DNA:RNA Hybrids Are Major Dinoflagellate Minicircle Molecular Types
Abstract
1. Introduction
2. Results
2.1. Minicircles Contain DNA:RNA Hybrids (DRHs)
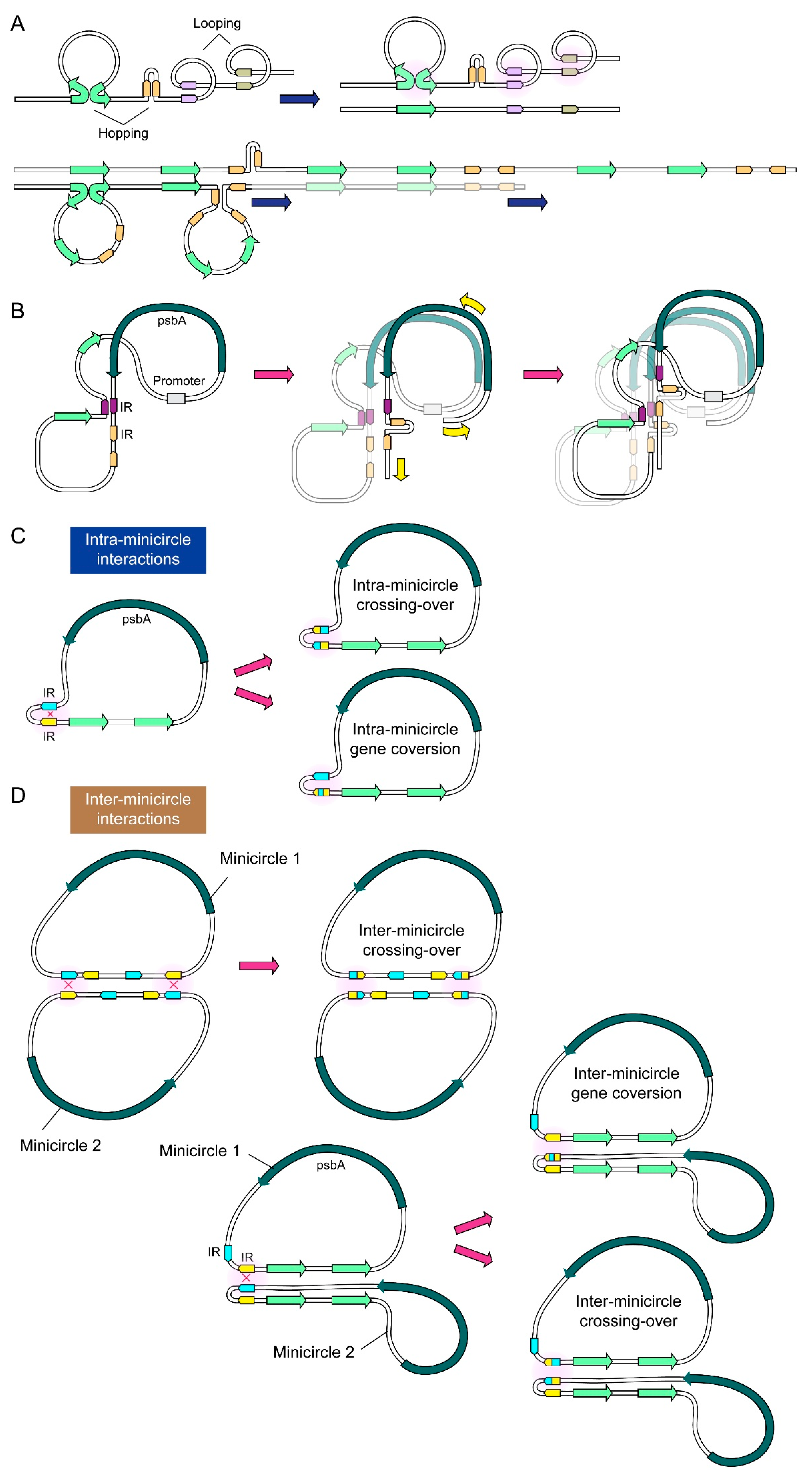
2.2. NCR Mediated Multiple Conversion/Crossing-over Events That Were Restricted after Incubation in the Dinoflagellate Cell Lysate
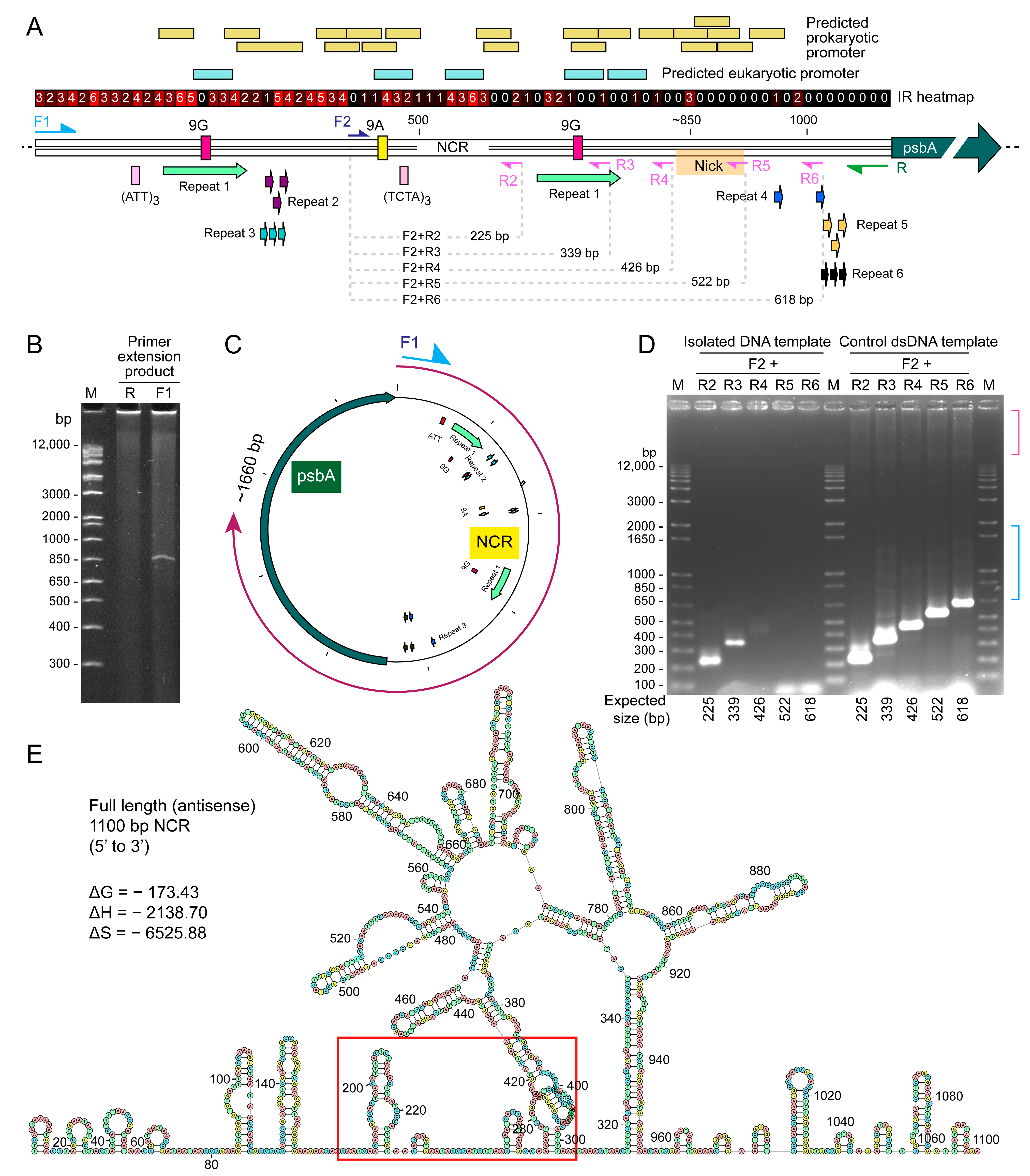
2.3. Distinct Sized PCR Products with Immobilized PCR Corresponded with Predicted Secondary Structure Locations and Potential Sites for Recombinations
2.4. Immobilized Template Generated Discrete Double Strand and Single Strand PCR Products
3. Discussion
4. Materials and Methods
4.1. Culture of Dinoflagellates
4.2. Nucleic Acid Extraction
4.3. Primer Extension Assay
4.4. Gel Retardation Assay and Immobilizated Minicircular DNA as PCR Template
4.5. In Silico Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Jarvis, P.; López-Juez, E. Biogenesis and homeostasis of chloroplasts and other plastids. Nat. Rev. Mol. Cell Biol. 2013, 14, 787–802. [Google Scholar] [CrossRef] [PubMed]
- Martin, G.; Baurens, F.C.; Cardi, C.; Aury, J.M.; D’Hont, A. The complete chloroplast genome of banana (Musa acuminata, Zingiberales): Insight into plastid monocotyledon evolution. PLoS ONE 2013, 8, e67350. [Google Scholar] [CrossRef] [PubMed]
- Nielsen, A.Z.; Mellor, S.B.; Vavitsas, K.; Wlodarczyk, A.J.; Gnanasekaran, T.; Perestrello Ramos, H.D.J.M.; King, B.C.; Bakowski, K.; Jensen, P.E. Extending the biosynthetic repertoires of cyanobacteria and chloroplasts. Plant J. 2016, 87, 87–102. [Google Scholar] [CrossRef] [PubMed]
- Song, W.; Ji, C.; Chen, Z.; Cai, H.; Wu, X.; Shi, C.; Wang, S. Comparative Analysis the Complete Chloroplast Genomes of Nine Musa Species: Genomic Features, Comparative Analysis, and Phylogenetic Implications. Front. Plant Sci. 2022, 13, 832884. [Google Scholar] [CrossRef] [PubMed]
- Onodera, K.-I.; Konishi, Y.; Taguchi, T.; Kiyoto, S.; Tominaga, A. Peridinin from the Marine Symbiotic Dinoflagellate, Symbiodinium sp., Regulates Eosinophilia in Mice. Mar. Drugs 2014, 12, 1773–1787. [Google Scholar] [CrossRef] [PubMed]
- Bustillos-Guzman, J.; Garate-Lizarraga, I.; Lopez-Cortes, D.; Hernandez-Sandoval, F. The use of pigment “fingerprints” in the study of harmful algal blooms. Rev. Biol. Trop. 2004, 52 (Suppl. S1), 17–26. [Google Scholar]
- Jeffrey, S.W.; Sielicki, M.; Haxo, F.T. Chloroplast Pigment Patterns in Dinoflagellates. J. Phycol. 1975, 11, 374–384. [Google Scholar] [CrossRef]
- Niedzwiedzki, D.M.; Jiang, J.; Lo, C.S.; Blankenship, R.E. Low-temperature spectroscopic properties of the peridinin-chlorophyll a-protein (PCP) complex from the coral symbiotic dinoflagellate Symbiodinium. J. Phys. Chem. B 2013, 117, 11091–11099. [Google Scholar] [CrossRef]
- Jeffrey, S.W.; Haxo, F.T. Photosynthetic Pigments of Symbiotic Dinoflagellates (Zooxanthellae) from Corals and Clams. Biol. Bull. 1968, 135, 149–165. [Google Scholar] [CrossRef]
- Zhang, Z.; Cavalier-Smith, T.; Green, B.R. Evolution of dinoflagellate unigenic minicircles and the partially concerted divergence of their putative replicon origins. Mol. Biol. Evol. 2002, 19, 489–500. [Google Scholar] [CrossRef]
- Zhang, Z.; Green, B.R.; Cavalier-Smith, T. Single gene circles in dinoflagellate chloroplast genomes. Nature 1999, 400, 155–159. [Google Scholar] [CrossRef]
- Mungpakdee, S.; Shinzato, C.; Takeuchi, T.; Kawashima, T.; Koyanagi, R.; Hisata, K.; Tanaka, M.; Goto, H.; Fujie, M.; Lin, S.; et al. Massive gene transfer and extensive RNA editing of a symbiotic dinoflagellate plastid genome. Genome Biol. Evol. 2014, 6, 1408–1422. [Google Scholar] [CrossRef]
- Koumandou, V.L.; Nisbet, R.E.; Barbrook, A.C.; Howe, C.J. Dinoflagellate chloroplasts—Where have all the genes gone? Trends Genet. 2004, 20, 261–267. [Google Scholar] [CrossRef]
- Lin, S. Genomic understanding of dinoflagellates. Res. Microbiol. 2011, 162, 551–569. [Google Scholar] [CrossRef]
- Asaf, S.; Khan, A.L.; Khan, M.A.; Shahzad, R.; Lubna; Kang, S.M.; Al-Harrasi, A.; Al-Rawahi, A.; Lee, I.-J. Complete chloroplast genome sequence and comparative analysis of loblolly pine (Pinus taeda L.) with related species. PLoS ONE 2018, 13, e0192966. [Google Scholar] [CrossRef]
- Yoshioka-Nishimura, M. Close Relationships Between the PSII Repair Cycle and Thylakoid Membrane Dynamics. Plant Cell Physiol. 2016, 57, 1115–1122. [Google Scholar] [CrossRef]
- Zhang, Y.; Zhang, A.; Li, X.; Lu, C. The Role of Chloroplast Gene Expression in Plant Responses to Environmental Stress. Int. J. Mol. Sci. 2020, 21, 6082. [Google Scholar] [CrossRef]
- Chang, S.S.; Prézelin, B.B.; Trench, R.K. Mechanisms of photoadaptation in three strains of the symbiotic dinoflagellate Symbiodinium microadriaticum. Mar. Biol. 1983, 76, 219–229. [Google Scholar] [CrossRef]
- Adir, N.; Zer, H.; Shochat, S.; Ohad, I. Photoinhibition—A historical perspective. Photosynth. Res. 2003, 76, 343–370. [Google Scholar] [CrossRef]
- Laatsch, T.; Zauner, S.; Stoebe-Maier, B.; Kowallik, K.V.; Maier, U.G. Plastid-derived single gene minicircles of the dinoflagellate Ceratium horridum are localized in the nucleus. Mol. Biol. Evol. 2004, 21, 1318–1322. [Google Scholar] [CrossRef]
- Owari, S.; Hayashi, A.; Ishida, K.-I. Subcellular localization of minicircle DNA in the dinoflagellate Amphidinium massartii. Phycol. Res. 2014, 62, 1–8. [Google Scholar] [CrossRef]
- Nassoury, N.; Wang, Y.; Morse, D. Brefeldin a inhibits circadian remodeling of chloroplast structure in the dinoflagellate Gonyaulax. Traffic 2005, 6, 548–561. [Google Scholar] [CrossRef]
- Trench, R.K.; Blank, R.J. Symbiodinium microadriaticum freudenthal, S. goreauii sp. Nov., S. kawagutii sp. nov. and S. pilosum sp. nov.: Gymnodinioid dinoflagellate symbionts of marine invertebrates 1. J. Phycol. 1987, 23, 469–481. [Google Scholar] [CrossRef]
- Nielsen, B.L.; Lu, Z.; Tewari, K.K. Characterization of the pea chloroplast DNA OriA region. Plasmid 1993, 30, 197–211. [Google Scholar] [CrossRef] [PubMed]
- Dang, Y.; Green, B.R. Long transcripts from dinoflagellate chloroplast minicircles suggest “rolling circle” transcription. J. Biol. Chem. 2010, 285, 5196–5203. [Google Scholar] [CrossRef]
- Shagin, D.A.; Rebrikov, D.V.; Kozhemyako, V.B.; Altshuler, I.M.; Shcheglov, A.S.; Zhulidov, P.A.; Bogdanova, E.A.; Staroverov, D.B.; Rasskazov, V.A.; Lukyanov, S. A novel method for SNP detection using a new duplex-specific nuclease from crab hepatopancreas. Genome Res. 2002, 12, 1935–1942. [Google Scholar] [CrossRef]
- Keller, W.; Crouch, R. Degradation of DNA RNA hybrids by ribonuclease H and DNA polymerases of cellular and viral origin. Proc. Natl. Acad. Sci. USA 1972, 69, 3360–3364. [Google Scholar] [CrossRef]
- Richardson, C.C.; Lehman, I.R.; Kornberg, A. A deoxyribonucleic acid phosphatase-exonuclease from Escherichia coli. ii. characterization of the exonuclease activity. J. Biol. Chem. 1964, 239, 251–258. [Google Scholar] [CrossRef]
- Anisimova, V.E.; Rebrikov, D.V.; Shagin, D.A.; Kozhemyako, V.B.; Menzorova, N.I.; Staroverov, D.B.; Ziganshin, R.; Vagner, L.L.; Rasskazov, V.A.; Lukyanov, S.A.; et al. Isolation, characterization and molecular cloning of duplex-specific nuclease from the hepatopancreas of the Kamchatka crab. BMC Biochem. 2008, 9, 14. [Google Scholar] [CrossRef]
- Leung, S.K.; Wong, J.T.W. The replication of plastid minicircles involves rolling circle intermediates. Nucleic. Acids Res. 2009, 37, 1991–2002. [Google Scholar] [CrossRef]
- Aguilera, A.; Gaillard, H. Transcription and recombination: When RNA meets DNA. Cold Spring Harb. Perspect. Biol. 2014, 6, a016543. [Google Scholar] [CrossRef]
- Kato-Inui, T.; Takahashi, G.; Hsu, S.; Miyaoka, Y. Clustered regularly interspaced short palindromic repeats (CRISPR)/CRISPR-associated protein 9 with improved proof-reading enhances homology-directed repair. Nucleic. Acids Res. 2018, 46, 4677–4688. [Google Scholar] [CrossRef]
- Smith, G.R. Meeting DNA palindromes head-to-head. Genes Dev. 2008, 22, 2612–2620. [Google Scholar] [CrossRef]
- Millard, A.D.; Gierga, G.; Clokie, M.R.; Evans, D.J.; Hess, W.R.; Scanlan, D.J. An antisense RNA in a lytic cyanophage links psbA to a gene encoding a homing endonuclease. ISME J. 2010, 4, 1121–1135. [Google Scholar] [CrossRef]
- Mulo, P.; Sicora, C.; Aro, E.M. Cyanobacterial psbA gene family: Optimization of oxygenic photosynthesis. Cell Mol. Life Sci. 2009, 66, 3697–3710. [Google Scholar] [CrossRef]
- Sakurai, I.; Stazic, D.; Eisenhut, M.; Vuorio, E.; Steglich, C.; Hess, W.R.; Aro, E.M. Positive regulation of psbA gene expression by cis-encoded antisense RNAs in Synechocystis sp. PCC 6803. Plant Physiol. 2012, 160, 1000–1010. [Google Scholar] [CrossRef]
- del Campo, E.M. Post-transcriptional control of chloroplast gene expression. Gene Regul. Syst. Biol. 2009, 3, 31–47. [Google Scholar] [CrossRef]
- Sharwood, R.E.; Hotto, A.M.; Bollenbach, T.J.; Stern, D.B. Overaccumulation of the chloroplast antisense RNA AS5 is correlated with decreased abundance of 5S rRNA in vivo and inefficient 5S rRNA maturation in vitro. RNA 2011, 17, 230–243. [Google Scholar] [CrossRef]
- Wu, M.; Lou, J.K.; Chang, D.Y.; Chang, C.H.; Nie, Z.Q. Structure and function of a chloroplast DNA replication origin of Chlamydomonas reinhardtii. Proc. Natl. Acad. Sci. USA 1986, 83, 6761–6765. [Google Scholar] [CrossRef]
- Kwok, A.C.M.; Zhang, F.; Ma, Z.; Chan, W.S.; Yu, V.C.; Tsang, J.S.H.; Wong, J.T.Y. Functional responses between PMP3 small membrane proteins and membrane potential. Environ. Microbiol. 2020, 22, 3066–3080. [Google Scholar] [CrossRef]
- Wang, Y.; Jensen, L.; Hojrup, P.; Morse, D. Synthesis and degradation of dinoflagellate plastid-encoded psbA proteins are light-regulated, not circadian-regulated. Proc. Natl. Acad. Sci. USA 2005, 102, 2844–2849. [Google Scholar] [CrossRef] [PubMed]
- Kabeya, Y.; Miyagishima, S.Y. Chloroplast DNA replication is regulated by the redox state independently of chloroplast division in Chlamydomonas reinhardtii. Plant Physiol. 2013, 161, 2102–2112. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.; MacCready, J.S.; Ducat, D.C.; Osteryoung, K.W. The Molecular Machinery of Chloroplast Division. Plant Physiol. 2018, 176, 138–151. [Google Scholar] [CrossRef] [PubMed]
- Nakanishi, H.; Suzuki, K.; Kabeya, Y.; Okazaki, K.; Miyagishima, S.Y. Conservation and differences of the Min system in the chloroplast and bacterial division site placement. Commun. Integr. Biol. 2009, 2, 400–402. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Morse, D. The plastid-encoded psbA gene in the dinoflagellate Gonyaulax is not encoded on a minicircle. Gene 2006, 371, 206–210. [Google Scholar] [CrossRef]
- Guillard, R.R.; Ryther, J.H. Studies of marine planktonic diatoms. I. Cyclotella nana Hustedt, and Detonula confervacea (cleve) Gran. Can. J. Microbiol. 1962, 8, 229–239. [Google Scholar] [CrossRef]
- Puchooa, D. A simple, rapid and efficient method for the extraction of genomic DNA from lychee (Litchi chinensis Sonn.). Afr. J. Biotechnol. 2004, 3, 253–255. [Google Scholar]
- Sambrook, J.; Russell, D.W. Molecular Cloning: A Laboratory Manual; Cold Spring Harbor Laboratory Press: Cold Spring Harbor, NY, USA, 2001. [Google Scholar]
- Brazda, V.; Kolomaznik, J.; Lysek, J.; Haronikova, L.; Coufal, J.; St’astny, J. Palindrome analyser—A new web-based server for predicting and evaluating inverted repeats in nucleotide sequences. Biochem. Biophys. Res. Commun. 2016, 478, 1739–1745. [Google Scholar] [CrossRef]
- Okonechnikov, K.; Golosova, O.; Fursov, M. Unipro UGENE: A unified bioinformatics toolkit. Bioinformatics 2012, 28, 1166–1167. [Google Scholar] [CrossRef]
- Reese, M.G. Application of a time-delay neural network to promoter annotation in the Drosophila melanogaster genome. Comput. Chem. 2001, 26, 51–56. [Google Scholar] [CrossRef]
- Markham, N.R.; Zuker, M. UNAFold: Software for Nucleic Acid Folding and Hybridization. In Bioinformatics, Volume II. Structure, Function and Applications, Number 453; Keith, J.M., Ed.; Humana Press: Totowa, NJ, USA, 2008; pp. 3–31. [Google Scholar]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kwok, A.C.M.; Leung, S.K.; Wong, J.T.Y. DNA:RNA Hybrids Are Major Dinoflagellate Minicircle Molecular Types. Int. J. Mol. Sci. 2023, 24, 9651. https://doi.org/10.3390/ijms24119651
Kwok ACM, Leung SK, Wong JTY. DNA:RNA Hybrids Are Major Dinoflagellate Minicircle Molecular Types. International Journal of Molecular Sciences. 2023; 24(11):9651. https://doi.org/10.3390/ijms24119651
Chicago/Turabian StyleKwok, Alvin Chun Man, Siu Kai Leung, and Joseph Tin Yum Wong. 2023. "DNA:RNA Hybrids Are Major Dinoflagellate Minicircle Molecular Types" International Journal of Molecular Sciences 24, no. 11: 9651. https://doi.org/10.3390/ijms24119651
APA StyleKwok, A. C. M., Leung, S. K., & Wong, J. T. Y. (2023). DNA:RNA Hybrids Are Major Dinoflagellate Minicircle Molecular Types. International Journal of Molecular Sciences, 24(11), 9651. https://doi.org/10.3390/ijms24119651





