Single Nematode Transcriptomic Analysis, Using Long-Read Technology, Reveals Two Novel Virulence Gene Candidates in the Soybean Cyst Nematode, Heterodera glycines
Abstract
1. Introduction
2. Results
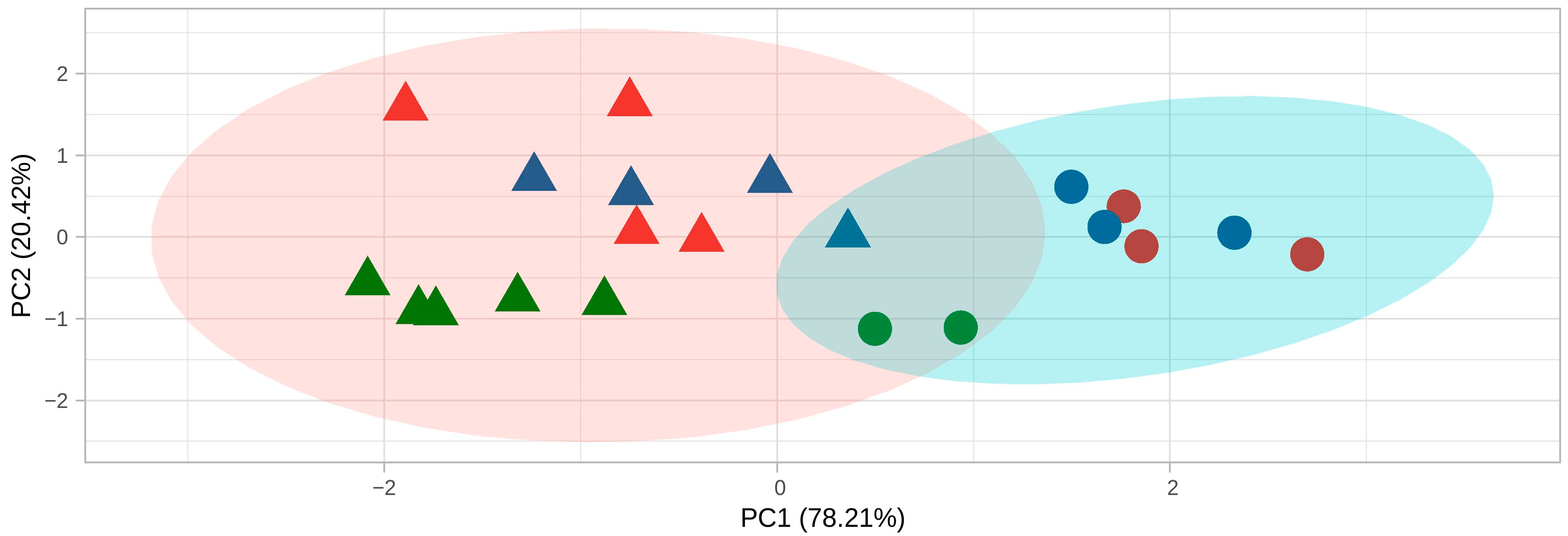
2.1. MinION Sequencing Efficiency and Transcript Variants Discovery
2.2. Reannotation of the SCN Genome
2.3. Differential Transcript Expression Analysis
3. Discussion
3.1. Differential Transcript Expression within Putative Effectors
3.2. Discovery of Novel Effectors Involved in Late-Stage Virulence in PI 88788
3.3. Genome-Wide Discovery of Novel Coding Transcripts
4. Materials and Methods
4.1. Plant and SCN Material
4.2. Hatching and Selection of Virulent SCN Individuals
4.3. Selection of Nematodes on Differential Plant Genotypes
4.4. RNA Extraction
4.5. Library Preparation
4.6. Read Processing and Novel Transcript Discovery
4.7. Differential Transcript Expression Analysis
4.8. Effector Gene Prediction and Gene Annotation
4.9. Validation of Putative Virulence Gene Candidate
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Bandara, A.Y.; Weerasooriya, D.K.; Bradley, C.A.; Allen, T.W.; Esker, P.D. Dissecting the economic impact of soybean diseases in the United States over two decades. PLoS ONE 2020, 15, e0231141. [Google Scholar] [CrossRef]
- Bradley, C.A.; Allen, T.W.; Sisson, A.J.; Bergstrom, G.C.; Bissonnette, K.M.; Bond, J.; Byamukama, E.; Chilvers, M.I.; Collins, A.A.; Damicone, J.P.; et al. Soybean yield loss estimates due to diseases in the United States and Ontario, Canada, from 2015 to 2019. Plant Health Prog. 2021, 22, 483–495. [Google Scholar] [CrossRef]
- Williamson, V.M.; Hussey, R.M. Nematode pathogenesis and resistance in plants. Plant Cell 1996, 8, 1735–1745. [Google Scholar] [CrossRef] [PubMed]
- Juvale, P.S.; Baum, T.J. “Cyst-ained” research into Heterodera parasitism. PLoS Pathog. 2018, 14, e1006791. [Google Scholar] [CrossRef]
- Vieira, P.; Gleason, C. Plant-parasitic nematode effectors—Insights into their diversity and new tools for their identification. Curr. Opin. Plant Biol. 2019, 50, 37–43. [Google Scholar] [CrossRef]
- Gao, B.; Allen, R.; Maier, T.; Davis, E.L.; Baum, T.J.; Hussey, R.S. Identification of Putative Parasitism Genes Expressed in the Esophageal Gland Cells of the Soybean Cyst Nematode Heterodera glycines. Mol. Plant Microbe Interact. 2001, 14, 1247–1254. [Google Scholar] [CrossRef]
- Gao, B.; Allen, R.; Maier, T.; Davis, E.L.; Baum, T.J.; Hussey, R.S. The Parasitome of the Phytonematode Heterodera glycines. Mol. Plant Microbe Interact. 2003, 16, 720–726. [Google Scholar] [CrossRef]
- Noon, J.B.; Hewezi, T.; Maier, T.R.; Simmons, C.; Wei, J.-Z.; Wu, G.; Llaca, V.; Deschamps, S.; Davis, E.L.; Mitchum, M.G.; et al. Eighteen New Candidate Effectors of the Phytonematode Heterodera glycines Produced Specifically in the Secretory Esophageal Gland Cells During Parasitism. Phytopathology 2015, 105, 1362–1372. [Google Scholar] [CrossRef] [PubMed]
- Maier, T.R.; Masonbrink, R.E.; Vijayapalani, P.; Gardner, M.; Howland, A.D.; Mitchum, M.G.; Baum, T.J. Esophageal Gland RNA-Seq Resource of a Virulent and Avirulent Population of the Soybean Cyst Nematode Heterodera glycines. Mol. Plant Microbe Interact. 2021, 34, 1084–1087. [Google Scholar] [CrossRef]
- Wang, X.; Allen, R.; Ding, X.; Goellner, M.; Maier, T.; de Boer, J.M.; Baum, T.J.; Hussey, R.S.; Davis, E.L. Signal Peptide-Selection of cDNA Cloned Directly from the Esophageal Gland Cells of the Soybean Cyst Nematode Heterodera glycines. Mol. Plant Microbe Interact. 2001, 14, 536–544. [Google Scholar] [CrossRef]
- Zhao, J.; Li, L.; Liu, Q.; Liu, P.; Li, S.; Yang, D.; Chen, Y.; Pagnotta, S.; Favery, B.; Abad, P.; et al. A MIF-like effector suppresses plant immunity and facilitates nematode parasitism by interacting with plant annexins. J. Exp. Bot. 2019, 70, 5943–5958. [Google Scholar] [CrossRef]
- Mitchum, M.G. Soybean Resistance to the Soybean Cyst Nematode Heterodera glycines: An Update. Phytopathology 2016, 106, 1444–1450. [Google Scholar] [CrossRef]
- Wang, J.; Niblack, T.L.; Tremain, J.A.; Wiebold, W.J.; Tylka, G.L.; Marett, C.C.; Noel, G.R.; Myers, O.; Schmidt, M.T. Soybean cyst nematode reduces soybean yield without causing obvious aboveground symptoms. Plant Dis. 2003, 87, 623–628. [Google Scholar] [CrossRef]
- Cook, D.E.; Lee, T.G.; Guo, X.; Melito, S.; Wang, K.; Bayless, A.M. Copy Number Variation of Multiple Genes at Rhg1 Mediates Nematode Resistance in Soybean. Science 2012, 338, 1206–1209. [Google Scholar] [CrossRef] [PubMed]
- Howland, A.; Monnig, N.; Mathesius, J.; Nathan, M.; Mitchum, M.G. Survey of Heterodera glycines Population Densities and Virulence Phenotypes During 2015–2016 in Missouri. Plant Dis. 2018, 102, 2407–2410. [Google Scholar] [CrossRef] [PubMed]
- Niblack, T.L.; Colgrove, A.L.; Colgrove, K.; Bond, J.P. Shift in Virulence of Soybean Cyst Nematode is Associated with Use of Resistance from PI 88788. Plant Health Prog. 2008, 9, 1475. [Google Scholar] [CrossRef]
- Bekal, S.; Domier, L.L.; Gonfa, B.; Lakhssassi, N.; Meksem, K.; Lambert, K.N. A snare-like protein and biotin are implicated in soybean cyst nematode virulence. PLoS ONE 2015, 10, e0145601. [Google Scholar] [CrossRef]
- Gardner, M.; Dhroso, A.; Johnson, N.; Davis, E.L.; Baum, T.J.; Korkin, D.; Michum, M.G. Novel global effector mining from the transcriptome of early life stages of the soybean cyst nematode Heterodera glycines. Sci. Rep. 2018, 8, 2505. [Google Scholar] [CrossRef]
- Ste-Croix, D.T.; St-Marseille, A.-F.G.; Lord, E.; Bélanger, R.R.; Brodeur, J.; Mimee, B. Genomic profiling of virulence in the soybean cyst nematode using single-nematode sequencing. Phytopathology 2021, 111, 137–148. [Google Scholar] [CrossRef]
- Lu, S.-W.; Tian, D.; Borchardt-Wier, H.B.; Wang, X. Alternative splicing: A novel mechanism of regulation identified in the chorismate mutase gene of the potato cyst nematode Globodera rostochiensis. Mol. Biochem. Parasitol. 2008, 162, 1–15. [Google Scholar] [CrossRef]
- Grutzmann, K.; Szafranski, K.; Pohl, M.; Voigt, K.; Petzold, A.; Schuster, S. Fungal Alternative Splicing is Associated with Multicellular Complexity and Virulence: A Genome-Wide Multi-Species Study. DNA Res. 2013, 21, 27–39. [Google Scholar] [CrossRef] [PubMed]
- Savory, E.A.; Zou, C.; Adhikari, B.N.; Hamilton, J.P.; Buell, C.R.; Shiu, S.-H.; Day, B. Alternative Splicing of a Multi-Drug Transporter from Pseudoperonospora cubensis Generates an RXLR Effector Protein That Elicits a Rapid Cell Death. PLoS ONE 2012, 7, e34701. [Google Scholar] [CrossRef] [PubMed]
- Hardwick, S.A.; Joglekar, A.; Flicek, P.; Frankish, A.; Tilgner, H.U. Getting the entire message: Progress in isoform sequencing. Front. Genet. 2019, 10, 709. [Google Scholar] [CrossRef] [PubMed]
- Steijger, T.; Abril, J.F.; Engström, P.G.; Kokocinski, F.; Hubbard, T.J.; Guigó, R.; Harrow, J.; Bertone, P. Assessment of transcript reconstruction methods for RNA-seq. Nat. Methods 2013, 10, 1177–1184. [Google Scholar] [CrossRef]
- Lian, Y.; Wei, H.; Wang, J.; Lei, C.; Li, H.; Li, J.; Wu, Y.; Wang, S.; Zhang, H.; Wang, T.; et al. Chromosome-level reference genome of X12, a highly virulent race of the soybean cyst nematode Heterodera glycines. Mol. Ecol. Resour. 2019, 19, 1637–1646. [Google Scholar] [CrossRef]
- Masonbrink, R.; Maier, T.R.; Muppirala, U.; Seetharam, A.S.; Lord, E.; Juvale, P.S.; Schmutz, J.; Johnson, N.T.; Korkin, D.; Mitchum, M.G.; et al. The genome of the soybean cyst nematode (Heterodera glycines) reveals complex patterns of duplications involved in the evolution of parasitism genes. BMC Genom. 2019, 20, 119. [Google Scholar] [CrossRef] [PubMed]
- Masonbrink, R.E.; Maier, T.R.; Hudson, M.; Severin, A.; Baum, T. A chromosomal assembly of the soybean cyst nematode genome. Mol. Ecol. Resour. 2021, 21, 2407–2422. [Google Scholar] [CrossRef] [PubMed]
- Serra, L.; Chang, D.; Macchietto, M.; Williams, K.; Murad, R.; Lu, D.; Dillman, A.; Mortazavi, A. Adapting the smart-seq2 protocol for robust single worm rna-seq. BIO-PROTOC 2018, 8, e2729. [Google Scholar] [CrossRef]
- Oikonomopoulos, S.; Wang, Y.C.; Djambazian, H.; Badescu, D.; Ragoussis, J. Benchmarking of the Oxford Nanopore MinION sequencing for quantitative and qualitative assessment of cDNA populations. Sci. Rep. 2016, 6, 31602. [Google Scholar] [CrossRef]
- Tilgner, H.; Grubert, F.; Sharon, D.; Snyder, M.P. Defining a personal, allele-specific, and single-molecule long-read transcriptome. Proc. Natl. Acad. Sci. USA 2014, 111, 9869–9874. [Google Scholar] [CrossRef]
- Grünberger, F.; Ferreira-Cerca, S.; Grohmann, D. Nanopore sequencing of RNA and cDNA molecules in Escherichia coli. RNA 2021, 28, 400–417. [Google Scholar] [CrossRef] [PubMed]
- Klink, V.P.; Hosseini, P.; MacDonald, M.H.; Alkharouf, N.W.; Matthews, B.F. Population-specific gene expression in the plant pathogenic nematode Heterodera glycines exists prior to infection and during the onset of a resistant or susceptible reaction in the roots of the Glycine max genotype Peking. BMC Genom. 2009, 10, 111. [Google Scholar] [CrossRef] [PubMed]
- Liu, S.; Kandoth, P.K.; Lakhssassi, N.; Kang, J.; Colantonio, V.; Heinz, R.; Yeckel, G.; Zhou, Z.; Bekal, S.; Dapprich, J.; et al. The soybean GmSNAP18 gene underlies two types of resistance to soybean cyst nematode. Nature Commun. 2017, 8, 14822. [Google Scholar] [CrossRef] [PubMed]
- Bayless, A.M.; Smith, J.M.; Song, J.; McMinn, P.H.; Teillet, A.; August, B.K.; Bent, A.F. Disease resistance through impairment of α-SNAP–NSF interaction and vesicular trafficking by soybean Rhg1. Proc. Natl. Acad. Sci. USA 2016, 113, E7375–E7382. [Google Scholar] [CrossRef] [PubMed]
- Dong, K.; Opperman, C.H. Genetic Analysis of Parasitism in the Soybean Cyst Nematode Heterodera glycines. Genetics 1997, 146, 1311–1318. [Google Scholar] [CrossRef]
- Da Rocha, M.; Bournaud, C.; Dazenière, J.; Thorpe, P.; Bailly-Bechet, M.; Pellegrin, C.; Péré, A.; Grynberg, P.; Perfus-Barbeoch, L.; Eves-van den Akker, S.; et al. Genome Expression Dynamics Reveal the Parasitism Regulatory Landscape of the Root-Knot Nematode Meloidogyne incognita and a Promoter Motif Associated with Effector Genes. Genes 2021, 12, 771. [Google Scholar] [CrossRef]
- Tritten, L.; Ballesteros, C.; Beech, R.; Geary, T.G.; Moreno, Y. Mining nematode protein secretomes to explain lifestyle and host specificity. PLoS Negl. Trop. Dis. 2021, 15, e0009828. [Google Scholar] [CrossRef]
- Saverwyns, H.; Visser, A.; Van Durme, J.; Power, D.; Morgado, I.; Kennedy, M.W.; Knox, D.P.; Schymkowitz, J.; Rousseau, F.; Gevaert, K.; et al. Analysis of the transthyretin-like (TTL) gene family in Ostertagia ostertagi—Comparison with other strongylid nematodes and Caenorhabditis elegans. Int. J. Parasitol. 2008, 38, 1545–1556. [Google Scholar] [CrossRef]
- Lin, B.; Zhuo, K.; Chen, S.; Hu, L.; Sun, L.; Wang, X.; Zhang, L.; Liao, J. A novel nematode effector suppresses plant immunity by activating host reactive oxygen species-scavenging system. New Phytol. 2015, 209, 1159–1173. [Google Scholar] [CrossRef]
- Espada, M.; Vicente, C.; Branco, J.; Mota, M.; Vieira, P. The role of Pratylenchus penetrans transthyretin-like protein in parasitism. In Proceedings of the International Symposium of Crop Protection, Gent, Bélgica, 18 May 2021. [Google Scholar]
- Kandoth, P.K.; Ithal, N.; Recknor, J.; Maier, T.; Nettleton, D.; Baum, T.J.; Mitchum, M.G. The Soybean Rhg1 Locus for Resistance to the Soybean Cyst Nematode Heterodera glycines Regulates the Expression of a Large Number of Stress- and Defense-Related Genes in Degenerating Feeding Cells. Plant Physiol. 2011, 155, 1960–1975. [Google Scholar] [CrossRef]
- Kirschke, H. Cathepsins. In xPharm: The Comprehensive Pharmacology Reference; Elsevier: Amsterdam, The Netherlands, 2007; pp. 1–4. [Google Scholar] [CrossRef]
- Li, Y.; Wang, K.; Xie, H.; Wang, D.-W.; Xu, C.-L.; Huang, X.; Wu, W.-J.; Li, D.-L. Cathepsin B Cysteine Proteinase is Essential for the Development and Pathogenesis of the Plant Parasitic Nematode Radopholus similis. Int. J. Mol. Sci. 2015, 11, 1073–1087. [Google Scholar] [CrossRef] [PubMed]
- Shinya, R.; Kirino, H.; Morisaka, H.; Takeuchi-Kaneko, Y.; Futai, K.; Ueda, M. Comparative Secretome and Functional Analyses Reveal Glycoside Hydrolase Family 30 and Cysteine Peptidase as Virulence Determinants in the Pinewood Nematode Bursaphelenchus xylophilus. Front. Plant Sci. 2021, 12, 640459. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.-L.; Cheng, X.; Ding, S.-W.; Wang, D.-W.; Chen, C.; Xu, C.-L.; Xie, H. Molecular identification and functional characterization of the cathepsin B gene (Ab-cb-1) in the plant parasitic nematode Aphelenchoides besseyi. PLoS ONE 2018, 13, e0199935. [Google Scholar] [CrossRef]
- Wang, K.; Li, Y.; Huang, X.; Wang, D.; Xu, C.; Xie, H. The cathepsin S cysteine proteinase of the burrowing nematode Radopholus similis is essential for the reproduction and invasion. Cell Biosci. 2016, 6, 39. [Google Scholar] [CrossRef]
- Xue, Q.; Wu, X.-Q.; Zhang, W.-J.; Deng, L.-N.; Wu, M.-M. Cathepsin L-like Cysteine Proteinase Genes Are Associated with the Development and Pathogenicity of Pine Wood Nematode, Bursaphelenchus xylophilus. Int. J. Mol. Sci. 2019, 20, 215. [Google Scholar] [CrossRef] [PubMed]
- Dutta, T.K.; Papolu, P.K.; Banakar, P.; Choudhary, D.; Sirohi, A.; Rao, U. Tomato transgenic plants expressing hairpin construct of a nematode protease gene conferred enhanced resistance to root-knot nematodes. Front. Microbiol. 2015, 6, 260. [Google Scholar] [CrossRef]
- Chen, C.; Chen, Y.; Jian, H.; Yang, D.; Dai, Y.; Pan, L.; Shi, F.; Yang, S.; Lui, Q. Large-Scale Identification and Characterization of Heterodera avenae Putative Effectors Suppressing or Inducing Cell Death in Nicotiana benthamiana. Front. Plant Sci. 2018, 8, 2062. [Google Scholar] [CrossRef]
- Mathew, R. Genomics and Management of the Burrowing Nematode, Radopholus similis. Ph.D. Thesis, North Carolina State University, Raleigh, NC, USA, 2020. [Google Scholar]
- Vieira, P.; Shao, J.; Vijayapalani, P.; Maier, T.R.; Pellegrin, C.; Eves-van den Akker, S.; Baum, T.J.; Nemchinov, L.G. A new esophageal gland transcriptome reveals signatures of large scale de novo effector birth in the root lesion nematode Pratylenchus penetrans. BMC Genom. 2020, 21, 738. [Google Scholar] [CrossRef]
- Hashmi, S.; Zhang, J.; Oksov, Y.; Lustigman, S. The Caenorhabditis elegans Cathepsin Z-like Cysteine Protease, Ce-CPZ-1, Has a Multifunctional Role during the Worms’ Development. J. Biol. Chem. 2004, 279, 6035–6045. [Google Scholar] [CrossRef]
- Lustigman, S.; McKerrow, J.H.; Shah, K.; Lui, J.; Huima, T.; Hough, M.; Brotman, B. Cloning of a Cysteine Protease Required for the Molting of Onchocerca volvulus Third Stage Larvae. J. Biol. Chem. 1996, 271, 30181–30189. [Google Scholar] [CrossRef]
- Yadati, T.; Houben, T.; Bitorina, A.; Shiri-Sverdlov, R. The ins and outs of cathepsins: Physiological function and role in disease management. Cells 2020, 9, 1679. [Google Scholar] [CrossRef]
- Qin, X.; Xue, B.; Tian, H.; Fang, C.; Yu, J.; Chen, C.; Xue, Q.; Jones, J.; Wang, X. An unconventionally secreted effector from the root knot nematode Meloidogyne incognita, Mi-ISC-1, promotes parasitism by disrupting salicylic acid biosynthesis in host plants. Mol. Plant Pathol. 2021, 23, 516–529. [Google Scholar] [CrossRef] [PubMed]
- Morillon, A.; Gautheret, D. Bridging the gap between reference and real transcriptomes. Genome Biol. 2019, 20, 112. [Google Scholar] [CrossRef] [PubMed]
- Abubucker, S.; McNulty, S.N.; Rosa, B.A.; Mitreva, M. Identification and characterization of alternative splicing in parasitic nematode transcriptomes. Parasites Vectors 2014, 7, 151. [Google Scholar] [CrossRef] [PubMed]
- Ramani, A.K.; Calarco, J.A.; Pan, Q.; Mavandadi, S.; Wang, Y.; Nelson, A.C.; Lee, L.J.; Morris, Q.; Blencowe, B.J.; Zhen, M.; et al. Genome-wide analysis of alternative splicing in Caenorhabditis elegans. Genome Res. 2010, 21, 342–348. [Google Scholar] [CrossRef]
- C. elegans Sequencing Consortium. Genome Sequence of the Nematode C. elegans: A Platform for Investigating Biology. Science 1998, 282, 2012–2018. [CrossRef]
- van Steenbrugge, J.J.M.; van den Elsen, S.; Holterman, M.; Sterken, M.G.; Thorpe, P.; Goverse, A.; Smant, G.; Helder, J. Comparative genomics of two inbred lines of the potato cyst nematode Globodera rostochiensis reveals disparate effector family-specific diversification patterns. BMC Genom. 2021, 22, 611. [Google Scholar] [CrossRef] [PubMed]
- Bhargava, V.; Head, S.R.; Ordoukhanian, P.; Mercola, M.; Subramaniam, S. Technical variations in low-input rna-seq methodologies. Sci. Rep. 2014, 4, 3678. [Google Scholar] [CrossRef]
- Gendron St-Marseille, A.-F.; Lord, E.; Véronneau, P.-Y.; Brodeur, J.; Mimee, B. Genome Scans Reveal Homogenization and Local Adaptations in Populations of the Soybean Cyst Nematode. Front. Plant Sci. 2018, 9, 987. [Google Scholar] [CrossRef]
- Niblack, T.L.; Arelli, P.R.; Noel, G.R.; Opperman, C.H.; Orf, J.H.; Schmitt, D.P.; Shannon, J.G.; Tylka, G.L. A Revised Classification Scheme for Genetically Diverse Populations of Heterodera glycines. J. Nematol. 2002, 34, 279–288. [Google Scholar]
- Audette, C.; Bélanger, R.R.; Mimee, B. Coinfection of soybean plants with Phytophthora sojae and soybean cyst nematode does not alter the efficacy of resistance genes. Plant Path 2020, 69, 1437–1444. [Google Scholar] [CrossRef]
- Eisenback, J. Techniques for measuring nematode development and egg production. In Laboratory Techniques in Nematode Ecology; Wheeler, T., Ed.; Society of Nematologists: Hyattsville, MD, USA, 2000; pp. 1–4. [Google Scholar]
- Handoo, Z.A.; Anand, S.C. Biological manifestation of resistance to soybean cyst nematode development in ‘Hartwig’ soybean. Crop Prot. 1993, 12, 371–372. [Google Scholar] [CrossRef]
- Chang, D.; Serra, L.; Lu, D.; Mortazavi, A.; Dillman, A. A revised adaptation of the smart-seq2 protocol for single-nematode rna-seq. In Methods in Molecular Biology; Springer: New York, NY, USA, 2020; pp. 79–99. [Google Scholar] [CrossRef]
- Lh3/seqtk: Toolkit for Processing Sequences in FASTA/Q Formats. Available online: https://github.com/lh3/seqtk (accessed on 12 September 2020).
- Danecek, P.; Bonfield, J.K.; Liddle, J.; Marshall, J.; Ohan, V.; Pollard, M.O.; Whitwham, A.; Keane, T.; McCarthy, S.A.; Davies, R.M.; et al. Twelve years of SAMtools and BCFtools. GigaScience 2021, 10, giab008. [Google Scholar] [CrossRef]
- Kovaka, S.; Zimin, A.V.; Pertea, G.M.; Razaghi, R.; Salzberg, S.L.; Pertea, M. Transcriptome assembly from long-read RNA-seq alignments with StringTie2. Genome Biol. 2019, 20, 278. [Google Scholar] [CrossRef] [PubMed]
- Camargo, A.P.; Sourkov, V.; Pereira, G.A.G.; Carazzolle, M.F. RNAsamba: Neural network-based assessment of the protein-coding potential of RNA sequences. NAR Genom. Bioinform. 2020, 2, lqz024. [Google Scholar] [CrossRef]
- Li, W.; Godzik, A. Cd-hit: A fast program for clustering and comparing large sets of protein or nucleotide sequences. Bioinformatics 2006, 22, 1658–1659. [Google Scholar] [CrossRef] [PubMed]
- Li, H. Minimap2: Pairwise alignment for nucleotide sequences. Bioinformatics 2018, 34, 3094–3100. [Google Scholar] [CrossRef]
- Tang, A.D.; Soulette, C.M.; van Baren, M.J.; Hart, K.; Hrabeta-Robinson, E.; Wu, C.J.; Brooks, A.N. Full-length transcript characterization of SF3B1 mutation in chronic lymphocytic leukemia reveals downregulation of retained introns. Nat. Commun. 2020, 11, 1438. [Google Scholar] [CrossRef] [PubMed]
- Robinson, M.D.; McCarthy, D.J.; Smyth, G.K. edgeR: A Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 2009, 26, 139–140. [Google Scholar] [CrossRef]
- Almagro Armenteros, J.J.; Tsirigos, K.D.; Sønderby, C.K.; Petersen, T.N.; Winther, O.; Brunak, S.; von Heijne, G.; Nielson, H. SignalP 5.0 improves signal peptide predictions using deep neural networks. Nat. Biotechnol. 2019, 37, 420–423. [Google Scholar] [CrossRef]
- Bendtsen, J.D.; Kiemer, L.; Fausbøll, A.; Brunak, S. Non-classical protein secretion in bacteria. BMC Microbiol. 2005, 5, 58. [Google Scholar] [CrossRef] [PubMed]
- Cohen, M.J.; Chirico, W.J.; Lipke, P.N. Through the back door: Unconventional protein secretion. Cell Surf. 2020, 6, 100045. [Google Scholar] [CrossRef]
- Reindl, M.; Hänsch, S.; Weidtkamp-Peters, S.; Schipper, K. A potential lock-type mechanism for unconventional secretion in fungi. Int. J. Mol. Sci. 2019, 20, 460. [Google Scholar] [CrossRef]
- Krogh, A.; Larsson, B.; von Heijne, G.; Sonnhammer, E.L.L. Predicting transmembrane protein topology with a hidden markov model: Application to complete genomes. JMB 2001, 305, 567–580. [Google Scholar] [CrossRef]
- Almagro Armenteros, J.J.; Sønderby, C.K.; Sønderby, S.K.; Nielsen, H.; Winther, O. DeepLoc: Prediction of protein subcellular localization using deep learning. Bioinformatics 2017, 33, 4049. [Google Scholar] [CrossRef]
- Sabeh, M.; Duceppe, M.-O.; St-Arnaud, M.; Mimee, B. Transcriptome-wide selection of a reliable set of reference genes for gene expression studies in potato cyst nematodes (Globodera spp.). PLoS ONE 2018, 13, e0193840. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Dobin, A.; Davis, C.A.; Schlesinger, F.; Drenkow, J.; Zaleski, C.; Jha, S.; Batut, P.; Chaisson, M.; Gingeras, T.R. STAR: Ultrafast universal RNA-seq aligner. Bioinformatics 2012, 29, 15–21. [Google Scholar] [CrossRef]
- Patro, R.; Duggal, G.; Love, M.I.; Irizarry, R.A.; Kingsford, C. Salmon provides fast and bias-aware quantification of transcript expression. Nat. Methods 2017, 14, 417–419. [Google Scholar] [CrossRef]


| Population Name | Original Field Location | Female Index | HG-Type | Experiment |
|---|---|---|---|---|
| QCSTJ5 | Blenheim, Ontario, Canada | 16.17.5.0- | 1.2- | Sequencing |
| QCSTJ6 | Bothwell, Ontario, Canada | 11.6.7.11- | 1.4- | Sequencing—qPCR |
| QCSTJ15 | Kingsville, Ontario, Canada | 8.18.3.4- | 2- | Sequencing—qPCR |
| QCSTJ-US2 | Brown County, Ohio, USA | 1.19.-.0- | 2- | qPCR |
| Transcript id | Log2FC | Adj. p-Val. | Localization | SP 1 | SecP 2 | GE 3 | Blastx Annotations | E-Value |
|---|---|---|---|---|---|---|---|---|
| Hetgly01325.t1 | 3.873 | 7.72 × 10−13 | Cytoplasm|Nucleus | 0.00 | 0.90 | Y | KAI1731729.1 Protein kinase domain-containing protein [Ditylenchus destructor] | 1.24 × 10−152 |
| Hetgly02290.t1 | 2.852 | 2.34 × 10−12 | Extracellular | 1.00 | 0.70 | N | --NA-- | --NA-- |
| Hetgly04490.t1 | 3.243 | 8.65 × 10−16 | Lysosome/Vacuole | 1.00 | 0.74 | Y | KAF7636717.1 Glyco_hydro_35 domain-containing protein [Meloidogyne graminicola] | 0.0 |
| Hetgly04491.t1 | 4.034 | 4.20 × 10−11 | Extracellular | 1.00 | 0.90 | Y | CAM84511.1 transthyretin-like protein 2 precursor [Radopholus similis] | 7.48 × 10−61 |
| Hetgly06729.t1 | 2.931 | 9.62 × 10−14 | Nucleus | 0.00 | 0.81 | Y | KAH7714403.1 U1 small nuclear ribonucleoprotein A [Aphelenchus avenae] | 4.53 × 10−93 |
| Hetgly06908.t1 | 4.213 | 5.23 × 10−13 | Extracellular | 1.00 | 0.69 | N | --NA-- | --NA-- |
| Hetgly07025.t1 | 4.347 | 2.86 × 10−15 | Mitochondrion | 0.00 | 0.85 | Y | KAH7694343.1 Protein MRPL-14 b [Aphelenchus avenae] | 9.09 × 10−62 |
| Hetgly07480.t1 | 2.642 | 2.76 × 10−11 | Extracellular | 1.00 | 0.87 | N | CAD2152915.1 unnamed protein product [Meloidogyne enterolobii] | 4.15 × 10−33 |
| Hetgly08734.t1 | 3.360 | 5.16 × 10−11 | Extracellular | 1.00 | 0.93 | N | --NA-- | --NA-- |
| Hetgly08995.t1 | 2.261 | 1.95 × 10−11 | Mitochondrion | 0.00 | 0.76 | Y | KAF7638520.1 Acyl carrier protein [Meloidogyne graminicola] | 2.08 × 10−26 |
| Hetgly09989.t1 | 2.669 | 3.05 × 10−13 | Nucleus | 0.00 | 0.79 | Y | --NA-- | --NA-- |
| Hetgly10002.t1 | 2.684 | 8.59 × 10−11 | Nucleus | 0.00 | 0.77 | Y | KAF7637276.1 RRM domain-containing protein [Meloidogyne graminicola] | 2.34 × 10−119 |
| Hetgly15196.t1 | 3.234 | 1.07 × 10−13 | Endoplasmic reticulum | 1.00 | 0.48 | Y | KAI1725617.1 thioredoxin domain-containing protein [Ditylenchus destructor] | 0.0 |
| Hetgly15644.t1 | 3.647 | 5.74 × 10−13 | Peroxisome | 0.00 | 0.78 | Y | KAI1705849.1 SCP-2 sterol transfer family domain-containing protein [Ditylenchus destructor] | 0.0 |
| Hetgly15709.t1 | 2.608 | 1.18 × 10−11 | Cytoplasm | 0.00 | 0.81 | N | KAH7727199.1 catalase B [Aphelenchus avenae] | 1.27 × 10−29 |
| Hetgly17937.t1 | 4.284 | 9.62 × 10−14 | Cytoplasm | 0.00 | 0.77 | Y | --NA-- | --NA-- |
| Hetgly18109.t1 | 4.526 | 5.09 × 10−13 | Mitochondrion | 0.00 | 0.91 | Y | KHN88347.1 28S ribosomal protein S18a, mitochondrial [Toxocara canis] | 3.00 × 10−60 |
| Hetgly18314.t1 | 3.235 | 1.14 × 10−11 | Cytoplasm | 0.00 | 0.79 | Y | KAH7727561.1 60S ribosomal protein L14 [Aphelenchus avenae] | 2.41 × 10−58 |
| Hetgly20098.t1 | 3.274 | 7.98 × 10−11 | Cytoplasm | 0.00 | 0.82 | Y | KAH7730990.1 60S ribosomal protein L34 [Aphelenchus avenae] | 2.39 × 10−52 |
| Hetgly20289.t1 | 3.348 | 2.29 × 10−11 | Cytoplasm | 0.00 | 0.81 | Y | KAI1728922.1 ribosomal protein s19e domain-containing protein [Ditylenchus destructor] | 1.00 × 10−70 |
| Hetgly21523.t1 | 4.403 | 8.52 × 10−12 | Endoplasmic reticulum | 0.00 | 0.93 | N | --NA-- | --NA-- |
| Hetgly21677.t1 | 4.491 | 2.17 × 10−12 | Extracellular | 1.00 | 0.43 | N | KAF7634869.1 AAA domain-containing protein [Meloidogyne graminicola] | 1.02 × 10−41 |
| Hetgly21799.t1 | 3.238 | 1.63 × 10−11 | Nucleus | 0.00 | 0.75 | Y | CAD2202259.1 unnamed protein product [Meloidogyne enterolobii] | 9.50 × 10−44 |
| Hg11990.1 | 2.169 | 5.19 × 10−11 | Endoplasmic reticulum | 1.00 | 0.30 | Y | KAI1725554.1 hsp70 protein domain-containing protein [Ditylenchus destructor] | 2.11 × 10−139 |
| Hg11990.3 | 2.874 | 6.06 × 10−11 | Endoplasmic reticulum | 1.00 | 0.30 | Y | KAI1725554.1 hsp70 protein domain-containing protein [Ditylenchus destructor] | 1.36 × 10−40 |
| Hg16414.1 | 3.474 | 2.76 × 10−11 | Extracellular | 0.97 | 0.69 | Y | AVA09678.1 putative effector protein [Heterodera avenae] | 6.27 × 10−117 |
| Hg17279.2 | 3.423 | 1.01 × 10−11 | Cytoplasm|Nucleus | 0.00 | 0.82 | Y | KAI1717340.1 ribosomal protein l21e domain-containing protein [Ditylenchus destructor] | 2.49 × 10−91 |
| Hg17540.2 | 3.303 | 3.95 × 10−11 | Nucleus | 0.00 | 0.77 | Y | KAF7639258.1 40S ribosomal protein S6, partial [Meloidogyne graminicola] | 3.31 × 10−140 |
| Hg4745.1 | 3.307 | 1.36 × 10−13 | Cell membrane | 0.00 | 0.88 | Y | --NA-- | --NA-- |
| Hg835.1 | 2.835 | 3.88 × 10−12 | Nucleus | 0.00 | 0.76 | N | KAI1727318.1 CAF1 family ribonuclease domain-containing protein [Ditylenchus destructor] | 8.24 × 10−43 |
| Hg8859.1 | 4.145 | 8.37 × 10−20 | Extracellular | 0.00 | 0.76 | Y | AVA09687.1 putative effector protein [Heterodera avenae] | 0.0 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 His Majesty the King in Right of Canada; Licensee MDPI, Basel, Switzerland. This is an open access article distributed under the terms of the Creative Commons Attribution IGO License (CC BY) license (http://creativecommons.org/licenses/by/3.0/igo), which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
Share and Cite
Ste-Croix, D.T.; Bélanger, R.R.; Mimee, B. Single Nematode Transcriptomic Analysis, Using Long-Read Technology, Reveals Two Novel Virulence Gene Candidates in the Soybean Cyst Nematode, Heterodera glycines. Int. J. Mol. Sci. 2023, 24, 9440. https://doi.org/10.3390/ijms24119440
Ste-Croix DT, Bélanger RR, Mimee B. Single Nematode Transcriptomic Analysis, Using Long-Read Technology, Reveals Two Novel Virulence Gene Candidates in the Soybean Cyst Nematode, Heterodera glycines. International Journal of Molecular Sciences. 2023; 24(11):9440. https://doi.org/10.3390/ijms24119440
Chicago/Turabian StyleSte-Croix, Dave T., Richard R. Bélanger, and Benjamin Mimee. 2023. "Single Nematode Transcriptomic Analysis, Using Long-Read Technology, Reveals Two Novel Virulence Gene Candidates in the Soybean Cyst Nematode, Heterodera glycines" International Journal of Molecular Sciences 24, no. 11: 9440. https://doi.org/10.3390/ijms24119440
APA StyleSte-Croix, D. T., Bélanger, R. R., & Mimee, B. (2023). Single Nematode Transcriptomic Analysis, Using Long-Read Technology, Reveals Two Novel Virulence Gene Candidates in the Soybean Cyst Nematode, Heterodera glycines. International Journal of Molecular Sciences, 24(11), 9440. https://doi.org/10.3390/ijms24119440






