrossRef]
- Fernandez-Medarde, A.; Santos, E. Ras in Cancer and Developmental Diseases. Genes Cancer 2011, 2, 344–358. [Google Scholar] [CrossRef]
- Bernard, M.; Bancos, S.; Sime, P.; Phipps, R. Targeting Cyclooxygenase-2 in Hematological Malignancies: Rationale and Promise. Curr. Pharm. Des. 2008, 14, 2051–2060. [Google Scholar] [CrossRef]
- Santos, M.V.C.; Figueiredo, V.O.; Arcuri, A.S.A.; Costa-Amaral, I.C.; Gonçalves, E.S.; Larentis, A.L. Aspectos toxicológicos do benzeno, biomarcadores de exposição e conflitos de interesses. Rev. Bras. Saúde Ocup. 2017, 42, e13s. [Google Scholar] [CrossRef]
- Friedrich, K.; Gurgel, A.D.M.; Sarpa, M.; Bedor, C.N.G.; Siqueira, M.T.D.; Gurgel, I.G.D.; Augusto, L. Toxicologia crítica aplicada aos agrotóxicos–perspectivas em defesa da vida. Saúde Debate 2022, 46, 293–315. [Google Scholar] [CrossRef]
- Mendes, M.P.R.; Paiva, M.J.N.; Costa-Amaral, I.C.; Carvalho, L.V.B.; Figueiredo, V.O.; Gonçalves, E.S.; Larentis, A.L.; André, L.C. Metabolomic Study of Urine from Workers Exposed to Low Concentrations of Benzene by UHPLC-ESI-QToF-MS Reveals Potential Biomarkers Associated with Oxidative Stress and Genotoxicity. Metabolites 2022, 12, 978. [Google Scholar] [CrossRef]
- Stobiecka, M.; Chalupa, A. DNA Strand Replacement Mechanism in Molecular Beacons Encoded for the Detection of Cancer Biomarkers. J. Phys. Chem. B 2016, 120, 4782–4790. [Google Scholar] [CrossRef]
- Montojo, J.; Zuberi, K.; Rodriguez, H.; Kazi, F.; Wright, G.; Donaldson, S.L.; Morris, Q.; Bader, G.D. GeneMANIA Cytoscape plugIn: Fast gene function predictions on the desktop. Bioinformatics 2010, 26, 2927–2928. [Google Scholar] [CrossRef]
- Scardoni, G.; Petterlini, M.; Laudanna, C. Analyzing biological network parameters with CentiScaPe. Bioinformatics 2009, 25, 2857–2859. [Google Scholar] [CrossRef]
- Azevedo, H.; Bando, S.Y.; Bertonha, F.B.; Moreira-Filho, C.A. Redes de interação gênica e controle epigenético na transição saúde-doença. Rev. Med. 2015, 94, 223. [Google Scholar] [CrossRef]
- Newman, M.E.J. A measure of betweenness centrality based on random walks. Soc. Netw. 2005, 27, 39–54. [Google Scholar] [CrossRef]
- Yu, H.; Kim, P.M.; Sprecher, E.; Trifonov, V.; Gerstein, M. The Importance of Bottlenecks in Protein Networks: Correlation with Gene Essentiality and Expression Dynamics. PLoS Comput. Biol. 2007, 3, e59. [Google Scholar] [CrossRef] [PubMed]
- Vilela, M.; Chou, I.-C.; Vinga, S.; Vasconcelos, A.; Voit, E.O.; Almeida, J.S. Parameter optimization in S-system models. BMC Syst. Biol. 2008, 2, 35. [Google Scholar] [CrossRef] [PubMed]

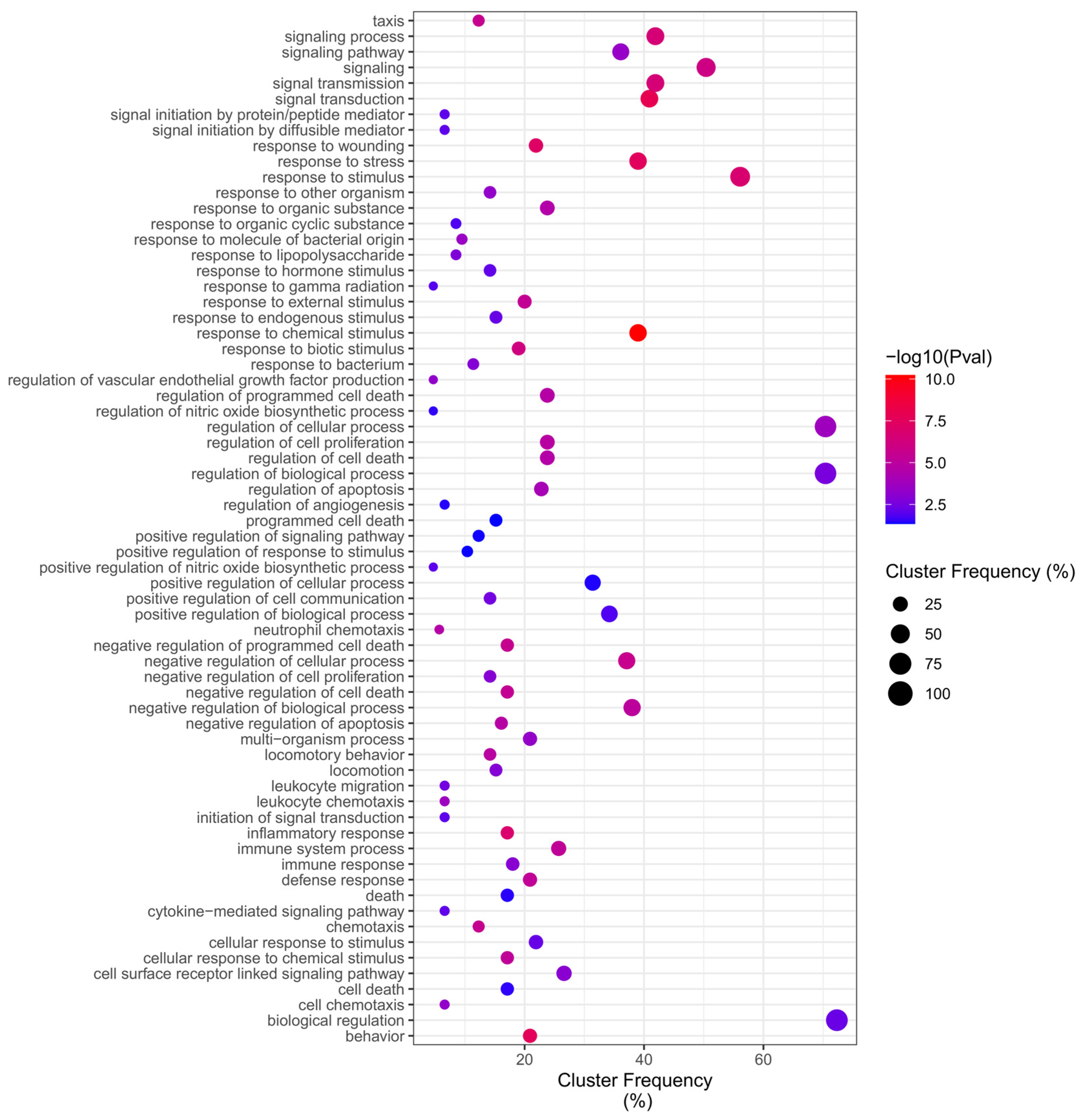
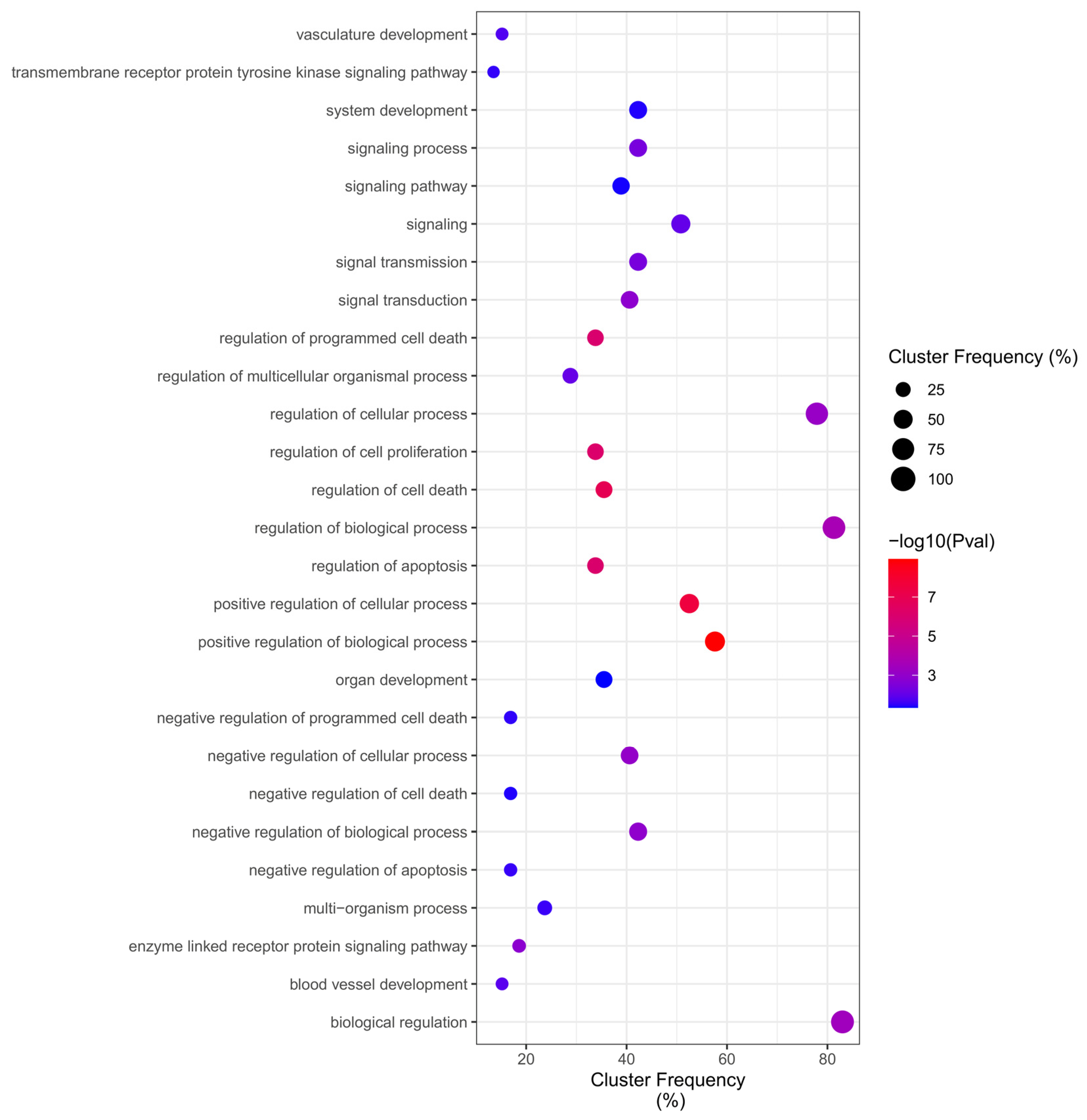
| Reference | Method | Exposure | Controls | DEGs |
|---|---|---|---|---|
| McHale et al. [47] | Microarray | 83 cases of benzene exposure ranging from <<1 to >10 ppm. | 42 | 16 genes with high expression (SERPINB2, TNFAIP6, IL1A, KCNJ2, PTX3, F3, CD44, CCL20, ACSL1, PTGS2 CLEC5A, IL1RN, PRG2, SLC2A6 GPR132, and PLAUR). |
| Bi et al. [48] | cDNA microarray | 7 women were diagnosed with benzene poisoning. | 7 | Top 40 genes with altered expression (PTGS2, BAI3, GCL, CYP4F3, MY047, TRA@, AD022, PRKCH, RASGRP1, FPR1, TGFBR3, GRO1, SEL1L, CSF2RB, IFITM1, STAT4, IFITM2, ABLIM, KIAA1382, SPTBN1, HBB, PRKDC, S100A10, ITGB2, TKT, VAMP8, FOSB, ASAHL, CDC37, SLC25A6, CLN2, ACTA2, CST3, HLA-DMB, ALDH2, LGALS2, LGALS1, ARHB, KLF4, and ATF3). |
| Xing et al. [49] | Microarray (RTPCR) | 11 | People in the same sector with no symptoms of benzene poisoning. | Decrease in the expression of p15 (CDKN2B) and p16 (CDKN2A). |
| Sarma et al. [50] | Microarray | Culture of HL-60 cells treated with IC50 concentrations of benzene, hydroquinone, and benzoquinone. | Culture of HL-60 cells treated with dimethyl sulfoxide. | Alteration in expression of 27 genes (CCL2, EGR1, GCLM, PMAIP1, SESN2, CD69, HERPUD1, HSPA8, RIT1, SERTAD1, SLC38A2, SLC7A11, DNAJB4, ANKFY1, ANLN, AR, ARHGAP19, CDCA2, DEPDC1, ELK1, FBXW9, HERC2, HTR5A, KIF20A, MKI67, MT1G, and MT1X). |
| Gao et al. [51] | cDNA microrray | 4 people were diagnosed with benzene poisoning, and 3 people from the same factory were exposed but had no symptoms. | 3 | Top 14 significant genes with altered expression (PIK3R1, PIK3CG, PIK3R2, GNAI3, SYK, PTPN6, KRAS, NRAS, PLCG2, NFKB1, LYN, SOCS4, HLA-DMA, and HLA-DMB). |
| Reference | Method | Exposure | Controls | DEGs |
|---|---|---|---|---|
| Anjitha et al. [52] | Microarray | Culture of human lymphocytes treated with three concentrations of malathion (50, 100, and 150 μg/mL). | Culture of human lymphocytes treated with DMSO (1%). | 57 DEGs (B4GALT1, BMI1, BTG1, C1QBP, CASP4, CBFB, CD14, CD5, CD36, CDK2, CEBPG, COL1A2, DDX11, DUSP1, EPHA4, EPS8L1, FGF6, FGFR1, FOXO4, FSHR, GNAI2, GPS2, GRAMD4, GSTP1, HLA-A, HLA-E, HTATIP2, IL13RA2, IRAK1, LGALS1, LPCAT4, LZTS2, MAP2K3, MICA, NINJ1, NME2, NTRK2, PAX1, PEBP1, PFN1, PHB, PLAU, PML, PRKD1, RAB11FIP3, SHC1, SOCS3, TEK, TFRC, TLR8, TPR, TRIM35, TUSC2, TWIST1, VHL, VPREB3, and WT1) |
| GeneMania Benzene Network | ||
|---|---|---|
| Symbol | Name | HGNC-ID |
| KLF4 | KLF transcription factor 4 | 6348 |
| SERTAD1 | SERTA domain containing 1 | 17932 |
| IL8 | Interleukin 8 | 6025 |
| JUN | Jun proto-oncogene, AP-1 transcription factor subunit | 6204 |
| KLF6 | KLF transcription factor 6 | 2235 |
| MT1H | Metallothionein 1H | 7400 |
| String Malathion | ||
| Symbol | Name | HGNC-ID |
| HRAS | Hras proto-oncogene, GTPase | 5173 |
| STAT3 | Signal transducer and activator of transcription 3 | 11364 |
| Network Categories | Datasets | Network Information | Data Source |
|---|---|---|---|
| Co-expression: | Gene expression | Two genes are linked if their expression levels are similar across conditions in a gene expression study. | Most of these data are collected from the Gene Expression Omnibus (GEO). |
| Physical Interaction: | Protein-protein interaction | Two gene products are linked if they were found to interact in a protein-protein interaction study. | These data are collected from primary studies found in protein interaction databases, including BioGRID and PathwayCommons. |
| Genetic interaction: | Genetic interaction | Two genes are functionally associated if the effects of perturbing one gene are found to be modified by perturbations to a second gene. | These data are collected from primary studies and BioGRID. |
| Shared protein domains: | Protein domain | Two gene products are linked if they have the same protein domain. | These data are collected from domain databases such as InterPro, SMART, and Pfam. |
| Co-localization: | Genes expressed in the same tissue or proteins found in the same location. | Two genes are linked if they are both expressed in the same tissue or if their gene products are both identified in the same cellular location. | |
| Pathway: | Pathway | Two gene products are linked if they participate in the same reaction within a pathway. | These data are collected from various sources, such as Reactome and BioCyc, via PathwayCommons. |
| Predicted: | Predicted functional relationships between genes, often protein interactions. | For instance, two proteins are predicted to interact if their orthologs are known to interact in another organism. In these cases, network names describe the original data source of experimentally measured interactions and the organism from which the interactions were mapped from (e.g. a mouse network predicted from a human network). | A major source of predicted data is mapping known functional relationships to another organism via orthology. These data are collected from various sources, such as BioGRID and I2D orthology. |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Santos, M.V.C.; Feltrin, A.S.; Costa-Amaral, I.C.; Teixeira, L.R.; Perini, J.A.; Martins, D.C., Jr.; Larentis, A.L. Network Analysis of Biomarkers Associated with Occupational Exposure to Benzene and Malathion. Int. J. Mol. Sci. 2023, 24, 9415. https://doi.org/10.3390/ijms24119415
Santos MVC, Feltrin AS, Costa-Amaral IC, Teixeira LR, Perini JA, Martins DC Jr., Larentis AL. Network Analysis of Biomarkers Associated with Occupational Exposure to Benzene and Malathion. International Journal of Molecular Sciences. 2023; 24(11):9415. https://doi.org/10.3390/ijms24119415
Chicago/Turabian StyleSantos, Marcus Vinicius C., Arthur S. Feltrin, Isabele C. Costa-Amaral, Liliane R. Teixeira, Jamila A. Perini, David C. Martins, Jr., and Ariane L. Larentis. 2023. "Network Analysis of Biomarkers Associated with Occupational Exposure to Benzene and Malathion" International Journal of Molecular Sciences 24, no. 11: 9415. https://doi.org/10.3390/ijms24119415
APA StyleSantos, M. V. C., Feltrin, A. S., Costa-Amaral, I. C., Teixeira, L. R., Perini, J. A., Martins, D. C., Jr., & Larentis, A. L. (2023). Network Analysis of Biomarkers Associated with Occupational Exposure to Benzene and Malathion. International Journal of Molecular Sciences, 24(11), 9415. https://doi.org/10.3390/ijms24119415







