TREX1 531C/T Polymorphism and Autoantibodies Associated with the Immune Status of HIV-1-Infected Individuals
Abstract
1. Introduction
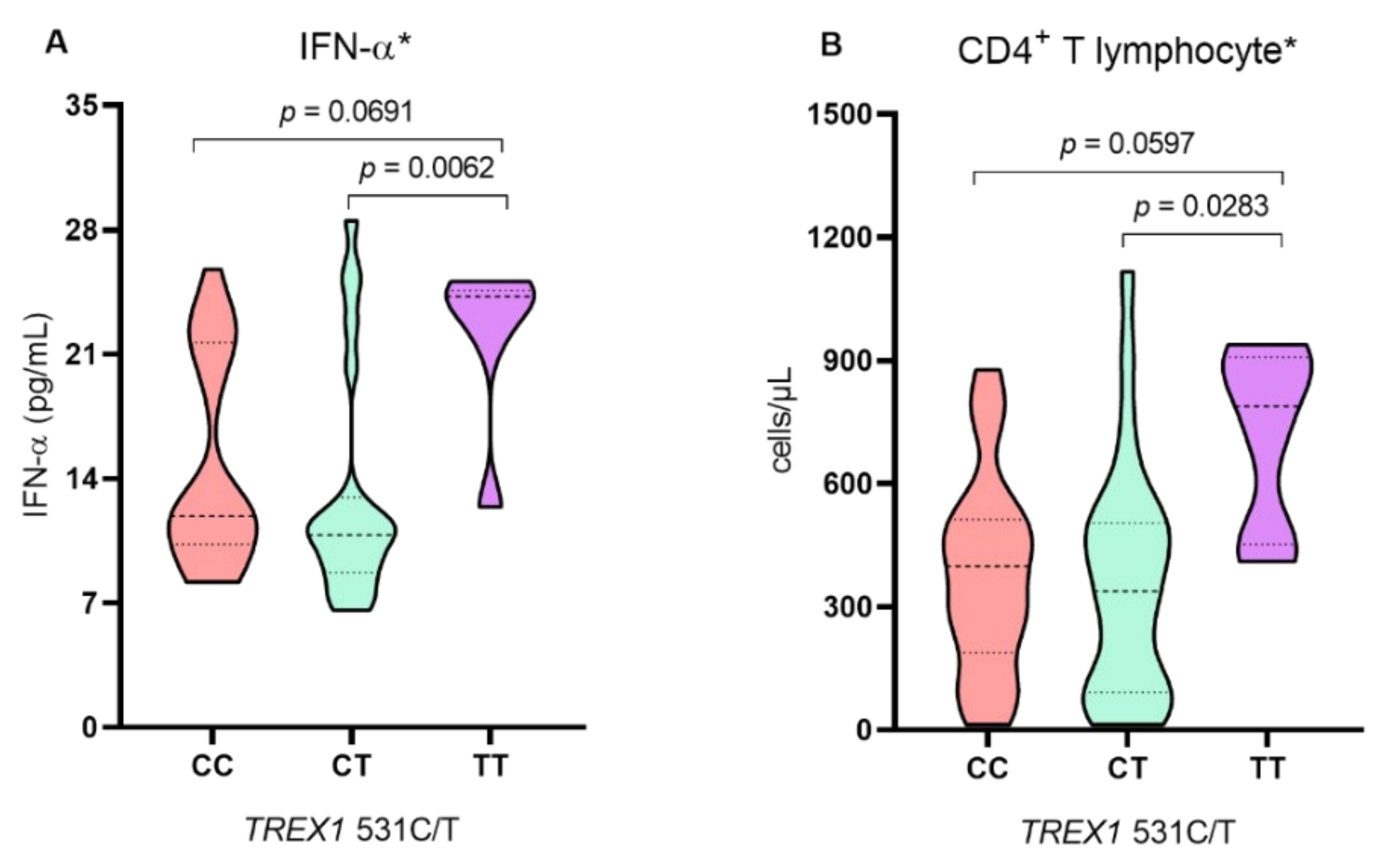
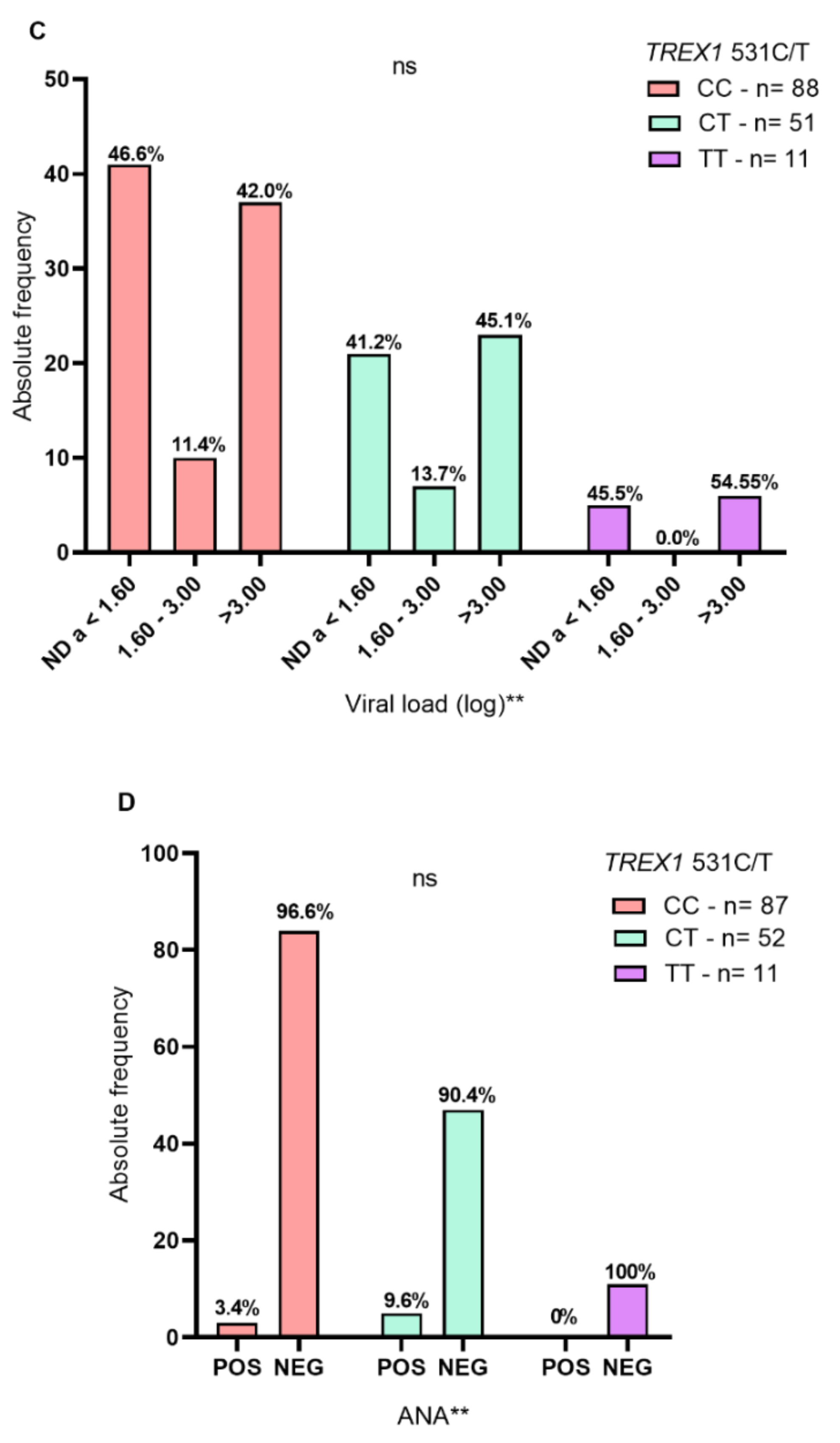
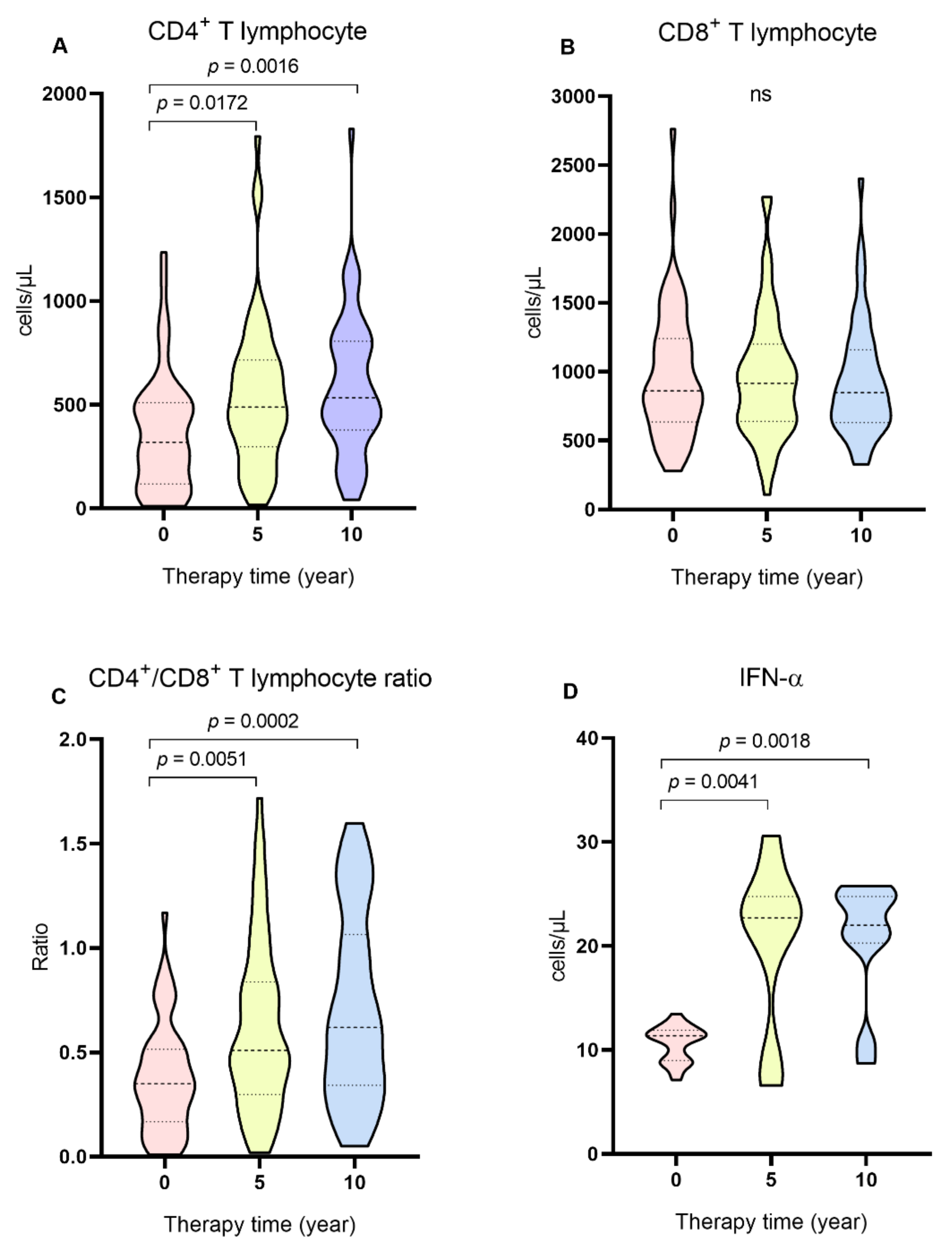
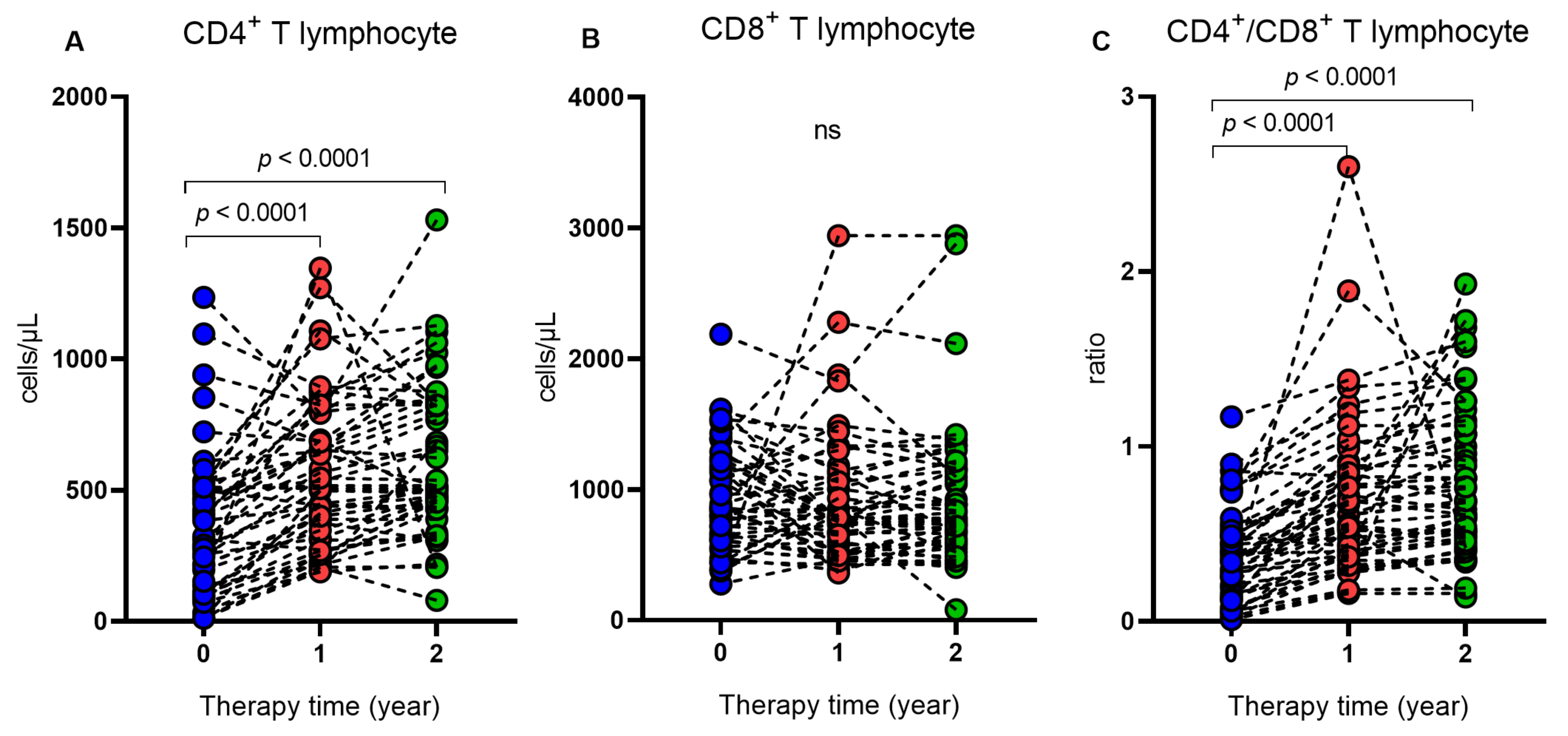
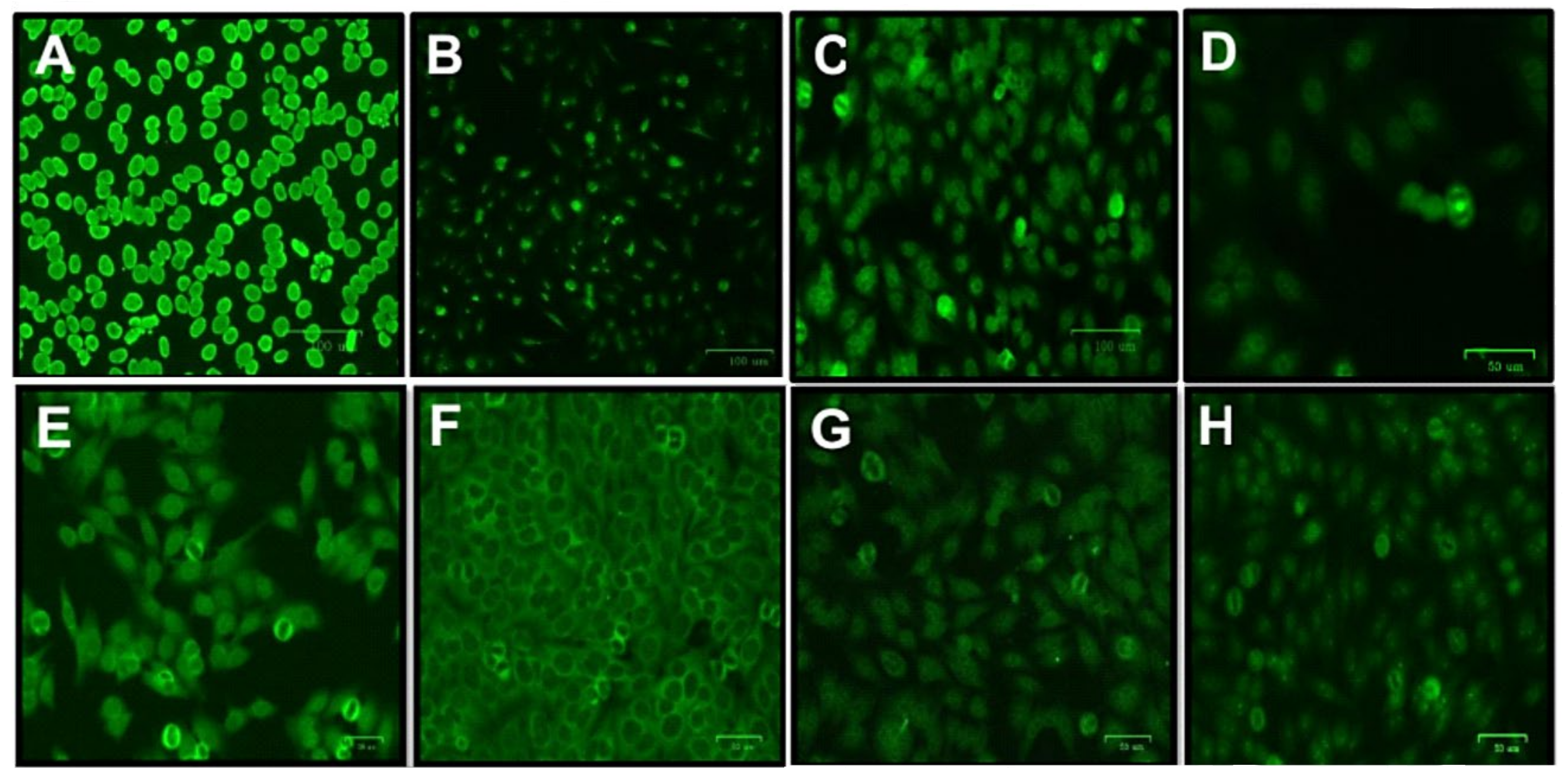
2. Results
3. Discussion
4. Materials and Methods
4.1. Type of Study, Population and Sample Collection
4.2. ANA Research in HEp-2 Cells
4.3. Plasma Dosage of IFN-α
4.4. DNA Extraction
4.5. TREX1 531C/T Genotyping (rs11797)
4.6. Quantification of TCD4+/TCD8+ Lymphocytes and HIV-1 Plasma Viral Load
4.7. Statistical Analysis
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- WHO. HIV. Key Factors. 2022. Available online: https://www.who.int/news-room/fact-sheets/detail/hiv-aids (accessed on 20 March 2023).
- Broder, S. The development of antiretroviral therapy and its impact on the HIV-1/AIDS pandemic. Antivir. Res. 2010, 85, 1–18. [Google Scholar] [CrossRef] [PubMed]
- Shah, S.; Hill, A. Risks of metabolic syndrome and diabetes with integrase inhibitor-based therapy. Curr. Opin. Infect. Dis. 2021, 34, 16–24. [Google Scholar] [CrossRef] [PubMed]
- de Souza, R.G.; Lima, S.S.; Pinto, A.C.; Souza, J.S.; Moura, T.C.F.; da Silva Graça Amoras, E.; Machado, L.F.A.; Guerreiro, J.F.; Vallinoto, A.C.R.; Queiroz, M.A.F.; et al. Change in Nutritional and Biochemical Status in People Living with HIV-1 on Antiretroviral Therapy. Viruses 2022, 14, 2573. [Google Scholar] [CrossRef] [PubMed]
- Zandman-Goddard, G.; Shoenfeld, Y. HIV and autoimmunity. Autoimmun. Rev. 2002, 1, 329–337. [Google Scholar] [CrossRef]
- Sedlácek, D.; Ulcová-Gallová, Z.; Milichovská, L.; Nováková, P.; Rokyta, Z. Seven antiphospholipid antibodies in HIV-positive patients: Correlation with clinical course and laboratory findings. Am. J. Reprod. Immunol. 2003, 50, 439–443. [Google Scholar] [CrossRef]
- Iordache, L.; Launay, O.; Bouchaud, O.; Jeantils, V.; Goujard, C.; Boue, F.; Cacoub, P.; Hanslik, T.; Mahr, A.; Lambotte, O.; et al. Autoimmune diseases in HIV-infected patients: 52 cases and literature review. Autoimmun. Rev. 2014, 13, 850–857. [Google Scholar] [CrossRef] [PubMed]
- Ma, W.T.; Chang, C.; Gershwin, M.E.; Lian, Z.X. Development of autoantibodies precedes clinical manifestations of autoimmune diseases: A comprehensive review. J. Autoimmun. 2017, 83, 95–112. [Google Scholar] [CrossRef]
- Zhao, Z.S.; Granucci, F.; Yeh, L.; Schaffer, P.A.; Cantor, H. Molecular mimicry by herpes simplex virus-type 1: Autoimmune disease after viral infection. Science 1998, 279, 1344–1347. [Google Scholar] [CrossRef]
- Stahl, D.; Lacroix-Desmazes, S.; Misra, N.; Karmochkine, M.; Kaveri, S.V.; Costagliola, D.; Sibrowski, W.; Kazatchkine, M.D.; ANRS SEROCO study group. Alterations of self-reactive antibody repertoires in HIV disease: An insight into the role of T cells in the selection of autoreactive B cells. Immunol. Lett. 2005, 99, 198–208. [Google Scholar] [CrossRef]
- Mazur, D.J.; Perrino, F.W. Structure and expression of the TREX1 and TREX2 3′ --> 5′ exonuclease genes. J. Biol. Chem. 2001, 276, 14718–14727. [Google Scholar] [CrossRef]
- Stetson, D.B.; Ko, J.S.; Heidmann, T.; Medzhitov, R. Trex1 prevents cell-intrinsic initiation of autoimmunity. Cell 2008, 134, 587–598. [Google Scholar] [CrossRef] [PubMed]
- Barber, G.N. STING-dependent cytosolic DNA sensing pathways. Trend. Immunol. 2012, 35, 88–93. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.G.; Lindahl, T.; Barnes, D.E. Trex1 exonuclease degrades ssDNA to prevent chronic checkpoint activation and autoimmune disease. Cell 2007, 131, 873–886. [Google Scholar] [CrossRef]
- Crow, Y.J. Type I interferonopathies: A novel set of inborn errors of immunity. Ann. N. Y. Acad. Sci. 2011, 1238, 91–98. [Google Scholar] [CrossRef]
- Simpson, S.R.; Rego, S.L.; Harvey, S.E.; Liu, M.; Hemphill, W.O.; Venkatadri, R.; Sharma, R.; Grayson, J.M.; Perrino, F.W. T Cells Produce IFN-α in the TREX1 D18N Model of Lupus-like Autoimmunity. J. Immunol. 2020, 204, 348–359. [Google Scholar] [CrossRef]
- Richards, A.; van den Maagdenberg, A.M.; Jen, J.C.; Kavanagh, D.; Bertram, P.; Spitzer, D.; Liszewski, M.K.; Barilla-Labarca, M.L.; Terwindt, G.M.; Kasai, Y.; et al. C-terminal truncations in human 3′-5′ DNA exonuclease TREX1 cause autosomal dominant retinal vasculopathy with cerebral leukodystrophy. Nat. Genet. 2007, 39, 1068–1070. [Google Scholar] [CrossRef]
- Rice, G.; Newman, W.G.; Dean, J.; Patrick, T.; Parmar, R.; Flintoff, K.; Robins, P.; Harvey, S.; Hollis, T.; O’Hara, A.; et al. Heterozygous mutations in TREX1 cause familial chilblain lupus and dominant Aicardi-Goutieres syndrome. Am. J. Hum. Genet. 2007, 80, 811–815. [Google Scholar] [CrossRef]
- Namjou, B.; Kothari, P.H.; Kelly, J.A.; Glenn, S.B.; Ojwang, J.O.; Adler, A.; Alarcón-Riquelme, M.E.; Gallant, C.J.; Boackle, S.A.; Criswell, L.A.; et al. Evaluation of the TREX1 gene in a large multiancestral lupus cohort. Genes Immun. 2011, 12, 270–279. [Google Scholar] [CrossRef]
- van Montfoort, N.; Olagnier, D.; Hiscott, J. Unmasking immune sensing of retroviruses: Interplay between innate sensors and host effectors. Cytokine Growth Factor Rev. 2014, 5, 657–668. [Google Scholar] [CrossRef]
- Yan, N.; Regalado-Magdos, A.D.; Stiggelbout, B.; Lee-Kirsch, M.A.; Lieberman, J. The cytosolic exonuclease TREX1 inhibits the innate immune response to human immunodeficiency virus type 1. Nat. Immunol. 2010, 11, 1005–1013. [Google Scholar] [CrossRef] [PubMed]
- Geijtenbeek, T.B. Host DNase TREX1 hides HIV from DNA sensors. Nat. Immunol. 2010, 11, 979–980. [Google Scholar] [CrossRef]
- Booiman, T.; Setiawan, L.C.; Kootstra, N.A. Genetic variation in Trex1 affects HIV-1 disease progression. AIDS 2014, 28, 2517–2521. [Google Scholar] [CrossRef] [PubMed]
- Buell, K.G.; Chung, C.; Chaudhry, Z.; Puri, A.; Nawab, K.; Ravindran, R.P. Lifelong antiretroviral therapy or HIV cure: The benefits for the individual patient. AIDS Care 2016, 28, 242–246. [Google Scholar] [CrossRef]
- Dionne, B. Key Principles of Antiretroviral Pharmacology. Infect. Dis. Clin. N. Am. 2019, 33, 787–805. [Google Scholar] [CrossRef] [PubMed]
- Pontillo, A.; Girardelli, M.; Catamo, E.; Duarte, A.J.; Crovella, S. Polymorphisms in TREX1 and susceptibility to HIV-1 infection. Int. J. Immunogenet. 2013, 40, 492–494. [Google Scholar] [CrossRef] [PubMed]
- Ivashkiv, L.B.; Donlin, L.T. Regulation of type I interferon responses. Nat. Rev. Immunol. 2014, 14, 36–49. [Google Scholar] [CrossRef]
- Nakano, K.; Watanabe, T. Tuning Rex rules HTLV-1 pathogenesis. Front. Immunol. 2022, 13, 959962. [Google Scholar] [CrossRef]
- Silva, D.C.; Amoras, E.D.S.G.; Moura, T.C.F.; Lopes, F.T.; Gomes, S.T.M.; Costa, C.A.D.; Sousa, M.S.; Ishak, R.; Vallinoto, A.C.R.; Queiroz, M.A.F. TREX1 531C>T Polymorphism is Associated with High Proviral Load Levels in HTLV-1-Infected Persons. Viruses 2019, 12, 7. [Google Scholar] [CrossRef]
- Meng, Z.; Du, L.; Hu, N.; Byrd, D.; Amet, T.; Desai, M.; Shepherd, N.; Lan, J.; Han, R.; Yu, Q. Antiretroviral Therapy Normalizes Autoantibody Profile of HIV Patients by Decreasing CD33⁺CD11b⁺HLA-DR⁺ Cells: A Cross-Sectional Study. Medicine 2016, 95, e3285. [Google Scholar] [CrossRef]
- Iordache, L.; Bengoufa, D.; Taulera, O.; Rami, A.; Lascoux-Combe, C.; Day, N.; Parrinello, M.; Sellier, P.O.; Molina, J.M.; Mahr, A. Nonorgan-specific autoantibodies in HIV-infected patients in the HAART era. Medicine 2017, 96, e6230. [Google Scholar] [CrossRef]
- Jubault, V.; Penfornis, A.; Schillo, F.; Hoen, B.; Izembart, M.; Timsit, J.; Kazatchkine, M.D.; Gilquin, J.; Viard, J.P. Sequential occurrence of thyroid autoantibodies and Graves’ disease after immune restoration in severely immunocompromised human immunodeficiency virus-1-infected patients. J. Clin. Endocrinol. Metab. 2000, 85, 4254–4257. [Google Scholar] [PubMed]
| Characteristics | ART-Naïve n = 100 n (%) | 5 Years ART n = 50 n (%) | 10 Years ART n = 50 n (%) |
|---|---|---|---|
| Sex | |||
| Female | 27 (27.0) | 22 (44.0) | 19 (38.0) |
| Male | 73 (73.0) | 28 (56.0) | 31 (62.0) |
| Age; mean (SD) | 29.8 (9.7) | 40.5 (10.2) | 44.6 (12.3) |
| Marital status | |||
| Married | 34 (34) | 15 (30) | 14 (28) |
| Single | 66 (66) | 28 (56) | 30 (60) |
| Divorced | 0 (0) | 4 (8) | 1 (2) |
| Widower | 0 (0) | 3 (6) | 5 (10) |
| Family income (minimum wage) | |||
| <1 | 16 (16) | 8 (16) | 7 (14) |
| 1 a 3 | 65 (65) | 32 (64) | 37 (74) |
| ≥4 | 14 (14) | 9 (18) | 5 (10) |
| No information | 5 (5) | 1 (2) | 1 (2) |
| Education | |||
| Elementary School | 16 (16) | 14 (28) | 14 (28) |
| High school | 65 (65) | 27 (54) | 28 (56) |
| University education | 19 (19) | 9 (18) | 8 (16) |
| Therapeutic scheme | |||
| Etravirine + Ritonavir | NA | 0 (0) | 1 (2) |
| Maraviroc + ritonavir + darunavir + dolutegravir | NA | 0 (0) | 1 (2) |
| Tenofovir + atazanavir + lamivudine + ritonavir | NA | 4 (8) | 2 (4) |
| Tenofovir + Atazanavir + ritonavir | NA | 2 (4) | 0 (0) |
| Tenofovir + lamivudine + efavirenz | NA | 24 (56) | 15 (30) |
| Tenofovir + lamivudine + dolutegravir | NA | 0 (0) | 0 (0) |
| Tenofovir + lamivudine + ritonavir + darunavir | NA | 1 (2) | 0 (0) |
| Tenofovir + lamivudine + lopinavir + ritonavir | NA | 0 (0) | 6 (12) |
| Tenofovir + abacavir + didanosine + lamivudine + zidovudine + efavirenz + nevirapine + etravirine | NA | 0 (0) | 2 (4) |
| Tenofovir + Lamivudine + atazanavir | NA | 0 (0) | 3 (6) |
| Tenofovir+ Raltegravir + Lamivudine | NA | 3 (6) | 0 (0) |
| Tenofovir + lamivudine + efavirenz + ritonavir + darunavir | NA | 0 (0) | 1 (2) |
| Tenofovir + lamivudine + Efavirenz + ritonavir + Darunavir | NA | 0 (0) | 1 (2) |
| Zidovudine + Lamivudine + lopnavir + ritonavir | NA | 3 (6) | 1 (2) |
| Zidovudine + lamivudine + efavirenz | NA | 12 (24) | 10 (20) |
| Zidovudine + didanosine + Efavirenz | NA | 0 (0) | 1 (2) |
| Zidovudine + lamivudine + atazanavir | NA | 0 (0) | 1 (2) |
| Zidovudine + lamivudine + atazanavir + ritonavir | NA | 0 (0) | 3 (6) |
| Zidovudine + lamivudine + nevirapine | NA | 0 (0) | 2 (4) |
| Zidovudine + lamivudine + daranavir + ritonavir + dolutegravir | 1 (2) | 0 (0) | |
| Change of therapy | |||
| No | NA | 28 (56) | 11 (22) |
| Yes | NA | 22 (44) | 39 (78) |
| Variables | HIV-1 n = 150 n (%) | Control n = 100 n (%) | p |
|---|---|---|---|
| TREX1531C/T Genotypes and alleles | |||
| CC | 88 (58.7) | 60 (60) | 0.8367 * |
| CT | 51 (34) | 31 (31) | |
| TT | 11 (7.3) | 9 (9) | |
| C | 0.76 | 0.75 | 0.9492 * |
| T | 0.24 | 0.25 | |
| ANA | |||
| Positive | 8 (5.3) | 1 (1) | 0.1231 * |
| Negative | 142 (94.7) | 99 (99) | |
| IFN-α, median (IQR) | 11.3 (12.2) | 22.7 (6.88) | <0.0001 ** |
| Variables | ART-Naïve n = 50 n (%) | 5 Years ART n = 50 n (%) | 10 Years ART n = 50 n (%) | p |
|---|---|---|---|---|
| ANA, n (%) | ||||
| Positive | 0 (0) | 4 (8) | 4 (8) | 0.0452 * |
| Negative | 50 (100) | 46 (92) | 46 (92) | |
| CD4+, median (IQR) | 319 (391.2) | 489.5 (418.7) | 534.5 (428.3) | 0.0012 ** |
| CD8+, median (IQR) | 862 (607.2) | 915.5 (560.2) | 848 (529.5) | 0.8685 ** |
| CD4+/CD8+, ratio (IQR) | 0.35 (0.3) | 0.51 (0.5) | 0.62 (0.7) | 0.0001 ** |
| Viral load, log10 | ||||
| ND a < 1.60 | 1 (2) | 31 (62) | 36 (72) | <0.0001 * |
| 1.60–3.00 | 1 (2) | 9 (18) | 5 (10) | |
| >3.00 | 48 (96) | 10 (20) | 9 (18) | |
| IFN-α,median (IQR) | 11.36 (2.91) | 22.7 (10.36) | 22.01 (4.47) | 0.0002 * |
| Variables | ART-Naïve n = 50 n (%) | 1 Year ART n = 50 n (%) | 2 Years ART n = 50 n (%) | p |
|---|---|---|---|---|
| ANA, n (%) | ||||
| Positive | 0 (0) | 3 (6) | 6 (12) | 0.0174 * |
| Negative | 50 (100) | 47 (94) | 44 (88) | |
| CD4+, median (IQR) | 314 (394) | 517 (410) | 509 (393.5) | <0.0001 ** |
| CD8+, median (IQR) | 852.5 (585) | 778 (579.7) | 782 (540.5) | 0.3966 ** |
| CD4+/CD8+, ratio | 0.35 (0.4) | 0.61 (0.5) | 0.69 (0.6) | <0.0001 ** |
| Viral load, log 10 | ||||
| ND a < 1.60 | 1 (2) | 42 (84) | 44 (88) | <0.0001 * |
| 1.60–3.00 | 1 (2) | 5 (10) | 5 (10) | |
| >3.00 | 48 (96) | 3 (6) | 1 (2) |
| Patient (ID) | Fluorescence Patterns | Therapeutic Scheme |
|---|---|---|
| Cross-sectional evaluation | ||
| 5 years ART | ||
| 27984 | Fine dotted cytoplasmic | Tenofovir + raltegravir + lamivudine |
| 27987 | Nucleolar | Tenofovir + raltegravir + lamivudine |
| 28273 | Numa (mitotic apparatus with positive poles) | Tenofovir + raltegravir + lamivudine |
| 28617 | Thick dotted nuclear | Zidovudine + lamivudine + darunavir + ritonavir + dolutegravir |
| 10 years ART | ||
| 26714 | Fine dotted nuclear | Tenofovir + lamivudine + efavirenz |
| 26733 | Homogeneous nuclear | Tenofovir + lamivudine + atazanavir |
| 26862 | Nucleolar | Tenofovir + Abacavir + didanosine + lamivudine + zidovudine + efavirenz + nevirapine + etravirine |
| 29947 | Thick dotted nuclear | Tenofovir + lamivudine + efavirenz |
| Longitudinal evaluation | ||
| 1 year ART | ||
| 26468 | Fine dotted nuclear | Tenofovir + lamivudine + efavirenz |
| 26541 | Fine dotted nuclear | Tenofovir + lamivudine + efavirenz |
| 26712 | Nucleolar with isolated dots | Tenofovir + ritonavir + atasanavir |
| 2 years ART | ||
| 26468 | Thick dotted nuclear | Tenofovir + lamivudine + efavirenz |
| 26471 | Fine dotted nuclear | Tenofovir + lamivudine + efavirenz |
| 26480 | Dotted nucleolar | Tenofovir + lamivudine + efavirenz |
| 26536 | Dotted nucleolar | Tenofovir + lamivudine + efavirenz |
| 26537 | Fine-speckled nuclear and Numa (mitotic apparatus) | Tenofovir + lamivudine + efavirenz |
| 26703 | Cytoplasmic | Tenofovir + lamivudine + efavirenz |
| Groups | Inclusion Criteria | Exclusion Criteria |
|---|---|---|
| ART-naïve | Age ≥ 18 years old, both sexes, never having used antiretrovirals. | Diagnosis of autoimmune disease, coinfection with Hepatitis B Virus, Hepatitis C Virus and Hepatitis D Virus and HTLV-1, use of corticosteroids and clinical diagnosis of AIDS. |
| 5 years ART | Age ≥ 18 years, both genders, having been on treatment (ARV) for a period of five years without interruptions | |
| 5 years ART | Age ≥ 18 years, both genders, having been on treatment (ARV) for a period of 10 years without interruptions. |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Queiroz, M.A.F.; Moura, T.C.F.; Bichara, C.D.A.; Lima, L.L.P.d.; Oliveira, A.Q.T.d.; Souza, R.G.d.; Gomes, S.T.M.; Amoras, E.d.S.G.; Vallinoto, A.C.R. TREX1 531C/T Polymorphism and Autoantibodies Associated with the Immune Status of HIV-1-Infected Individuals. Int. J. Mol. Sci. 2023, 24, 9660. https://doi.org/10.3390/ijms24119660
Queiroz MAF, Moura TCF, Bichara CDA, Lima LLPd, Oliveira AQTd, Souza RGd, Gomes STM, Amoras EdSG, Vallinoto ACR. TREX1 531C/T Polymorphism and Autoantibodies Associated with the Immune Status of HIV-1-Infected Individuals. International Journal of Molecular Sciences. 2023; 24(11):9660. https://doi.org/10.3390/ijms24119660
Chicago/Turabian StyleQueiroz, Maria Alice Freitas, Tuane Carolina Ferreira Moura, Carlos David Araújo Bichara, Lorena Leticia Peixoto de Lima, Allysson Quintino Tenório de Oliveira, Ranilda Gama de Souza, Samara Tatielle Monteiro Gomes, Ednelza da Silva Graça Amoras, and Antonio Carlos Rosário Vallinoto. 2023. "TREX1 531C/T Polymorphism and Autoantibodies Associated with the Immune Status of HIV-1-Infected Individuals" International Journal of Molecular Sciences 24, no. 11: 9660. https://doi.org/10.3390/ijms24119660
APA StyleQueiroz, M. A. F., Moura, T. C. F., Bichara, C. D. A., Lima, L. L. P. d., Oliveira, A. Q. T. d., Souza, R. G. d., Gomes, S. T. M., Amoras, E. d. S. G., & Vallinoto, A. C. R. (2023). TREX1 531C/T Polymorphism and Autoantibodies Associated with the Immune Status of HIV-1-Infected Individuals. International Journal of Molecular Sciences, 24(11), 9660. https://doi.org/10.3390/ijms24119660








