The Emerging Role of Autophagy-Associated lncRNAs in the Pathogenesis of Neurodegenerative Diseases
Abstract
1. Introduction
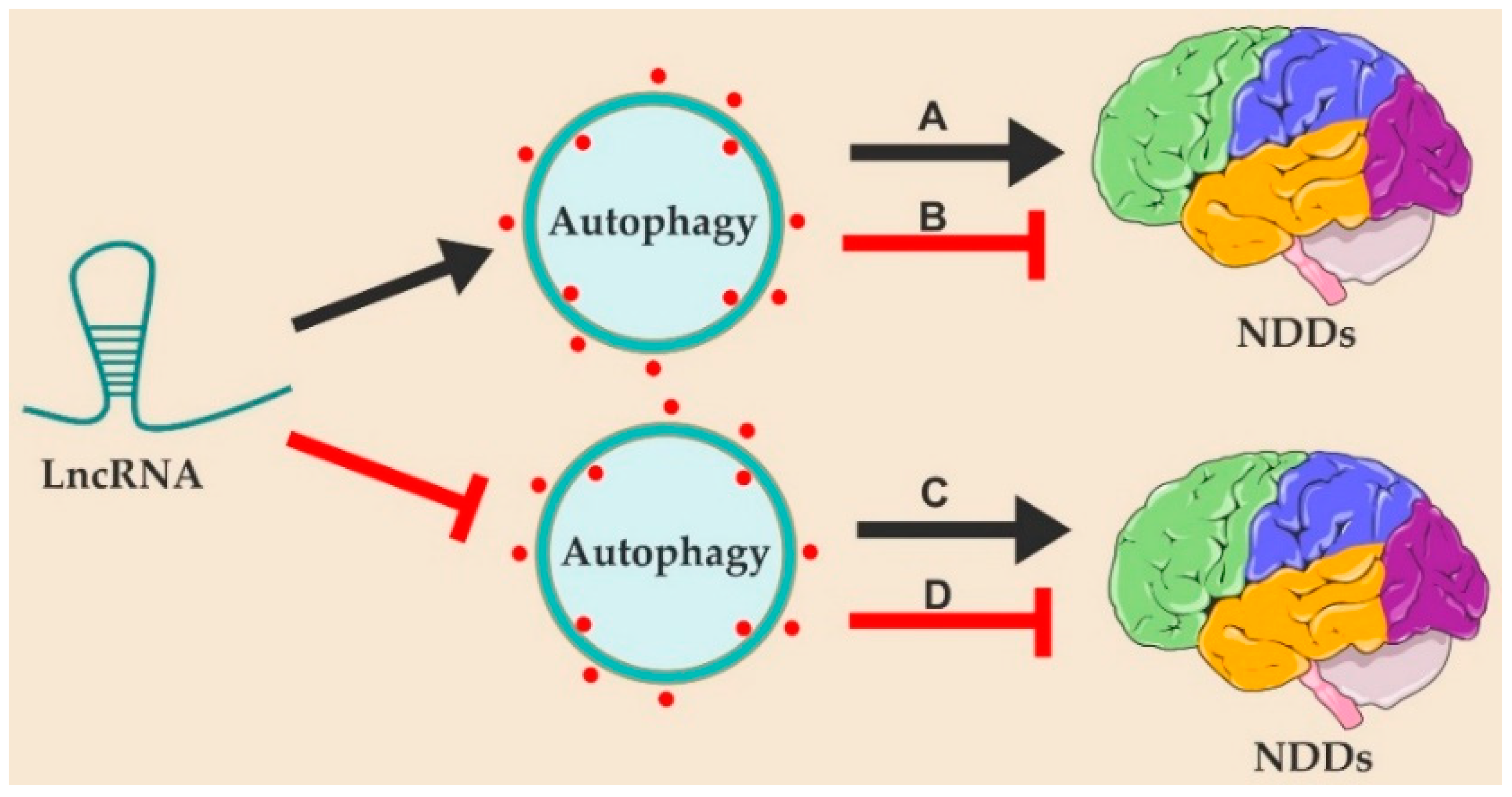
2. LncRNAs Regulate the Pathogenesis of Neurodegenerative Diseases by Modulating Autophagy
2.1. LncRNAs in the Pathogenesis of Neurodegenerative Diseases
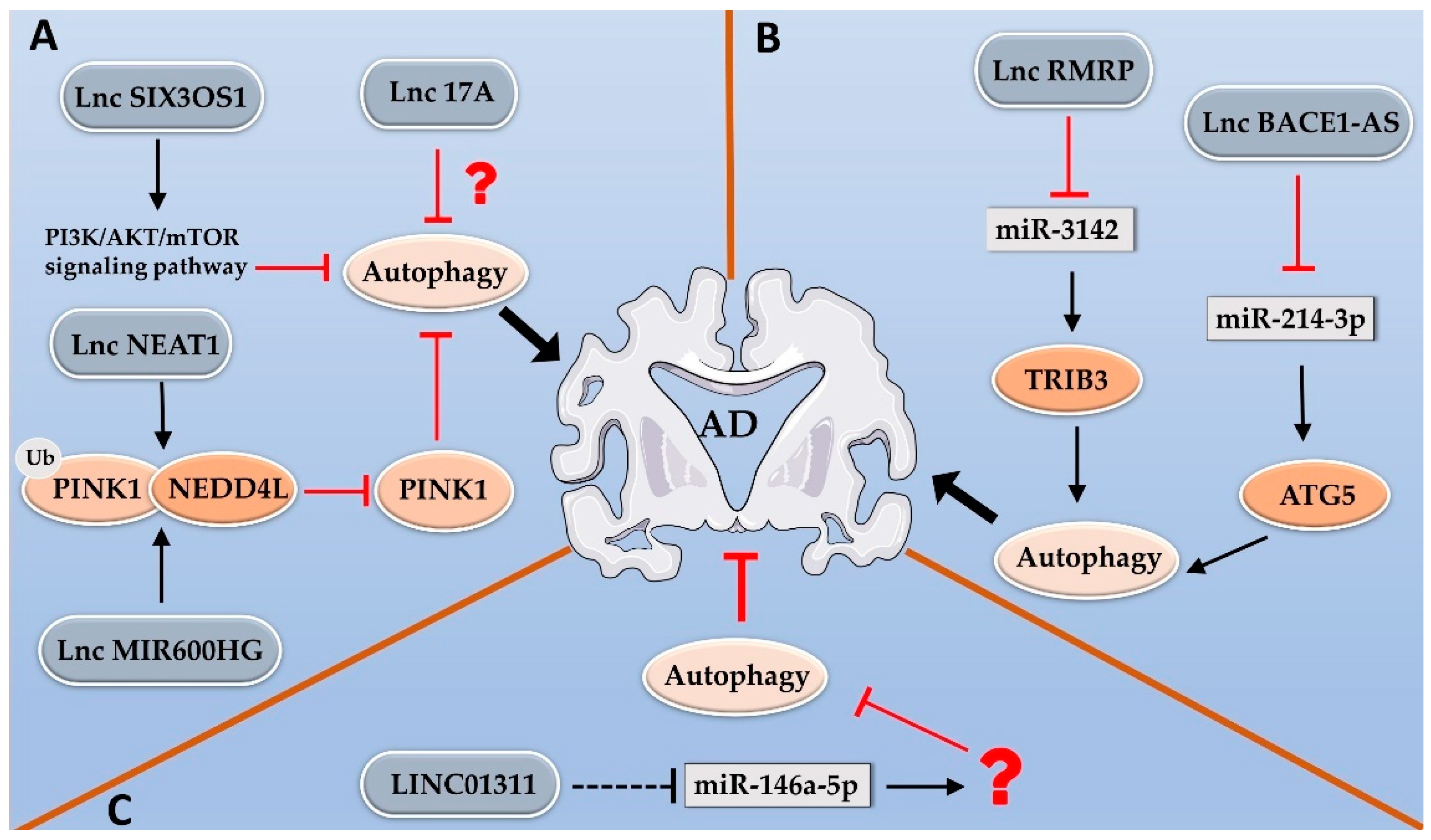
2.2. LncRNAs Regulate Alzheimer’s Disease by Modulating Autophagy
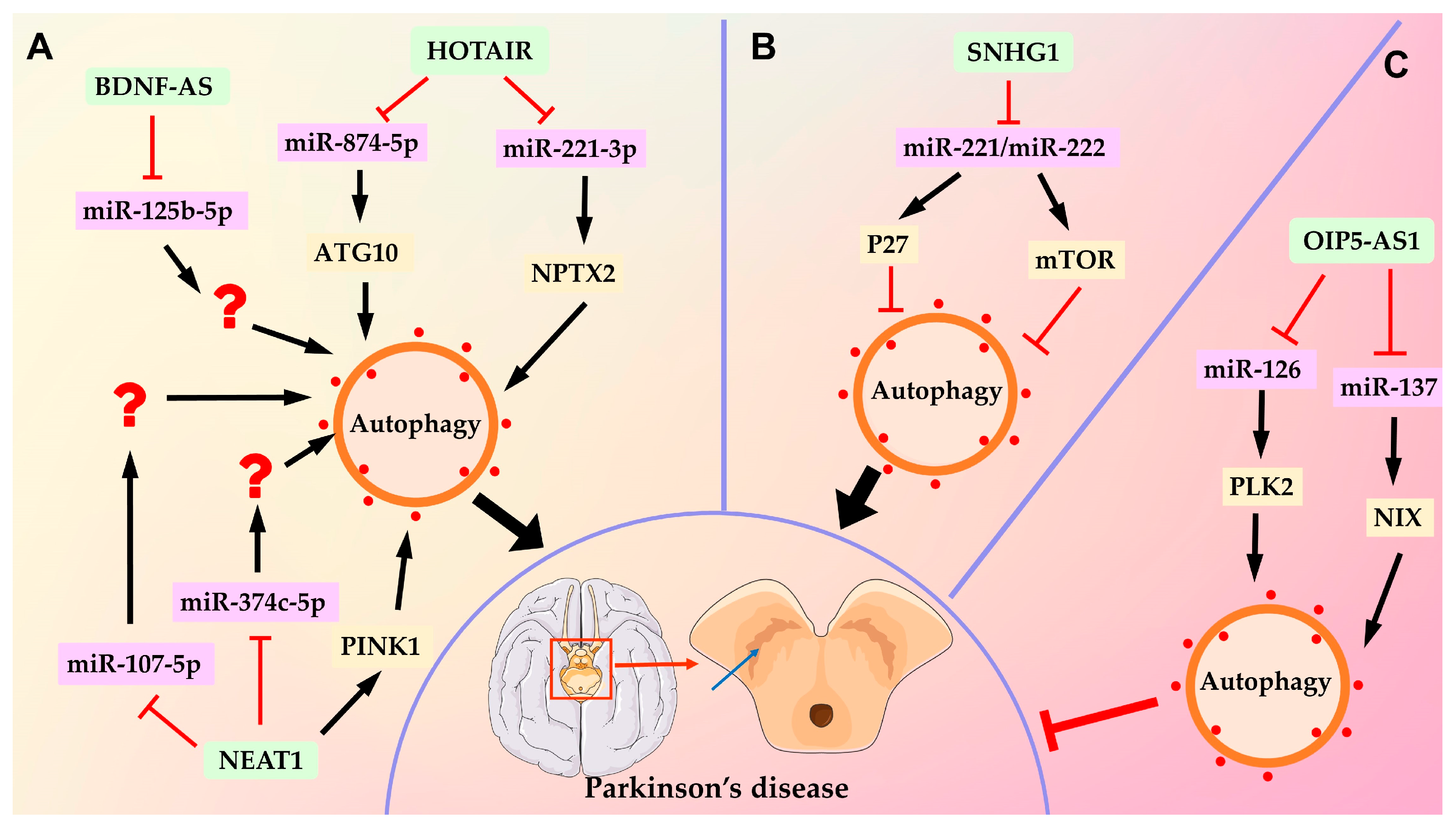
2.3. LncRNAs Regulate Parkinson’s Disease by Modulating Autophagy
3. Targeting Autophagy-Related lncRNAs as a Therapeutic Strategy for Neurodegenerative Diseases
4. Concluding Remarks
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Conflicts of Interest
References
- Quinn, J.J.; Chang, H.Y. Unique features of long non-coding RNA biogenesis and function. Nat. Rev. Genet. 2016, 17, 47–62. [Google Scholar] [CrossRef]
- Iyer, M.K.; Niknafs, Y.S.; Malik, R.; Singhal, U.; Sahu, A.; Hosono, Y.; Barrette, T.R.; Prensner, J.R.; Evans, J.R.; Zhao, S.; et al. The landscape of long noncoding RNAs in the human transcriptome. Nat. Genet. 2015, 47, 199–208. [Google Scholar] [CrossRef] [PubMed]
- Zhang, P.; Wu, W.; Chen, Q.; Chen, M. Non-Coding RNAs and their Integrated Networks. J. Integr. Bioinform. 2019, 16, 20190027. [Google Scholar] [CrossRef] [PubMed]
- Huang, Y. The novel regulatory role of lncRNA-miRNA-mRNA axis in cardiovascular diseases. J. Cell Mol. Med. 2018, 22, 5768–5775. [Google Scholar] [CrossRef] [PubMed]
- Bridges, M.C.; Daulagala, A.C.; Kourtidis, A. LNCcation: LncRNA localization and function. J. Cell Biol. 2021, 220, e202009045. [Google Scholar] [CrossRef]
- Mukherjee, N.; Calviello, L.; Hirsekorn, A.; de Pretis, S.; Pelizzola, M.; Ohler, U. Integrative classification of human coding and noncoding genes through RNA metabolism profiles. Nat. Struct. Mol. Biol. 2017, 24, 86–96. [Google Scholar] [CrossRef]
- Wang, K.C.; Chang, H.Y. Molecular mechanisms of long noncoding RNAs. Mol. Cell 2011, 43, 904–914. [Google Scholar] [CrossRef]
- Zhang, X.; Wang, W.; Zhu, W.; Dong, J.; Cheng, Y.; Yin, Z.; Shen, F. Mechanisms and Functions of Long Non-Coding RNAs at Multiple Regulatory Levels. Int. J. Mol. Sci. 2019, 20, 5573. [Google Scholar] [CrossRef]
- Jiang, N.; Zhang, X.; Gu, X.; Li, X.; Shang, L. Progress in understanding the role of lncRNA in programmed cell death. Cell Death Discov. 2021, 7, 30. [Google Scholar] [CrossRef]
- Riva, P.; Ratti, A.; Venturin, M. The Long Non-Coding RNAs in Neurodegenerative Diseases: Novel Mechanisms of Pathogenesis. Curr. Alzheimer Res. 2016, 13, 1219–1231. [Google Scholar] [CrossRef]
- Wu, P.; Zuo, X.; Deng, H.; Liu, X.; Liu, L.; Ji, A. Roles of long noncoding RNAs in brain development, functional diversification and neurodegenerative diseases. Brain Res. Bull. 2013, 97, 69–80. [Google Scholar] [CrossRef]
- Levine, B.; Kroemer, G. Biological Functions of Autophagy Genes: A Disease Perspective. Cell 2019, 176, 11–42. [Google Scholar] [CrossRef] [PubMed]
- Choi, A.M.; Ryter, S.W.; Levine, B. Autophagy in human health and disease. N Engl J Med 2013, 368, 651–662. [Google Scholar] [CrossRef] [PubMed]
- Li, Q.; Liu, Y.; Sun, M. Autophagy and Alzheimer’s Disease. Cell Mol. Neurobiol. 2017, 37, 377–388. [Google Scholar] [CrossRef] [PubMed]
- Maher, P. The Potential of Flavonoids for the Treatment of Neurodegenerative Diseases. Int. J. Mol. Sci. 2019, 20, 3056. [Google Scholar] [CrossRef]
- Aguzzi, A.; O’Connor, T. Protein aggregation diseases: Pathogenicity and therapeutic perspectives. Nat. Rev. Drug Discov. 2010, 9, 237–248. [Google Scholar] [CrossRef]
- Rubinsztein, D.C.; Codogno, P.; Levine, B. Autophagy modulation as a potential therapeutic target for diverse diseases. Nat. Rev. Drug Discov. 2012, 11, 709–730. [Google Scholar] [CrossRef]
- Nixon, R.A. The role of autophagy in neurodegenerative disease. Nat. Med. 2013, 19, 983–997. [Google Scholar] [CrossRef]
- Park, H.; Kang, J.H.; Lee, S. Autophagy in Neurodegenerative Diseases: A Hunter for Aggregates. Int. J. Mol. Sci. 2020, 21, 3369. [Google Scholar] [CrossRef]
- Tung, Y.T.; Wang, B.J.; Hu, M.K.; Hsu, W.M.; Lee, H.; Huang, W.P.; Liao, Y.F. Autophagy: A double-edged sword in Alzheimer’s disease. J. Biosci. 2012, 37, 157–165. [Google Scholar] [CrossRef]
- Briggs, J.A.; Wolvetang, E.J.; Mattick, J.S.; Rinn, J.L.; Barry, G. Mechanisms of Long Non-coding RNAs in Mammalian Nervous System Development, Plasticity, Disease, and Evolution. Neuron 2015, 88, 861–877. [Google Scholar] [CrossRef] [PubMed]
- Zhou, X.; Xu, J. Identification of Alzheimer’s disease-associated long noncoding RNAs. Neurobiol. Aging 2015, 36, 2925–2931. [Google Scholar] [CrossRef]
- Ni, Y.; Huang, H.; Chen, Y.; Cao, M.; Zhou, H.; Zhang, Y. Investigation of Long Non-coding RNA Expression Profiles in the Substantia Nigra of Parkinson’s Disease. Cell Mol. Neurobiol. 2017, 37, 329–338. [Google Scholar] [CrossRef]
- Johnson, R. Long non-coding RNAs in Huntington’s disease neurodegeneration. Neurobiol. Dis. 2012, 46, 245–254. [Google Scholar] [CrossRef]
- Gagliardi, S.; Zucca, S.; Pandini, C.; Diamanti, L.; Bordoni, M.; Sproviero, D.; Arigoni, M.; Olivero, M.; Pansarasa, O.; Ceroni, M.; et al. Long non-coding and coding RNAs characterization in Peripheral Blood Mononuclear Cells and Spinal Cord from Amyotrophic Lateral Sclerosis patients. Sci. Rep. 2018, 8, 2378. [Google Scholar] [CrossRef] [PubMed]
- Dong, L.X.; Zhang, Y.Y.; Bao, H.L.; Liu, Y.; Zhang, G.W.; An, F.M. LncRNA NEAT1 promotes Alzheimer’s disease by down regulating micro-27a-3p. Am. J. Transl. Res. 2021, 13, 8885–8896. [Google Scholar] [PubMed]
- Ke, S.; Yang, Z.; Yang, F.; Wang, X.; Tan, J.; Liao, B. Long Noncoding RNA NEAT1 Aggravates Abeta-Induced Neuronal Damage by Targeting miR-107 in Alzheimer’s Disease. Yonsei. Med. J. 2019, 60, 640–650. [Google Scholar] [CrossRef]
- Ma, P.Z.; Li, Y.L.; Zhang, W.; Fang, F.Q.; Sun, J.; Liu, M.Z.; Li, K.; Dong, L.F. Long Non-coding RNA MALAT1 Inhibits Neuron Apoptosis and Neuroinflammation While Stimulates Neurite Outgrowth and Its Correlation with MiR-125b Mediates PTGS2, CDK5 and FOXQ1 in Alzheimer’s Disease. Curr. Alzheimer Res. 2019, 16, 596–612. [Google Scholar] [CrossRef]
- Chanda, K.; Jana, N.R.; Mukhopadhyay, D. Long non-coding RNA MALAT1 protects against Abeta(1–42) induced toxicity by regulating the expression of receptor tyrosine kinase EPHA2 via quenching miR-200a/26a/26b in Alzheimer’s disease. Life Sci. 2022, 302, 120652. [Google Scholar] [CrossRef]
- Duan, R.; Wang, S.Y.; Wei, B.; Deng, Y.; Fu, X.X.; Gong, P.Y.; Yan, E.; Sun, X.J.; Cao, H.M.; Shi, J.Q.; et al. Angiotensin-(1–7) Analogue AVE0991 Modulates Astrocyte-Mediated Neuroinflammation via lncRNA SNHG14/miR-223-3p/NLRP3 Pathway and offers Neuroprotection in a Transgenic Mouse Model of Alzheimer’s Disease. J. Inflamm. Res. 2021, 14, 7007–7019. [Google Scholar] [CrossRef]
- He, Y.; Qiang, Y. Mechanism of Autonomic Exercise Improving Cognitive Function of Alzheimer’s Disease by Regulating lncRNA SNHG14. Am. J. Alzheimers Dis. Other Demen. 2021, 36, 15333175211027681. [Google Scholar] [CrossRef] [PubMed]
- Lu, J.; Liu, L.; Chen, J.; Zhi, J.; Li, J.; Li, L.; Jiang, Z. LncRNA HOTAIR in exercise-induced neuro-protective function in Alzheimer’s disease. Folia Neuropathol. 2022, 60, 414–420. [Google Scholar] [CrossRef] [PubMed]
- Lu, J.; Liu, L.; Chen, J.; Zhi, J.; Li, J.; Li, L.; Jiang, Z. The Involvement of lncRNA HOTAIR/miR-130a-3p Axis in the Regulation of Voluntary Exercise on Cognition and Inflammation of Alzheimer’s Disease. Am. J. Alzheimers Dis. Other Demen. 2022, 37, 15333175221091424. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Wang, R. Deregulated lncRNA MAGI2-AS3 in Alzheimer’s disease attenuates amyloid-beta induced neurotoxicity and neuroinflammation by sponging miR-374b-5p. Exp. Gerontol. 2021, 144, 111180. [Google Scholar] [CrossRef] [PubMed]
- Yan, X.W.; Liu, H.J.; Hong, Y.X.; Meng, T.; Du, J.; Chang, C. lncRNA XIST induces Abeta accumulation and neuroinflammation by the epigenetic repression of NEP in Alzheimer’s disease. J. Neurogenet. 2022, 36, 11–20. [Google Scholar] [CrossRef]
- Yue, D.; Guanqun, G.; Jingxin, L.; Sen, S.; Shuang, L.; Yan, S.; Minxue, Z.; Ping, Y.; Chong, L.; Zhuobo, Z.; et al. Silencing of long noncoding RNA XIST attenuated Alzheimer’s disease-related BACE1 alteration through miR-124. Cell Biol. Int. 2020, 44, 630–636. [Google Scholar] [CrossRef]
- Chen, X.; Ren, G.; Li, Y.; Chao, W.; Chen, S.; Li, X.; Xue, S. Level of LncRNA GAS5 and Hippocampal Volume are Associated with the Progression of Alzheimer’s Disease. Clin. Interv. Aging 2022, 17, 745–753. [Google Scholar] [CrossRef]
- Ding, Y.; Luan, W.; Shen, X.; Wang, Z.; Cao, Y. LncRNA BDNF-AS as ceRNA regulates the miR-9-5p/BACE1 pathway affecting neurotoxicity in Alzheimer’s disease. Arch. Gerontol. Geriatr. 2022, 99, 104614. [Google Scholar] [CrossRef]
- Liu, N.X.; Li, Q.H. LncRNA BC200 regulates neuron apoptosis and neuroinflammation via PI3K/AKT pathway in Alzheimer’s disease. J. Biol. Regul. Homeost. Agents 2020, 34, 2255–2261. [Google Scholar]
- Zhang, Y.Y.; Bao, H.L.; Dong, L.X.; Liu, Y.; Zhang, G.W.; An, F.M. Silenced lncRNA H19 and up-regulated microRNA-129 accelerates viability and restrains apoptosis of PC12 cells induced by Abeta(25–35) in a cellular model of Alzheimer’s disease. Cell Cycle 2021, 20, 112–125. [Google Scholar] [CrossRef]
- Yi, J.; Chen, B.; Yao, X.; Lei, Y.; Ou, F.; Huang, F. Upregulation of the lncRNA MEG3 improves cognitive impairment, alleviates neuronal damage, and inhibits activation of astrocytes in hippocampus tissues in Alzheimer’s disease through inactivating the PI3K/Akt signaling pathway. J. Cell Biochem. 2019, 120, 18053–18065. [Google Scholar] [CrossRef] [PubMed]
- Gao, Y.; Zhang, N.; Lv, C.; Li, N.; Li, X.; Li, W. lncRNA SNHG1 Knockdown Alleviates Amyloid-beta-Induced Neuronal Injury by Regulating ZNF217 via Sponging miR-361-3p in Alzheimer’s Disease. J. Alzheimers Dis. 2020, 77, 85–98. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Cai, M.; Lou, Y.; Zhang, S.; Liu, X. ZBTB20-AS1 promoted Alzheimer’s disease progression through ZBTB20/GSK-3beta/Tau pathway. Biochem. Biophys. Res. Commun. 2023, 640, 88–96. [Google Scholar] [CrossRef] [PubMed]
- Wang, Q.; Ge, X.; Zhang, J.; Chen, L. Effect of lncRNA WT1-AS regulating WT1 on oxidative stress injury and apoptosis of neurons in Alzheimer’s disease via inhibition of the miR-375/SIX4 axis. Aging 2020, 12, 23974–23995. [Google Scholar] [CrossRef]
- Gu, C.; Chen, C.; Wu, R.; Dong, T.; Hu, X.; Yao, Y.; Zhang, Y. Long Noncoding RNA EBF3-AS Promotes Neuron Apoptosis in Alzheimer’s Disease. DNA Cell Biol. 2018, 37, 220–226. [Google Scholar] [CrossRef]
- Yan, Y.; Yan, H.; Teng, Y.; Wang, Q.; Yang, P.; Zhang, L.; Cheng, H.; Fu, S. Long non-coding RNA 00507/miRNA-181c-5p/TTBK1/MAPT axis regulates tau hyperphosphorylation in Alzheimer’s disease. J. Gene Med. 2020, 22, e3268. [Google Scholar] [CrossRef]
- Li, X.; Wang, S.W.; Li, X.L.; Yu, F.Y.; Cong, H.M. Retraction Note: Knockdown of long non-coding RNA TUG1 depresses apoptosis of hippocampal neurons in Alzheimer’s disease by elevating microRNA-15a and repressing ROCK1 expression. Inflamm. Res. 2023, 72, 5. [Google Scholar] [CrossRef]
- Li, Y.; Jin, L.; Wang, F.; Ren, L.; Pen, R.; Bo, G.; Wang, L. Epigenetic axis of SNHG19/miR-137/TNFAIP1 modulates amyloid beta peptide 25–35-induced SH-SY5Y cytotoxicity. Epigenomics 2022, 14, 187–198. [Google Scholar] [CrossRef]
- Zhou, B.; Li, L.; Qiu, X.; Wu, J.; Xu, L.; Shao, W. Long non-coding RNA ANRIL knockdown suppresses apoptosis and pro-inflammatory cytokines while enhancing neurite outgrowth via binding microRNA-125a in a cellular model of Alzheimer’s disease. Mol. Med. Rep. 2020, 22, 1489–1497. [Google Scholar] [CrossRef]
- Jiang, Q.; Shan, K.; Qun-Wang, X.; Zhou, R.M.; Yang, H.; Liu, C.; Li, Y.J.; Yao, J.; Li, X.M.; Shen, Y.; et al. Long non-coding RNA-MIAT promotes neurovascular remodeling in the eye and brain. Oncotarget 2016, 7, 49688–49698. [Google Scholar] [CrossRef]
- Zhang, L.; Fang, Y.; Cheng, X.; Lian, Y.J.; Xu, H.L. Silencing of Long Noncoding RNA SOX21-AS1 Relieves Neuronal Oxidative Stress Injury in Mice with Alzheimer’s Disease by Upregulating FZD3/5 via the Wnt Signaling Pathway. Mol. Neurobiol. 2019, 56, 3522–3537. [Google Scholar] [CrossRef] [PubMed]
- Guan, F.; Gao, Q.; Dai, X.; Li, L.; Bao, R.; Gu, J. LncRNA RP11-59J16.2 aggravates apoptosis and increases tau phosphorylation by targeting MCM2 in AD. Front. Genet. 2022, 13, 824495. [Google Scholar] [CrossRef]
- Wang, J.; Zhou, T.; Wang, T.; Wang, B. Suppression of lncRNA-ATB prevents amyloid-beta-induced neurotoxicity in PC12 cells via regulating miR-200/ZNF217 axis. Biomed. Pharmacother. 2018, 108, 707–715. [Google Scholar] [CrossRef] [PubMed]
- Gu, R.; Liu, R.; Wang, L.; Tang, M.; Li, S.R.; Hu, X. LncRNA RPPH1 attenuates Abeta(25–35)-induced endoplasmic reticulum stress and apoptosis in SH-SY5Y cells via miR-326/PKM2. Int. J. Neurosci. 2021, 131, 425–432. [Google Scholar] [CrossRef]
- Cai, Y.; Sun, Z.; Jia, H.; Luo, H.; Ye, X.; Wu, Q.; Xiong, Y.; Zhang, W.; Wan, J. Rpph1 Upregulates CDC42 Expression and Promotes Hippocampal Neuron Dendritic Spine Formation by Competing with miR-330-5p. Front. Mol. Neurosci. 2017, 10, 27. [Google Scholar] [CrossRef] [PubMed]
- Yamanaka, Y.; Faghihi, M.A.; Magistri, M.; Alvarez-Garcia, O.; Lotz, M.; Wahlestedt, C. Antisense RNA controls LRP1 Sense transcript expression through interaction with a chromatin-associated protein, HMGB2. Cell Rep. 2015, 11, 967–976. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Zhang, M.; Liu, H. LncRNA17A regulates autophagy and apoptosis of SH-SY5Y cell line as an in vitro model for Alzheimer’s disease. Biosci. Biotechnol. Biochem. 2019, 83, 609–621. [Google Scholar] [CrossRef]
- Liu, Q.; Ling, Z.; Zhang, J.; Yu, H.; Wang, Y.; Xue, Y.; Wang, C.; Zhao, J.; Cao, J.; Duan, S.; et al. lncRNA MIR600HG Knockdown Alleviates Cognitive Impairment in Alzheimer’s Disease Through NEDD4L Mediated PINK1 Degradation. J. Alzheimers Dis. 2022, 85, 1783–1794. [Google Scholar] [CrossRef]
- Tang, Z.B.; Chen, H.P.; Zhong, D.; Song, J.H.; Cao, J.W.; Zhao, M.Q.; Han, B.C.; Duan, Q.; Sheng, X.M.; Yao, J.L.; et al. LncRNA RMRP accelerates autophagy-mediated neurons apoptosis through miR-3142/TRIB3 signaling axis in alzheimer’s disease. Brain Res. 2022, 1785, 147884. [Google Scholar] [CrossRef]
- Zhou, Y.; Ge, Y.; Liu, Q.; Li, Y.X.; Chao, X.; Guan, J.J.; Diwu, Y.C.; Zhang, Q. LncRNA BACE1-AS Promotes Autophagy-Mediated Neuronal Damage Through The miR-214-3p/ATG5 Signalling Axis in Alzheimer’s Disease. Neuroscience 2021, 455, 52–64. [Google Scholar] [CrossRef]
- Fan, Y.; Zhang, J.; Zhuang, X.; Geng, F.; Jiang, G.; Yang, X. Epigenetic transcripts of LINC01311 and hsa-miR-146a-5p regulate neural development in a cellular model of Alzheimer’s disease. IUBMB Life 2021, 73, 916–926. [Google Scholar] [CrossRef] [PubMed]
- Jia, Y.M.; Zhu, C.F.; She, Z.Y.; Wu, M.M.; Wu, Y.Y.; Zhou, B.Y.; Zhang, N. Effects on Autophagy of Moxibustion at Governor Vessel Acupoints in APP/PS1double-Transgenic Alzheimer’s Disease Mice through the lncRNA Six3os1/miR-511-3p/AKT3 Molecular Axis. Evid. Based Complement Alternat. Med. 2022, 2022, 3881962. [Google Scholar] [CrossRef] [PubMed]
- Boros, F.A.; Vecsei, L.; Klivenyi, P. NEAT1 on the Field of Parkinson’s Disease: Offense, Defense, or a Player on the Bench? J. Parkinsons Dis. 2021, 11, 123–138. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Lu, Z. Long non-coding RNA NEAT1 mediates the toxic of Parkinson’s disease induced by MPTP/MPP+ via regulation of gene expression. Clin. Exp. Pharmacol. Physiol. 2018, 45, 841–848. [Google Scholar] [CrossRef]
- Lv, K.; Liu, Y.; Zheng, Y.; Dai, S.; Yin, P.; Miao, H. Long non-coding RNA MALAT1 regulates cell proliferation and apoptosis via miR-135b-5p/GPNMB axis in Parkinson’s disease cell model. Biol. Res. 2021, 54, 10. [Google Scholar] [CrossRef] [PubMed]
- Cai, L.J.; Tu, L.; Huang, X.M.; Huang, J.; Qiu, N.; Xie, G.H.; Liao, J.X.; Du, W.; Zhang, Y.Y.; Tian, J.Y. LncRNA MALAT1 facilitates inflammasome activation via epigenetic suppression of Nrf2 in Parkinson’s disease. Mol. Brain 2020, 13, 130. [Google Scholar] [CrossRef] [PubMed]
- Liu, W.; Zhang, Q.; Zhang, J.; Pan, W.; Zhao, J.; Xu, Y. Long non-coding RNA MALAT1 contributes to cell apoptosis by sponging miR-124 in Parkinson disease. Cell Biosci. 2017, 7, 19. [Google Scholar] [CrossRef]
- Huang, H.; Zheng, S.; Lu, M. Downregulation of lncRNA MEG3 is involved in Parkinson’s disease. Metab. Brain Dis. 2021, 36, 2323–2328. [Google Scholar] [CrossRef]
- Liu, L.; Zhou, T.; Li, T.; Liang, Z.; Luo, X. LncRNA DLX6-AS1 promotes microglial inflammatory response in Parkinson’s disease by regulating the miR-223-3p/NRP1 axis. Behav. Brain Res. 2022, 431, 113923. [Google Scholar] [CrossRef]
- Zhang, Y.; Xia, Q.; Lin, J. LncRNA H19 Attenuates Apoptosis in MPTP-Induced Parkinson’s Disease Through Regulating miR-585-3p/PIK3R3. Neurochem. Res. 2020, 45, 1700–1710. [Google Scholar] [CrossRef]
- Jiang, J.; Piao, X.; Hu, S.; Gao, J.; Bao, M. LncRNA H19 diminishes dopaminergic neuron loss by mediating microRNA-301b-3p in Parkinson’s disease via the HPRT1-mediated Wnt/beta-catenin signaling pathway. Aging 2020, 12, 8820–8836. [Google Scholar] [CrossRef]
- Zhai, K.; Liu, B.; Gao, L. Long-Noncoding RNA TUG1 Promotes Parkinson’s Disease via Modulating MiR-152-3p/PTEN Pathway. Hum. Gene. Ther. 2020, 31, 1274–1287. [Google Scholar] [CrossRef]
- Chen, C.; Zhang, S.; Wei, Y.; Sun, X. LncRNA RMST Regulates Neuronal Apoptosis and Inflammatory Response via Sponging miR-150-5p in Parkinson’s Disease. Neuroimmunomodulation 2022, 29, 55–62. [Google Scholar] [CrossRef]
- Ma, X.; Wang, Y.; Yin, H.; Hua, L.; Zhang, X.; Xiao, J.; Yuan, Q.; Wang, S.; Liu, Y.; Zhang, S.; et al. Down-regulated long non-coding RNA RMST ameliorates dopaminergic neuron damage in Parkinson’s disease rats via regulation of TLR/NF-kappaB signaling pathway. Brain Res. Bull. 2021, 174, 22–30. [Google Scholar] [CrossRef]
- Wang, C.; Zhang, H.; Li, J. LncRNA JHDM1D-AS1 Suppresses MPP +-Induced Neuronal Injury in Parkinson’s Disease via miR-134-5p/PIK3R3 Axis. Neurotox. Res. 2021, 39, 1771–1781. [Google Scholar] [CrossRef] [PubMed]
- Song, Q.; Geng, Y.; Li, Y.; Wang, L.; Qin, J. Long noncoding RNA NORAD regulates MPP+-induced Parkinson’s disease model cells. J. Chem. Neuroanat. 2019, 101, 101668. [Google Scholar] [CrossRef] [PubMed]
- Zhou, S.; Zhang, D.; Guo, J.; Chen, Z.; Chen, Y.; Zhang, J. Long non-coding RNA NORAD functions as a microRNA-204-5p sponge to repress the progression of Parkinson’s disease in vitro by increasing the solute carrier family 5 member 3 expression. IUBMB Life 2020, 72, 2045–2055. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Q.; Zhang, M.M.; Liu, M.; Tan, Z.G.; Qin, Q.L.; Jiang, Y.G. LncRNA XIST sponges miR-199a-3p to modulate the Sp1/LRRK2 signal pathway to accelerate Parkinson’s disease progression. Aging 2021, 13, 4115–4137. [Google Scholar] [CrossRef]
- Zhang, J.; Yang, Y.; Zhou, C.; Zhu, R.; Xiao, X.; Zhou, B.; Wan, D. LncRNA miR-17-92a-1 cluster host gene (MIR17HG) promotes neuronal damage and microglial activation by targeting the microRNA-153-3p/alpha-synuclein axis in Parkinson’s disease. Bioengineered 2022, 13, 4493–4516. [Google Scholar] [CrossRef]
- Li, Y.; Wang, H.; Zhan, L.; Li, Q.; Li, Y.; Wu, G.; Wei, H.; Dong, X. LncRNA FER1L4 promotes differentiation and inhibits proliferation of NSCs via miR-874-3p/Ascl2. Am. J. Transl. Res. 2022, 14, 2256–2266. [Google Scholar] [PubMed]
- Shen, Y.; Cui, X.; Hu, Y.; Zhang, Z.; Zhang, Z. LncRNA-MIAT regulates the growth of SHSY5Y cells by regulating the miR-34-5p-SYT1 axis and exerts a neuroprotective effect in a mouse model of Parkinson’s disease. Am. J. Transl. Res. 2021, 13, 9993–10013. [Google Scholar]
- Li, L.; Wang, H.; Li, H.; Lu, X.; Gao, Y.; Guo, X. Long noncoding RNA BACE1-antisense transcript plays a critical role in Parkinson’s disease via microRNA-214-3p/Cell death-inducing p53-target protein 1 axis. Bioengineered 2022, 13, 10889–10901. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Fang, J.; Zhou, Z.; Zhou, Q.; Sun, S.; Jin, Z.; Xi, Z.; Wei, J. Downregulation of lncRNA BACE1-AS improves dopamine-dependent oxidative stress in rats with Parkinson’s disease by upregulating microRNA-34b-5p and downregulating BACE1. Cell Cycle 2020, 19, 1158–1171. [Google Scholar] [CrossRef] [PubMed]
- Yang, P.; Lin, G.; Wang, M.; Chen, X.; Huang, J. Long non-coding RNA ANRIL interacts with microRNA-34a and microRNA-125a, and they all correlate with disease risk and severity of Parkinson’s disease. J. Clin. Lab. Anal. 2022, 36, e24037. [Google Scholar] [CrossRef] [PubMed]
- Yuan, X.; Wu, Y.; Lu, L.; Feng, J. Long noncoding RNA SNHG14 knockdown exerts a neuroprotective role in MPP(+)-induced Parkinson’s disease cell model through mediating miR-135b-5p/KPNA4 axis. Metab. Brain Dis. 2022, 37, 2363–2373. [Google Scholar] [CrossRef] [PubMed]
- Huo, X.; Wang, L.; Shao, J.; Zhou, C.; Ying, X.; Zhao, J.; Jin, X. LINC00667 regulates MPP(+)-induced neuronal injury in Parkinson’s disease. Ann. Clin. Transl. Neurol. 2022, 9, 707–721. [Google Scholar] [CrossRef]
- Li, J.; Sun, Z.; Song, L. LncRNA SNHG15 mediates 1-methyl-4-phenylpyridinium (MPP(+))-induced neuronal damage through targeting miR-29c-3p/SNCA axis. Neurol. Res. 2023, 45, 181–190. [Google Scholar] [CrossRef]
- Xu, X.; Zhuang, C.; Wu, Z.; Qiu, H.; Feng, H.; Wu, J. LincRNA-p21 Inhibits Cell Viability and Promotes Cell Apoptosis in Parkinson’s Disease through Activating alpha-Synuclein Expression. Biomed. Res. Int. 2018, 2018, 8181374. [Google Scholar] [CrossRef]
- Cai, L.; Tu, L.; Li, T.; Yang, X.; Ren, Y.; Gu, R.; Zhang, Q.; Yao, H.; Qu, X.; Wang, Q.; et al. Downregulation of lncRNA UCA1 ameliorates the damage of dopaminergic neurons, reduces oxidative stress and inflammation in Parkinson’s disease through the inhibition of the PI3K/Akt signaling pathway. Int. Immunopharmacol. 2019, 75, 105734. [Google Scholar] [CrossRef]
- Lian, H.; Wang, B.; Lu, Q.; Chen, B.; Yang, H. LINC00943 knockdown exerts neuroprotective effects in Parkinson’s disease through regulates CXCL12 expression by sponging miR-7-5p. Genes Genom. 2021, 43, 797–805. [Google Scholar] [CrossRef]
- Lun, P.; Ji, T.; Wan, D.H.; Liu, X.; Chen, X.D.; Yu, S.; Sun, P. HOTTIP downregulation reduces neuronal damage and microglial activation in Parkinson’s disease cell and mouse models. Neural. Regen. Res. 2022, 17, 887–897. [Google Scholar] [PubMed]
- Ma, J.; Sun, W.; Chen, S.; Wang, Z.; Zheng, J.; Shi, X.; Li, M.; Li, D.; Gu, Q. The long noncoding RNA GAS5 potentiates neuronal injury in Parkinson’s disease by binding to microRNA-150 to regulate Fosl1 expression. Exp. Neurol. 2022, 347, 113904. [Google Scholar] [CrossRef] [PubMed]
- Xu, W.; Zhang, L.; Geng, Y.; Liu, Y.; Zhang, N. Long noncoding RNA GAS5 promotes microglial inflammatory response in Parkinson’s disease by regulating NLRP3 pathway through sponging miR-223-3p. Int. Immunopharmacol. 2020, 85, 106614. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Wang, Z.; Hu, K.; Liu, H. Downregulation of long noncoding RNA SNHG7 protects against inflammation and apoptosis in Parkinson’s disease model by targeting the miR-425-5p/TRAF5/NF-kappaB axis. J. Biochem. Mol. Toxicol. 2021, 35, e22867. [Google Scholar] [CrossRef] [PubMed]
- Yan, L.; Li, L.; Lei, J. Long noncoding RNA small nucleolar RNA host gene 12/microRNA-138-5p/nuclear factor I/B regulates neuronal apoptosis, inflammatory response, and oxidative stress in Parkinson’s disease. Bioengineered 2021, 12, 12867–12879. [Google Scholar] [CrossRef]
- Peng, T.; Liu, X.; Wang, J.; Liu, Y.; Fu, Z.; Ma, X.; Li, J.; Sun, G.; Ji, Y.; Lu, J.; et al. Long noncoding RNA HAGLROS regulates apoptosis and autophagy in Parkinson’s disease via regulating miR-100/ATG10 axis and PI3K/Akt/mTOR pathway activation. Artif. Cells Nanomed. Biotechnol. 2019, 47, 2764–2774. [Google Scholar] [CrossRef]
- Shen, Y.; Cui, X.; Xu, N.; Hu, Y.; Zhang, Z. lncRNA PART1 mitigates MPP(+)-induced neuronal injury in SH-SY5Y cells via micRNA-106b-5p/MCL1 axis. Am. J. Transl. Res. 2021, 13, 8897–8908. [Google Scholar]
- Guo, Y.; Liu, Y.; Wang, H.; Liu, P. Long noncoding RNA SRY-box transcription factor 2 overlapping transcript participates in Parkinson’s disease by regulating the microRNA-942-5p/nuclear apoptosis-inducing factor 1 axis. Bioengineered 2021, 12, 8570–8582. [Google Scholar] [CrossRef]
- Cao, H.; Han, X.; Jia, Y.; Zhang, B. Inhibition of long non-coding RNA HOXA11-AS against neuroinflammation in Parkinson’s disease model via targeting miR-124-3p mediated FSTL1/NF-kappaB axis. Aging 2021, 13, 11455–11469. [Google Scholar] [CrossRef]
- Yang, X.; Zhang, Y.; Chen, Y.; He, X.; Qian, Y.; Xu, S.; Gao, C.; Mo, C.; Chen, S.; Xiao, Q. LncRNA HOXA-AS2 regulates microglial polarization via recruitment of PRC2 and epigenetic modification of PGC-1alpha expression. J. Neuroinflamm. 2021, 18, 197. [Google Scholar] [CrossRef]
- Lang, Y.; Li, Y.; Yu, H.; Lin, L.; Chen, X.; Wang, S.; Zhang, H. HOTAIR drives autophagy in midbrain dopaminergic neurons in the substantia nigra compacta in a mouse model of Parkinson’s disease by elevating NPTX2 via miR-221-3p binding. Aging 2020, 12, 7660–7678. [Google Scholar] [CrossRef] [PubMed]
- Zhao, J.; Li, H.; Chang, N. LncRNA HOTAIR promotes MPP+-induced neuronal injury in Parkinson’s disease by regulating the miR-874-5p/ATG10 axis. EXCLI J. 2020, 19, 1141–1153. [Google Scholar] [PubMed]
- Fan, Y.; Zhao, X.; Lu, K.; Cheng, G. LncRNA BDNF-AS promotes autophagy and apoptosis in MPTP-induced Parkinson’s disease via ablating microRNA-125b-5p. Brain Res. Bull. 2020, 157, 119–127. [Google Scholar] [CrossRef] [PubMed]
- Qian, C.; Ye, Y.; Mao, H.; Yao, L.; Sun, X.; Wang, B.; Zhang, H.; Xie, L.; Zhang, H.; Zhang, Y.; et al. Downregulated lncRNA-SNHG1 enhances autophagy and prevents cell death through the miR-221/222 /p27/mTOR pathway in Parkinson’s disease. Exp. Cell Res. 2019, 384, 111614. [Google Scholar] [CrossRef]
- Zhao, Y.; Xie, Y.; Yao, W.Y.; Wang, Y.Y.; Song, N. Long non-coding RNA Opa interacting protein 5-antisense RNA 1 promotes mitochondrial autophagy and protects SH-SY5Y cells from 1-methyl-4-phenylpyridine-induced damage by binding to microRNA-137 and upregulating NIX. Kaohsiung J. Med. Sci. 2022, 38, 207–217. [Google Scholar] [CrossRef]
- Song, Z.; Xie, B. LncRNA OIP5-AS1 reduces alpha-synuclein aggregation and toxicity by targeting miR-126 to activate PLK2 in human neuroblastoma SH-SY5Y cells. Neurosci. Lett. 2021, 740, 135482. [Google Scholar] [CrossRef]
- Chanda, K.; Das, S.; Chakraborty, J.; Bucha, S.; Maitra, A.; Chatterjee, R.; Mukhopadhyay, D.; Bhattacharyya, N.P. Altered Levels of Long NcRNAs Meg3 and Neat1 in Cell and Animal Models of Huntington’s Disease. RNA Biol. 2018, 15, 1348–1363. [Google Scholar] [CrossRef]
- Cheng, C.; Spengler, R.M.; Keiser, M.S.; Monteys, A.M.; Rieders, J.M.; Ramachandran, S.; Davidson, B.L. The long non-coding RNA NEAT1 is elevated in polyglutamine repeat expansion diseases and protects from disease gene-dependent toxicities. Hum. Mol. Genet. 2018, 27, 4303–4314. [Google Scholar] [CrossRef]
- Dong, X.; Cong, S. DNM3OS regulates GAPDH expression and influences the molecular pathogenesis of Huntington’s disease. J. Cell Mol. Med. 2021, 25, 9066–9071. [Google Scholar] [CrossRef]
- Francelle, L.; Galvan, L.; Gaillard, M.C.; Petit, F.; Bernay, B.; Guillermier, M.; Bonvento, G.; Dufour, N.; Elalouf, J.M.; Hantraye, P.; et al. Striatal long noncoding RNA Abhd11os is neuroprotective against an N-terminal fragment of mutant huntingtin in vivo. Neurobiol. Aging 2015, 36, 1601.e7–1601.e16. [Google Scholar] [CrossRef]
- Nishimoto, Y.; Nakagawa, S.; Okano, H. NEAT1 lncRNA and amyotrophic lateral sclerosis. Neurochem. Int. 2021, 150, 105175. [Google Scholar] [CrossRef] [PubMed]
- Rey, F.; Maghraby, E.; Messa, L.; Esposito, L.; Barzaghini, B.; Pandini, C.; Bordoni, M.; Gagliardi, S.; Diamanti, L.; Raimondi, M.T.; et al. Identification of a novel pathway in sporadic Amyotrophic Lateral Sclerosis mediated by the long non-coding RNA ZEB1-AS1. Neurobiol. Dis. 2023, 178, 106030. [Google Scholar] [CrossRef] [PubMed]
- Li, K.; Wang, Z. lncRNA NEAT1: Key player in neurodegenerative diseases. Ageing Res. Rev. 2023, 86, 101878. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Hamblin, M.H.; Yin, K.J. The long noncoding RNA Malat1: Its physiological and pathophysiological functions. RNA Biol. 2017, 14, 1705–1714. [Google Scholar] [CrossRef]
- Abrishamdar, M.; Jalali, M.S.; Rashno, M. MALAT1 lncRNA and Parkinson’s Disease: The role in the Pathophysiology and Significance for Diagnostic and Therapeutic Approaches. Mol. Neurobiol. 2022, 59, 5253–5262, Erratum in Mol. Neurobiol. 2022, 59, 5263. [Google Scholar] [CrossRef] [PubMed]
- Feng, Y.S.; Tan, Z.X.; Wu, L.Y.; Dong, F.; Zhang, F. The involvement of NLRP3 inflammasome in the treatment of Alzheimer’s disease. Ageing Res. Rev. 2020, 64, 101192. [Google Scholar] [CrossRef]
- Martin, J.P.; Anders, W.; Maelenn, M.G.; Gemma, C.A.; Yu-Tzu, W.; Matthew, P. World Alzheimer Report 2015—The Global Impact of Dementia: An Analysis of Prevalence, Incidence, Cost and Trends; Alzheimer’s Disease International: London, UK, 2015. [Google Scholar]
- Fang, E.F.; Hou, Y.; Palikaras, K.; Adriaanse, B.A.; Kerr, J.S.; Yang, B.; Lautrup, S.; Hasan-Olive, M.M.; Caponio, D.; Dan, X.; et al. Mitophagy inhibits amyloid-beta and tau pathology and reverses cognitive deficits in models of Alzheimer’s disease. Nat. Neurosci. 2019, 22, 401–412. [Google Scholar] [CrossRef]
- Khan, S.; Barve, K.H.; Kumar, M.S. Recent Advancements in Pathogenesis, Diagnostics and Treatment of Alzheimer’s Disease. Curr. Neuropharmacol. 2020, 18, 1106–1125. [Google Scholar] [CrossRef]
- Huang, Z.; Zhao, J.; Wang, W.; Zhou, J.; Zhang, J. Depletion of LncRNA NEAT1 Rescues Mitochondrial Dysfunction Through NEDD4L-Dependent PINK1 Degradation in Animal Models of Alzheimer’s Disease. Front. Cell Neurosci. 2020, 14, 28. [Google Scholar] [CrossRef]
- Cao, Y.; Li, Q.; Liu, L.; Wu, H.; Huang, F.; Wang, C.; Lan, Y.; Zheng, F.; Xing, F.; Zhou, Q.; et al. Modafinil protects hippocampal neurons by suppressing excessive autophagy and apoptosis in mice with sleep deprivation. Br. J. Pharmacol. 2019, 176, 1282–1297. [Google Scholar] [CrossRef]
- Liu, T.; Huang, Y.; Chen, J.; Chi, H.; Yu, Z.; Wang, J.; Chen, C. Attenuated ability of BACE1 to cleave the amyloid precursor protein via silencing long noncoding RNA BACE1—AS expression. Mol. Med. Rep. 2014, 10, 1275–1281. [Google Scholar] [CrossRef] [PubMed]
- Nair, V.D.; Ge, Y. Alterations of miRNAs reveal a dysregulated molecular regulatory network in Parkinson’s disease striatum. Neurosci. Lett. 2016, 629, 99–104. [Google Scholar] [CrossRef] [PubMed]
- Lyu, Y.; Bai, L.; Qin, C. Long noncoding RNAs in neurodevelopment and Parkinson’s disease. Animal Model Exp. Med. 2019, 2, 239–251. [Google Scholar] [CrossRef] [PubMed]
- Simon, D.K.; Tanner, C.M.; Brundin, P. Parkinson Disease Epidemiology, Pathology, Genetics, and Pathophysiology. Clin. Geriatr. Med. 2020, 36, 1–12. [Google Scholar] [CrossRef]
- Hou, X.; Watzlawik, J.O.; Fiesel, F.C.; Springer, W. Autophagy in Parkinson’s Disease. J. Mol. Biol. 2020, 432, 2651–2672. [Google Scholar] [CrossRef]
- Cacabelos, R. Parkinson’s Disease: From Pathogenesis to Pharmacogenomics. Int. J. Mol. Sci. 2017, 18, 551. [Google Scholar] [CrossRef]
- Luk, K.C.; Kehm, V.; Carroll, J.; Zhang, B.; O’Brien, P.; Trojanowski, J.Q.; Lee, V.M. Pathological alpha-synuclein transmission initiates Parkinson-like neurodegeneration in nontransgenic mice. Science 2012, 338, 949–953. [Google Scholar] [CrossRef]
- Yan, W.; Chen, Z.Y.; Chen, J.Q.; Chen, H.M. LncRNA NEAT1 promotes autophagy in MPTP-induced Parkinson’s disease through stabilizing PINK1 protein. Biochem. Biophys. Res. Commun. 2018, 496, 1019–1024. [Google Scholar] [CrossRef]
- Dong, L.I.; Zheng, Y.; Gao, L.; Luo, X. lncRNA NEAT1 prompts autophagy and apoptosis in MPTP-induced Parkinson’s disease by impairing miR-374c-5p. Acta Biochim. Biophys. Sin. 2021, 53, 870–882. [Google Scholar] [CrossRef]
- Dong, L.; Zheng, Y.; Luo, X. lncRNA NEAT1 promotes autophagy of neurons in mice by impairing miR-107-5p. Bioengineered 2022, 13, 12261–12274. [Google Scholar] [CrossRef]
- Kraus, T.F.J.; Haider, M.; Spanner, J.; Steinmaurer, M.; Dietinger, V.; Kretzschmar, H.A. Altered Long Noncoding RNA Expression Precedes the Course of Parkinson’s Disease—A Preliminary Report. Mol. Neurobiol. 2017, 54, 2869–2877. [Google Scholar] [CrossRef] [PubMed]
- Safari, M.R.; Komaki, A.; Arsang-Jang, S.; Taheri, M.; Ghafouri-Fard, S. Expression Pattern of Long Non-coding RNAs in Schizophrenic Patients. Cell Mol. Neurobiol. 2019, 39, 211–221. [Google Scholar] [CrossRef] [PubMed]
- Gammon, K. Neurodegenerative disease: Brain windfall. Nature 2014, 515, 299–300. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Yang, X.; Song, Y.Q.; Tu, J. Autophagy in Alzheimer’s disease pathogenesis: Therapeutic potential and future perspectives. Ageing Res. Rev. 2021, 72, 101464. [Google Scholar] [CrossRef] [PubMed]
- Long, J.M.; Holtzman, D.M. Alzheimer Disease: An Update on Pathobiology and Treatment Strategies. Cell 2019, 179, 312–339. [Google Scholar] [CrossRef]
- Kumar, D.; Md Ashraf, G.; Bilgrami, A.L.; Imtaiyaz Hassan, M. Emerging therapeutic developments in neurodegenerative diseases: A clinical investigation. Drug Discov. Today 2022, 27, 103305. [Google Scholar] [CrossRef]
- Sun, J.; Roy, S. Gene-based therapies for neurodegenerative diseases. Nat. Neurosci. 2021, 24, 297–311. [Google Scholar] [CrossRef]
- De Gioia, R.; Biella, F.; Citterio, G.; Rizzo, F.; Abati, E.; Nizzardo, M.; Bresolin, N.; Comi, G.P.; Corti, S. Neural Stem Cell Transplantation for Neurodegenerative Diseases. Int. J. Mol. Sci. 2020, 21, 3103. [Google Scholar] [CrossRef]
- Triaca, V.; Ruberti, F.; Canu, N. NGF and the Amyloid Precursor Protein in Alzheimer’s Disease: From Molecular Players to Neuronal Circuits. Adv. Exp. Med. Biol. 2021, 1331, 145–165. [Google Scholar] [PubMed]
- Rafii, M.S.; Tuszynski, M.H.; Thomas, R.G.; Barba, D.; Brewer, J.B.; Rissman, R.A.; Siffert, J.; Aisen, P.S.; Team, A.N.S. Adeno-Associated Viral Vector (Serotype 2)-Nerve Growth Factor for Patients with Alzheimer Disease: A Randomized Clinical Trial. JAMA Neurol. 2018, 75, 834–841. [Google Scholar] [CrossRef]
- Sudhakar, V.; Richardson, R.M. Gene Therapy for Neurodegenerative Diseases. Neurotherapeutics 2019, 16, 166–175. [Google Scholar] [CrossRef] [PubMed]
- Macdonald, R.; Barnes, K.; Hastings, C.; Mortiboys, H. Mitochondrial abnormalities in Parkinson’s disease and Alzheimer’s disease: Can mitochondria be targeted therapeutically? Biochem. Soc. Trans. 2018, 46, 891–909. [Google Scholar] [CrossRef] [PubMed]
- Deng, Z.; Dong, Y.; Zhou, X.; Lu, J.H.; Yue, Z. Pharmacological modulation of autophagy for Alzheimer’s disease therapy: Opportunities and obstacles. Acta Pharm. Sin. B 2022, 12, 1688–1706. [Google Scholar] [CrossRef] [PubMed]
- Lu, J.; Wu, M.; Yue, Z. Autophagy and Parkinson’s Disease. Adv. Exp. Med. Biol. 2020, 1207, 21–51. [Google Scholar] [PubMed]
- Policarpo, R.; Sierksma, A.; De Strooper, B.; d’Ydewalle, C. From Junk to Function: LncRNAs in CNS Health and Disease. Front. Mol. Neurosci. 2021, 14, 714768. [Google Scholar] [CrossRef]
- Roberts, T.C.; Langer, R.; Wood, M.J.A. Advances in oligonucleotide drug delivery. Nat. Rev. Drug Discov. 2020, 19, 673–694. [Google Scholar] [CrossRef]
- Grafals-Ruiz, N.; Rios-Vicil, C.I.; Lozada-Delgado, E.L.; Quinones-Diaz, B.I.; Noriega-Rivera, R.A.; Martinez-Zayas, G.; Santana-Rivera, Y.; Santiago-Sanchez, G.S.; Valiyeva, F.; Vivas-Mejia, P.E. Brain Targeted Gold Liposomes Improve RNAi Delivery for Glioblastoma. Int. J. Nanomed. 2020, 15, 2809–2828. [Google Scholar] [CrossRef]
- Bernat, V.; Disney, M.D. RNA Structures as Mediators of Neurological Diseases and as Drug Targets. Neuron 2015, 87, 28–46. [Google Scholar] [CrossRef]
- Wu, Y.Y.; Kuo, H.C. Functional roles and networks of non-coding RNAs in the pathogenesis of neurodegenerative diseases. J. Biomed. Sci. 2020, 27, 49. [Google Scholar] [CrossRef]
- Matsui, M.; Corey, D.R. Non-coding RNAs as drug targets. Nat. Rev. Drug Discov. 2017, 16, 167–179. [Google Scholar] [CrossRef]
| Alzheimer’s Disease | Parkinson’s Disease | ||
|---|---|---|---|
| NEAT1 [26,27] MALAT1 [28,29] SNHG14 [30,31] HOTAIR [32,33] MAGI2-AS3 [34] XIST [35,36] GAS5 [37] BDNF-AS [38] BC200 [39] H19 [40] MEG3 [41] SNHG1 [42] ZBTB20-AS1 [43] WT1-AS [44] EBF3-AS [45] | 00507 [46] TUG1 [47] SNHG19 [48] ANRIL [49] MIAT [50] SOX21-AS1 [51] RP11-59J16.2 [52] ATB [53] RPPH1 [54,55] LRP1-AS [56] 17A [57] MIR600HG [58] RMRP [59] BACE1-AS [60] LINC-01311 [61] SIX3OS1 [62] | NEAT1 [63,64] MALAT1 [65,66,67] MEG3 [68] DLX6-AS1 [69] H19 [70,71] TUG1 [72] RMST [73,74] JHDM1D-AS1 [75] NORAD [76,77] XIST [78] MIR17HG [79] FER1L4 [80] MIAT [81] BACE1-AS [82,83] ANRIL [84] SNHG14 [85] Linc-00667 [86] | SNHG15 [87] LINCRNA-P21 [88] UCA1 [89] LINC-00943 [90] HOTTIP [91] GAS5 [92,93] SNHG7 [94] SNHG12 [95] HAGLROS [96] PART1 [97] SOX2-OT [98] HOXA11-AS [99] HOXA-AS2 [100] HOTAIR [101,102] BDNF-AS [103] SNHG1 [104] OIP5-AS1 [105,106] |
| Huntington’s Disease | Amyotrophic Lateral Sclerosis | ||
| NEAT1 [107,108] | DNM3OS [109] ABHD11OS [110] | NEAT1 [111] | ZEB1-AS1 [112] |
| MEG3 [107] | |||
| Neurodegenerative Diseases | LncRNA | Role in Autophagy | Role in NDDs |
|---|---|---|---|
| Alzheimer’s disease | 17A [57] | Inhibit | Promote |
| NEAT1 [120] | |||
| MIR600HG [58] | |||
| SIX3OS1 [62] | |||
| RMRP [59] | Activate | Promote | |
| BACE1-AS [60] | |||
| LINC-01311 [61] | Inhibit | Suppress | |
| Parkinson’s disease | NEAT1 [129,130,131] | Activate | Promote |
| HOTAIR [101,102] | |||
| BDNF-AS [103] | |||
| SNHG1 [104] | Inhibit | Promote | |
| OIP5-AS1 [105,106] | Activate | Suppress |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Jiang, Y.; Xu, N. The Emerging Role of Autophagy-Associated lncRNAs in the Pathogenesis of Neurodegenerative Diseases. Int. J. Mol. Sci. 2023, 24, 9686. https://doi.org/10.3390/ijms24119686
Jiang Y, Xu N. The Emerging Role of Autophagy-Associated lncRNAs in the Pathogenesis of Neurodegenerative Diseases. International Journal of Molecular Sciences. 2023; 24(11):9686. https://doi.org/10.3390/ijms24119686
Chicago/Turabian StyleJiang, Yapei, and Naihan Xu. 2023. "The Emerging Role of Autophagy-Associated lncRNAs in the Pathogenesis of Neurodegenerative Diseases" International Journal of Molecular Sciences 24, no. 11: 9686. https://doi.org/10.3390/ijms24119686
APA StyleJiang, Y., & Xu, N. (2023). The Emerging Role of Autophagy-Associated lncRNAs in the Pathogenesis of Neurodegenerative Diseases. International Journal of Molecular Sciences, 24(11), 9686. https://doi.org/10.3390/ijms24119686





