SPINK2 Protein Expression Is an Independent Adverse Prognostic Marker in AML and Is Potentially Implicated in the Regulation of Ferroptosis and Immune Response
Abstract
1. Introduction
2. Results
2.1. Identification of SPINK2 and Assessment of Its Protein Expression in AML Patients
2.2. Mutational and Clinicopathological Associations of SPINK2 in AML
2.3. High SPINK2 Expression Contributes to Therapy Resistance in AML
2.4. High SPINK2 Expression Refines Current Prognostic Stratification and Is an Independent Adverse Prognostic Marker
2.5. Transcriptome Analysis Reveals a Potential Link between SPINK2 and Ferroptosis-Related Genes
2.6. Identification and Testing of SPINK2 Small-Molecule Inhibitor (SMI)
2.7. Genetic and Pharmacologic Modulation of SPINK2 Expression Influences Sensitivity to Erastin, a Ferroptosis Inducer
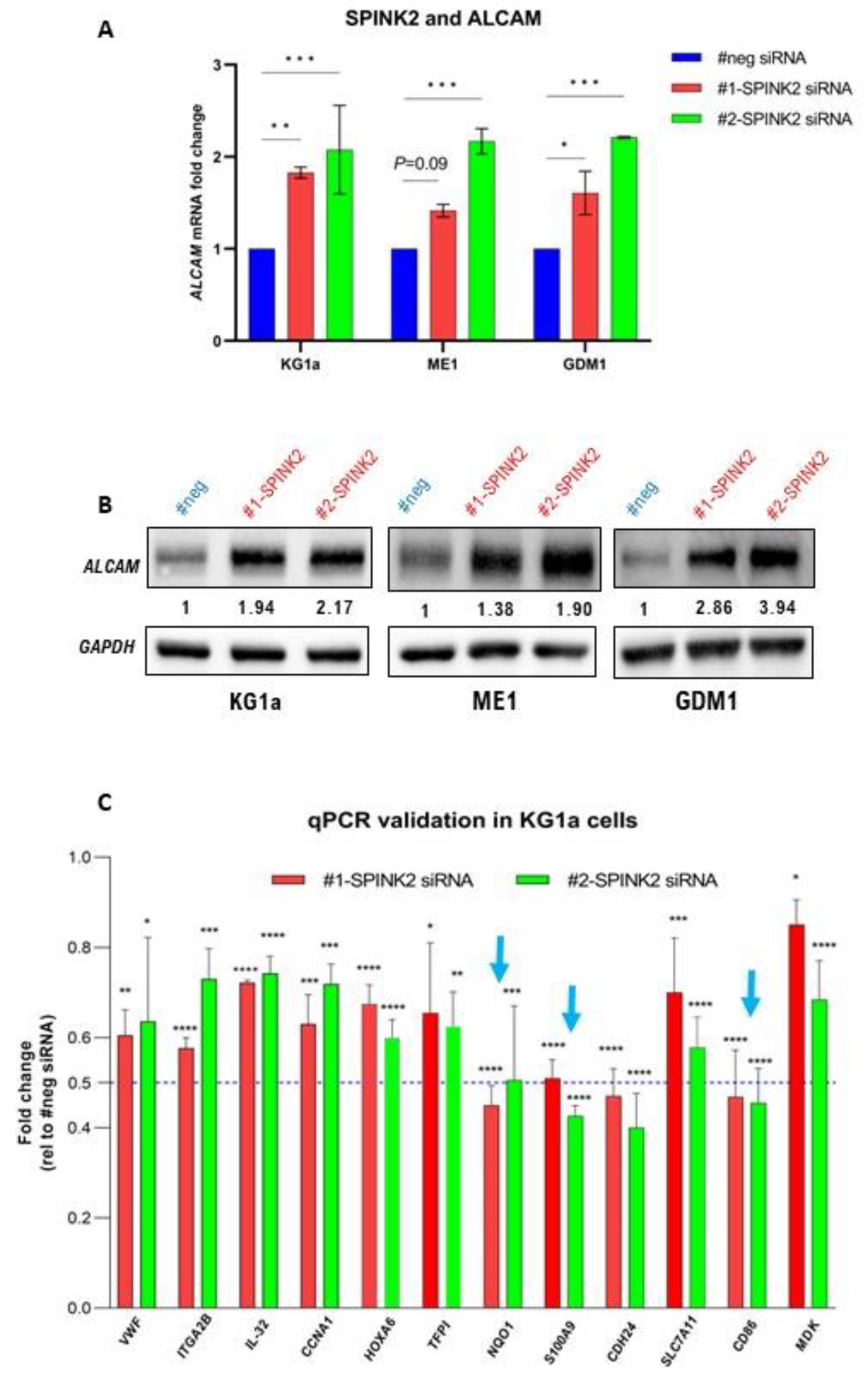
2.8. SPINK2 Modulation Affects Expression of Immune-Response-Related Genes in LSC-Like Cells
3. Discussion
4. Materials and Methods
4.1. Antibodies and Drugs
4.2. Immunohistochemistry (IHC)
4.3. In-House Adult AML Patient Dataset and Exclusion Criteria for Survival Analysis
4.4. Definition of Clinical End-Points
4.5. SPINK2 IHC Score Calculation and Prognostic Cut-Off Determination
4.6. RNA Extraction, Quantitative Polymerase Chain Reaction (qPCR)
4.7. Targeted Next-Generation DNA Sequencing
4.8. Cell Lines and Cell Culture
4.9. RNA Interference
4.10. Lentiviral Transduction
4.11. Transcriptome Sequencing
4.12. Biochemical Assays
4.13. Western Blotting
4.14. Drug Treatment and Cell Viability Assays
4.15. Statistical Analyses
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Ferrara, F.; Schiffer, C.A. Acute myeloid leukaemia in adults. Lancet 2013, 381, 484–495. [Google Scholar] [CrossRef] [PubMed]
- Estey, E.H. Acute myeloid leukemia: 2019 update on risk-stratification and management. Am. J. Hematol. 2018, 93, 1267–1291. [Google Scholar] [CrossRef] [PubMed]
- Roussel, X.; Daguindau, E.; Berceanu, A.; Desbrosses, Y.; Warda, W.; Da Rocha, M.N.; Trad, R.; Deconinck, E.; Deschamps, M.; Ferrand, C. Acute Myeloid Leukemia: From Biology to Clinical Practices Through Development and Pre-Clinical Therapeutics. Front. Oncol. 2020, 10, 599933. [Google Scholar] [CrossRef]
- Döhner, H.; Wei, A.H.; Appelbaum, F.R.; Craddock, C.; DiNardo, C.D.; Dombret, H.; Ebert, B.L.; Fenaux, P.; Godley, L.A.; Hasserjian, R.P.; et al. Diagnosis and management of AML in adults: 2022 recommendations from an international expert panel on behalf of the ELN. Blood 2022, 140, 1345–1377. [Google Scholar] [CrossRef] [PubMed]
- Roloff, G.W.; Odenike, O.; Bajel, A.; Wei, A.H.; Foley, N.; Uy, G.L. Contemporary Approach to Acute Myeloid Leukemia Therapy in 2022. Am. Soc. Clin. Oncol. Educ. Book 2022, 42, 1–16. [Google Scholar] [CrossRef] [PubMed]
- Kantarjian, H.; Kadia, T.; DiNardo, C.; Daver, N.; Borthakur, G.; Jabbour, E.; Garcia-Manero, G.; Konopleva, M.; Ravandi, F. Acute myeloid leukemia: Current progress and future directions. Blood Cancer J. 2021, 11, 41. [Google Scholar] [CrossRef]
- Knorr, K.L.B.; Goldberg, A.D. Leukemia stem cell gene expression signatures contribute to acute myeloid leukemia risk stratification. Haematologica 2020, 105, 533–536. [Google Scholar] [CrossRef]
- Terwijn, M.; Zeijlemaker, W.; Kelder, A.; Rutten, A.P.; Snel, A.N.; Scholten, W.J.; Pabst, T.; Verhoef, G.; Löwenberg, B.; Zweegman, S.; et al. Leukemic Stem Cell Frequency: A Strong Biomarker for Clinical Outcome in Acute Myeloid Leukemia. PLoS ONE 2014, 9, e107587. [Google Scholar] [CrossRef]
- Pollyea, D.A.; Jordan, C.T. Therapeutic targeting of acute myeloid leukemia stem cells. Blood 2017, 129, 1627–1635. [Google Scholar] [CrossRef]
- Laverdière, I.; Boileau, M.; Neumann, A.L.; Frison, H.; Mitchell, A.; Ng, S.W.K.; Wang, J.C.Y.; Minden, M.D.; Eppert, K. Leukemic stem cell signatures identify novel therapeutics targeting acute myeloid leukemia. Blood Cancer J. 2018, 8, 52. [Google Scholar] [CrossRef]
- Stelmach, P.; Trumpp, A. Leukemic stem cells and therapy resistance in acute myeloid leukemia. Haematologica 2023, 108, 353–366. [Google Scholar] [CrossRef]
- Khaldoyanidi, S.K.; Hindoyan, A.; Stein, A.; Subklewe, M. Leukemic stem cells as a target for eliminating acute myeloid leukemia: Gaps in translational research. Crit. Rev. Oncol. 2022, 175, 103710. [Google Scholar] [CrossRef]
- Chen, X.; Zhao, L.; Yu, T.; Zeng, J.; Chen, M. SPINK2 is a prognostic biomarker related to immune infiltration in acute myeloid leukemia. Am. J. Transl. Res. 2022, 14, 197–210. [Google Scholar] [PubMed]
- Xue, C.; Zhang, J.; Zhang, G.; Xue, Y.; Zhang, G.; Wu, X. Elevated SPINK2 gene expression is a predictor of poor prognosis in acute myeloid leukemia. Oncol. Lett. 2019, 18, 2877–2884. [Google Scholar] [CrossRef] [PubMed]
- Eshibona, N.; Livesey, M.; Christoffels, A.; Bendou, H. Investigation of distinct gene expression profile patterns that can improve the classification of intermediate-risk prognosis in AML patients. Front. Genet. 2023, 14, 1131159. [Google Scholar] [CrossRef]
- Barresi, V.; Di Bella, V.; Andriano, N.; Privitera, A.P.; Bonaccorso, P.; La Rosa, M.; Iachelli, V.; Spampinato, G.; Pulvirenti, G.; Scuderi, C.; et al. NUP-98 Rearrangements Led to the Identification of Candidate Biomarkers for Primary Induction Failure in Pediatric Acute Myeloid Leukemia. Int. J. Mol. Sci. 2021, 22, 4575. [Google Scholar] [CrossRef]
- Stockwell, B.R. Ferroptosis turns 10: Emerging mechanisms, physiological functions, and therapeutic applications. Cell 2022, 185, 2401–2421. [Google Scholar] [CrossRef]
- Rhodes, D.R.; Yu, J.; Shanker, K.; Deshpande, N.; Varambally, R.; Ghosh, D.; Barrette, T.; Pander, A.; Chinnaiyan, A.M. ONCOMINE: A Cancer Microarray Database and Integrated Data-Mining Platform. Neoplasia 2004, 6, 1–6. [Google Scholar] [CrossRef] [PubMed]
- Barrett, T.; Wilhite, S.E.; Ledoux, P.; Evangelista, C.; Kim, I.F.; Tomashevsky, M.; Marshall, K.A.; Phillippy, K.H.; Sherman, P.M.; Holko, M.; et al. NCBI GEO: Archive for functional genomics data sets—Update. Nucleic Acids Res. 2012, 41, D991–D995. [Google Scholar] [CrossRef] [PubMed]
- Grimwade, D.; Hills, R.K.; Moorman, A.V.; Walker, H.; Chatters, S.; Goldstone, A.H.; Wheatley, K.; Harrison, C.J.; Burnett, A.K.; on behalf of the National Cancer Research Institute Adult Leukaemia Working Group. Refinement of cytogenetic classification in acute myeloid leukemia: Determination of prognostic significance of rare recurring chromosomal abnormalities among 5876 younger adult patients treated in the United Kingdom Medical Research Council trials. Blood 2010, 116, 354–365. [Google Scholar] [CrossRef]
- Cancer Genome Atlas Research Network; Ley, T.J.; Miller, C.; Ding, L.; Raphael, B.J.; Mungall, A.J.; Robertson, A.G.; Hoadley, K.; Triche, T.J.J.; Laird, P.W.; et al. Genomic and epigenomic landscapes of adult de novo acute myeloid leukemia. N. Engl. J. Med. 2013, 368, 2059–2074. [Google Scholar]
- Verhaak, R.G.; Wouters, B.J.; Erpelinck, C.A.; Abbas, S.; Beverloo, H.B.; Lugthart, S.; Lowenberg, B.; Delwel, R.; Valk, P.J. Prediction of molecular subtypes in acute myeloid leukemia based on gene expression profiling. Haematologica 2009, 94, 131–134. [Google Scholar] [CrossRef]
- Tyner, J.W.; Tognon, C.E.; Bottomly, D.; Wilmot, B.; Kurtz, S.E.; Savage, S.L.; Long, N.; Schultz, A.R.; Traer, E.; Abel, M.; et al. Functional genomic landscape of acute myeloid leukaemia. Nat. Cell Biol. 2018, 562, 526–531. [Google Scholar] [CrossRef] [PubMed]
- Balgobind, B.V.; Van den Heuvel-Eibrink, M.M.; De Menezes, R.X.; Reinhardt, D.; Hollink, I.H.I.M.; Arentsen-Peters, S.T.J.C.M.; van Wering, E.R.; Kaspers, G.J.; Cloos, J.; de Bont, E.S.; et al. Evaluation of gene expression signatures predictive of cytogenetic and molecular subtypes of pediatric acute myeloid leukemia. Haematologica 2011, 96, 221–230. [Google Scholar] [CrossRef] [PubMed]
- Bolouri, H.; Farrar, J.E.; Triche, T., Jr.; Ries, R.E.; Lim, E.L.; Alonzo, T.A.; Ma, Y.; Moore, R.; Mungall, A.J.; Marra, M.A.; et al. The molecular landscape of pediatric acute myeloid leukemia reveals recurrent structural alterations and age-specific mutational interactions. Nat. Med. 2018, 24, 103–112. [Google Scholar] [CrossRef] [PubMed]
- Ng, S.W.K.; Mitchell, A.; Kennedy, J.A.; Chen, W.C.; McLeod, J.; Ibrahimova, N.; Arruda, A.; Popescu, A.; Gupta, V.; Schimmer, A.D.; et al. A 17-gene stemness score for rapid determination of risk in acute leukaemia. Nature 2016, 540, 433–437. [Google Scholar] [CrossRef]
- Gentles, A.J.; Plevritis, S.K.; Majeti, R.; Alizadeh, A.A. Association of a Leukemic Stem Cell Gene Expression Signature with Clinical Outcomes in Acute Myeloid Leukemia. JAMA 2010, 304, 2706–2715. [Google Scholar] [CrossRef]
- Eppert, K.; Takenaka, K.; Lechman, E.R.; Waldron, L.; Nilsson, B.; van Galen, P.; Metzeler, K.H.; Poeppl, A.; Ling, V.; Beyene, J.; et al. Stem cell gene expression programs influence clinical outcome in human leukemia. Nat. Med. 2011, 17, 1086–1093. [Google Scholar] [CrossRef]
- Subramanian, A.; Tamayo, P.; Mootha, V.K.; Mukherjee, S.; Ebert, B.L.; Gillette, M.A.; Paulovich, A.; Pomeroy, S.L.; Golub, T.R.; Lander, E.S.; et al. Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. USA 2005, 102, 15545–15550. [Google Scholar] [CrossRef]
- Koppula, P.; Zhuang, L.; Gan, B. Cystine transporter SLC7A11/xCT in cancer: Ferroptosis, nutrient dependency, and cancer therapy. Protein Cell 2021, 12, 599–620. [Google Scholar] [CrossRef]
- Shi, Z.-Z.; Tao, H.; Fan, Z.-W.; Song, S.-J.; Bai, J. Prognostic and Immunological Role of Key Genes of Ferroptosis in Pan-Cancer. Front. Cell Dev. Biol. 2021, 9, 748925. [Google Scholar] [CrossRef] [PubMed]
- Zhao, X.; Li, Y.; Wu, H. A novel scoring system for acute myeloid leukemia risk assessment based on the expression levels of six genes. Int. J. Mol. Med. 2018, 42, 1495–1507. [Google Scholar] [CrossRef] [PubMed]
- Yin, Z.; Li, F.; Zhou, Q.; Zhu, J.; Liu, Z.; Huang, J.; Shen, H.; Ou, R.; Zhu, Y.; Zhang, Q.; et al. A ferroptosis-related gene signature and immune infiltration patterns predict the overall survival in acute myeloid leukemia patients. Front. Mol. Biosci. 2022, 9, 959738. [Google Scholar] [CrossRef]
- Yu, Y.; Xie, Y.; Cao, L.; Yang, L.; Yang, M.; Lotze, M.T.; Zeh, H.J.; Kang, R.; Tang, D. The ferroptosis inducer erastin enhances sensitivity of acute myeloid leukemia cells to chemotherapeutic agents. Mol. Cell. Oncol. 2015, 2, e1054549. [Google Scholar] [CrossRef]
- Birsen, R.; Larrue, C.; Decroocq, J.; Johnson, N.; Guiraud, N.; Gotanegre, M.; Cantero-Aguilar, L.; Grignano, E.; Huynh, T.; Fontenay, M.; et al. APR-246 induces early cell death by ferroptosis in acute myeloid leukemia. Haematologica 2021, 107, 403–416. [Google Scholar] [CrossRef]
- Narr, A.; Alborzinia, H.; Donato, E.; Vogel, F.; Boch, T.; Leppä, A.-M.; Waclawiczek, A.; Renders, S.; Schulze, A.; Trumpp, A. S120: Acute Myeloid Leukemia Represents a Ferroptosis-Sensitive Cancer Entity Raising the Possibility for Novel Targeting Strategies. Hemasphere 2022, 6, 21–22. [Google Scholar] [CrossRef]
- Jiang, L.; Kon, N.; Li, T.; Wang, S.-J.; Su, T.; Hibshoosh, H.; Baer, R.; Gu, W. Ferroptosis as a p53-mediated activity during tumour suppression. Nature 2015, 520, 57–62. [Google Scholar] [CrossRef]
- Liu, Y.; Gu, W. p53 in ferroptosis regulation: The new weapon for the old guardian. Cell Death Differ. 2022, 29, 895–910. [Google Scholar] [CrossRef]
- Kucab, J.E.; Hollstein, M.; Arlt, V.M.; Phillips, D.H. Nutlin-3a selects for cells harbouring TP53 mutations. Int. J. Cancer 2017, 140, 877–887. [Google Scholar] [CrossRef] [PubMed]
- Zhang, F.; Tao, Y.; Zhang, Z.; Guo, X.; An, P.; Shen, Y.; Wu, Q.; Yu, Y.; Wang, F. Metalloreductase Steap3 coordinates the regulation of iron homeostasis and inflammatory responses. Haematologica 2012, 97, 1826–1835. [Google Scholar] [CrossRef] [PubMed]
- Lespagnol, A.; Duflaut, D.; Beekman, C.; Blanc, L.; Fiucci, G.; Marine, J.-C.; Vidal, M.; Amson, R.; Telerman, A. Exosome secretion, including the DNA damage-induced p53-dependent secretory pathway, is severely compromised in TSAP6/Steap3-null mice. Cell Death Differ. 2008, 15, 1723–1733. [Google Scholar] [CrossRef]
- Chen, X.; Yu, C.; Kang, R.; Tang, D. Iron Metabolism in Ferroptosis. Front. Cell Dev. Biol. 2020, 8, 590226. [Google Scholar] [CrossRef]
- Li, Q.; Shah, S. Structure-Based Virtual Screening. Methods Mol. Biol. 2017, 1558, 111–124. [Google Scholar]
- Li, H.; Leung, K.-S.; Wong, M.-H. idock: A multithreaded virtual screening tool for flexible ligand docking. In Proceedings of the 2012 IEEE Symposium on Computational Intelligence in Bioinformatics and Computational Biology (CIBCB), San Diego, CA, USA, 9–12 May 2012; pp. 77–84. [Google Scholar]
- Cheng, L.Y.; Du, Y.; Yu, X.; Zhi, Y.C.; Wang, L.; Zheng, Y.F.; Xiu, H.L. STEAP3 Affects Ferroptosis and Progression of Renal Cell Carcinoma Through the p53/xCT Pathway. Technol. Cancer Res. Treat. 2022, 21, 15330338221078728. [Google Scholar]
- Sato, M.; Kusumi, R.; Hamashima, S.; Kobayashi, S.; Sasaki, S.; Komiyama, Y.; Izumikawa, T.; Conrad, M.; Bannai, S.; Sato, H. The ferroptosis inducer erastin irreversibly inhibits system x(c)- and synergizes with cisplatin to increase cisplatin’s cytotoxicity in cancer cells. Sci. Rep. 2018, 8, 968. [Google Scholar] [CrossRef]
- Hanahan, D. Hallmarks of Cancer: New Dimensions. Cancer Discov. 2022, 12, 31–46. [Google Scholar] [CrossRef]
- Tettamanti, S.; Pievani, A.; Biondi, A.; Dotti, G.; Serafini, M. Catch me if you can: How AML and its niche escape immunotherapy. Leukemia 2021, 36, 13–22. [Google Scholar] [CrossRef] [PubMed]
- Paczulla, A.M.; Rothfelder, K.; Raffel, S.; Konantz, M.; Steinbacher, J.; Wang, H.; Tandler, C.; Mbarga, M.; Schaefer, T.; Falcone, M.; et al. Absence of NKG2D ligands defines leukaemia stem cells and mediates their immune evasion. Nat. Cell Biol. 2019, 572, 254–259. [Google Scholar] [CrossRef] [PubMed]
- Zimmerman, A.W.; Joosten, B.; Torensma, R.; Parnes, J.R.; van Leeuwen, F.N.; Figdor, C.G. Long-term engagement of CD6 and ALCAM is essential for T-cell proliferation induced by dendritic cells. Blood 2006, 107, 3212–3220. [Google Scholar] [CrossRef] [PubMed]
- Elsayed, A.H.; Rafiee, R.; Cao, X.; Raimondi, S.; Downing, J.R.; Ribeiro, R.; Fan, Y.; Gruber, T.A.; Baker, S.; Klco, J.; et al. A six-gene leukemic stem cell score identifies high risk pediatric acute myeloid leukemia. Leukemia 2019, 34, 735–745. [Google Scholar] [CrossRef]
- Lamba, J.K.; Cao, X.; Raimondi, S.C.; Rafiee, R.; Downing, J.R.; Shi, L.; Gruber, T.; Ribeiro, R.C.; Rubnitz, J.E.; Pounds, S.B. Integrated epigenetic and genetic analysis identifies markers of prognostic significance in pediatric acute myeloid leukemia. Oncotarget 2018, 9, 26711–26723. [Google Scholar] [CrossRef]
- Nehme, A.; Dakik, H.; Picou, F.; Cheok, M.; Preudhomme, C.; Dombret, H.; Lambert, J.; Gyan, E.; Pigneux, A.; Récher, C.; et al. Horizontal meta-analysis identifies common deregulated genes across AML subgroups providing a robust prognostic signature. Blood Adv. 2020, 4, 5322–5335. [Google Scholar] [CrossRef]
- Mehner, C.; Radisky, E.S. Bad Tumors Made Worse: SPINK1. Front. Cell Dev. Biol. 2019, 7, 10. [Google Scholar] [CrossRef]
- Hoefnagel, J.J.; Dijkman, R.; Basso, K.; Jansen, P.M.; Hallermann, C.; Willemze, R.; Tensen, C.P.; Vermeer, M. Distinct types of primary cutaneous large B-cell lymphoma identified by gene expression profiling. Blood 2005, 105, 3671–3678. [Google Scholar] [CrossRef]
- Chen, T.; Lee, T.-R.; Liang, W.-G.; Chang, W.-S.W.; Lyu, P.-C. Identification of trypsin-inhibitory site and structure determination of human SPINK2 serine proteinase inhibitor. Proteins Struct. Funct. Bioinform. 2009, 77, 209–219. [Google Scholar] [CrossRef]
- Shyu, R.-Y.; Wang, C.-H.; Wu, C.-C.; Wang, L.-K.; Chen, M.-L.; Kuo, C.-Y.; Lee, M.-C.; Lin, Y.-Y.; Tsai, F.-M. Tazarotene-Induced Gene 1 (TIG1) Interacts with Serine Protease Inhibitor Kazal-Type 2 (SPINK2) to Inhibit Cellular Invasion of Testicular Carcinoma Cells. BioMed Res. Int. 2019, 2019, 6171065. [Google Scholar] [CrossRef]
- Kherraf, Z.E.; Christou-Kent, M.; Karaouzene, T.; Amiri-Yekta, A.; Martinez, G.; Vargas, A.S.; Lambert, E.; Borel, C.; Dorphin, B.; Aknin-Seifer, I. SPINK2 deficiency causes infertility by inducing sperm defects in heterozygotes and azoospermia in homozygotes. EMBO Mol. Med. 2017, 9, 1132–1149. [Google Scholar] [CrossRef] [PubMed]
- He, X.; Gonzalez, V.; Tsang, A.; Thompson, J.; Tsang, T.C.; Harris, D.T. Differential Gene Expression Profiling of CD34+CD133+Umbilical Cord Blood Hematopoietic Stem Progenitor Cells. Stem Cells Dev. 2005, 14, 188–198. [Google Scholar] [CrossRef] [PubMed]
- Naldini, M.M.; Casirati, G.; Zonari, E.; DeSantis, G.; Cammarata, A.; Beretta, S.; Merelli, I.; Ciceri, F.; Gentner, B. Dissecting Ex Vivo Expansion of Mobilized Peripheral Blood Hematopoietic Stem and Progenitor Cells By Single Cell RNA Sequencing. Blood 2018, 132, 3343. [Google Scholar] [CrossRef]
- Döhner, K.; Paschka, P. Intermediate-risk acute myeloid leukemia therapy: Current and future. Hematol. Am. Soc. Hematol. Educ. Program 2014, 2014, 34–43. [Google Scholar] [CrossRef]
- Papaemmanuil, E.; Gerstung, M.; Bullinger, L.; Gaidzik, V.I.; Paschka, P.; Roberts, N.D.; Potter, N.E.; Heuser, M.; Thol, F.; Bolli, N.; et al. Genomic Classification and Prognosis in Acute Myeloid Leukemia. N. Engl. J. Med. 2016, 374, 2209–2221. [Google Scholar] [CrossRef] [PubMed]
- Lindsley, R.C.; Mar, B.G.; Mazzola, E.; Grauman, P.V.; Shareef, S.; Allen, S.L.; Pigneux, A.; Wetzler, M.; Stuart, R.K.; Erba, H.P.; et al. Acute myeloid leukemia ontogeny is defined by distinct somatic mutations. Blood 2015, 125, 1367–1376. [Google Scholar] [CrossRef]
- Pollyea, D.A.; Stevens, B.M.; Jones, C.L.; Winters, A.; Pei, S.; Minhajuddin, M.; D’alessandro, A.; Culp-Hill, R.; Riemondy, K.A.; Gillen, A.E.; et al. Venetoclax with azacitidine disrupts energy metabolism and targets leukemia stem cells in patients with acute myeloid leukemia. Nat. Med. 2018, 24, 1859–1866. [Google Scholar] [CrossRef]
- Jordan, C.T. Can we selectively target AML stem cells? Best Pract. Res. Clin. Haematol. 2019, 32, 101100. [Google Scholar] [CrossRef] [PubMed]
- Walter, R.B.; Appelbaum, F.R.; Estey, E.H.; Bernstein, I.D. Acute myeloid leukemia stem cells and CD33-targeted immunotherapy. Blood 2012, 119, 6198–6208. [Google Scholar] [CrossRef]
- Buckley, S.A.; Wood, B.L.; Othus, M.; Hourigan, C.S.; Ustun, C.; Linden, M.A.; DeFor, T.E.; Malagola, M.; Anthias, C.; Valkova, V.; et al. Minimal residual disease prior to allogeneic hematopoietic cell transplantation in acute myeloid leukemia: A meta-analysis. Haematologica 2017, 102, 865–873. [Google Scholar] [CrossRef] [PubMed]
- Jiang, X.; Stockwell, B.R.; Conrad, M. Ferroptosis: Mechanisms, biology and role in disease. Nat. Rev. Mol. Cell Biol. 2021, 22, 266–282. [Google Scholar] [CrossRef]
- Lei, G.; Zhuang, L.; Gan, B. Targeting ferroptosis as a vulnerability in cancer. Nat. Rev. Cancer 2022, 22, 381–396. [Google Scholar] [CrossRef]
- Cosialls, E.; El Hage, R.; Dos Santos, L.; Gong, C.; Mehrpour, M.; Hamaï, A. Ferroptosis: Cancer Stem Cells Rely on Iron until “to Die for” It. Cells 2021, 10, 2981. [Google Scholar] [CrossRef]
- Sato, H.; Tamba, M.; Ishii, T.; Bannai, S. Cloning and Expression of a Plasma Membrane Cystine/Glutamate Exchange Transporter Composed of Two Distinct Proteins. J. Biol. Chem. 1999, 274, 11455–11458. [Google Scholar] [CrossRef]
- Jones, C.L.; Stevens, B.M.; D’alessandro, A.; Culp-Hill, R.; Reisz, J.A.; Pei, S.; Gustafson, A.; Khan, N.; DeGregori, J.; Pollyea, D.A.; et al. Cysteine depletion targets leukemia stem cells through inhibition of electron transport complex II. Blood 2019, 134, 389–394. [Google Scholar] [CrossRef]
- Bowen, M.A.; Patel, D.D.; Li, X.; Modrell, B.; Malacko, A.R.; Wang, W.C.; Marquardt, H.; Neubauer, M.S.; Pesando, J.M.; Francke, U. Cloning, mapping, and characterization of activated leukocyte-cell adhesion molecule (ALCAM), a CD6 ligand. J. Exp. Med. 1995, 181, 2213–2220. [Google Scholar] [CrossRef] [PubMed]
- Gimferrer, I.; Calvo, M.; Mittelbrunn, M.; Farnós, M.; Sarrias, M.R.; Enrich, C.; Vives, J.; Sánchez-Madrid, F.; Lozano, F. Relevance of CD6-mediated interactions in T cell activation and proliferation. J. Immunol. 2004, 173, 2262–2270. [Google Scholar] [CrossRef]
- Riet, J.T.; Helenius, J.; Strohmeyer, N.; Cambi, A.; Figdor, C.; Muller, D.J. Dynamic coupling of ALCAM to the actin cortex strengthens cell adhesion to CD6. J. Cell Sci. 2014, 127, 1595–1606. [Google Scholar] [CrossRef]
- Freitas, R.F.; Basto, A.; Almeida, S.C.; Santos, R.F.; Gonçalves, C.M.; Corria-Osorio, J.; Carvalho, T.; Carmo, A.; Oliveira, V.G.; Leon, K.; et al. Modulation of CD4 T cell function via CD6-targeting. Ebiomedicine 2019, 47, 427–435. [Google Scholar] [CrossRef] [PubMed]
- Archana, S.S.; Selvaraju, S.; Binsila, B.K.; Arangasamy, A.; Krawetz, S.A. Immune regulatory molecules as modifiers of semen and fertility: A review. Mol. Reprod. Dev. 2019, 86, 1485–1504. [Google Scholar] [CrossRef]
- Liao, P.; Wang, W.; Wang, W.; Kryczek, I.; Li, X.; Bian, Y.; Sell, A.; Wei, S.; Grove, S.; Johnson, J.K.; et al. CD8+ T cells and fatty acids orchestrate tumor ferroptosis and immunity via ACSL4. Cancer Cell 2022, 40, 365–378.e6. [Google Scholar] [CrossRef]
- Wang, W.; Green, M.; Choi, J.E.; Gijón, M.; Kennedy, P.D.; Johnson, J.K.; Liao, P.; Lang, X.; Kryczek, I.; Sell, A.; et al. CD8+ T cells regulate tumour ferroptosis during cancer immunotherapy. Nature 2019, 569, 270–274. [Google Scholar] [CrossRef] [PubMed]
- Xu, C.; Gu, X.; Padmanabhan, R.; Wu, Z.; Peng, Q.; DiCarlo, J.; Wang, Y. smCounter2: An accurate low-frequency variant caller for targeted sequencing data with unique molecular identifiers. Bioinformatics 2018, 35, 1299–1309. [Google Scholar] [CrossRef]
- Wang, K.; Li, M.; Hakonarson, H. ANNOVAR: Functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res. 2010, 38, e164. [Google Scholar] [CrossRef]
- Metzeler, K.H.; Herold, T.; Rothenberg-Thurley, M.; Amler, S.; Sauerland, M.C.; Görlich, D.; Schneider, S.; Konstandin, N.P.; Dufour, A.; Bräundl, K.; et al. Spectrum and prognostic relevance of driver gene mutations in acute myeloid leukemia. Blood 2016, 128, 686–698. [Google Scholar] [CrossRef]
- Chakravarty, D.; Gao, J.; Phillips, S.; Kundra, R.; Zhang, H.; Wang, J.; Rudolph, J.E.; Yaeger, R.; Soumerai, T.; Nissan, M.H.; et al. OncoKB: A Precision Oncology Knowledge Base. JCO Precis. Oncol. 2017, 2017, PO.17.00011. [Google Scholar] [CrossRef]
- Kircher, M.; Witten, D.M.; Jain, P.; O’Roak, B.J.; Cooper, G.M.; Shendure, J. A general framework for estimating the relative pathogenicity of human genetic variants. Nat. Genet. 2014, 46, 310–315. [Google Scholar] [CrossRef] [PubMed]
- Sim, N.-L.; Kumar, P.; Hu, J.; Henikoff, S.; Schneider, G.; Ng, P.C. SIFT web server: Predicting effects of amino acid substitutions on proteins. Nucleic Acids Res. 2012, 40, W452–W457. [Google Scholar] [CrossRef] [PubMed]
- A Adzhubei, I.; Schmidt, S.; Peshkin, L.; Ramensky, V.E.; Gerasimova, A.; Bork, P.; Kondrashov, A.S.; Sunyaev, S.R. A method and server for predicting damaging missense mutations. Nat. Methods 2010, 7, 248–249. [Google Scholar] [CrossRef] [PubMed]
- Schwarz, J.M.; Cooper, D.N.; Schuelke, M.; Seelow, D. MutationTaster2: Mutation prediction for the deep-sequencing age. Nat. Methods 2014, 11, 361–362. [Google Scholar] [CrossRef] [PubMed]
- Choi, Y.; Chan, A.P. PROVEAN web server: A tool to predict the functional effect of amino acid substitutions and indels. Bioinformatics 2015, 31, 2745–2747. [Google Scholar] [CrossRef]
- Falini, B.; Mecucci, C.; Tiacci, E.; Alcalay, M.; Rosati, R.; Pasqualucci, L.; La Starza, R.; Diverio, D.; Colombo, E.; Santucci, A.; et al. Cytoplasmic Nucleophosmin in Acute Myelogenous Leukemia with a Normal Karyotype. N. Engl. J. Med. 2005, 352, 254–266. [Google Scholar] [CrossRef] [PubMed]
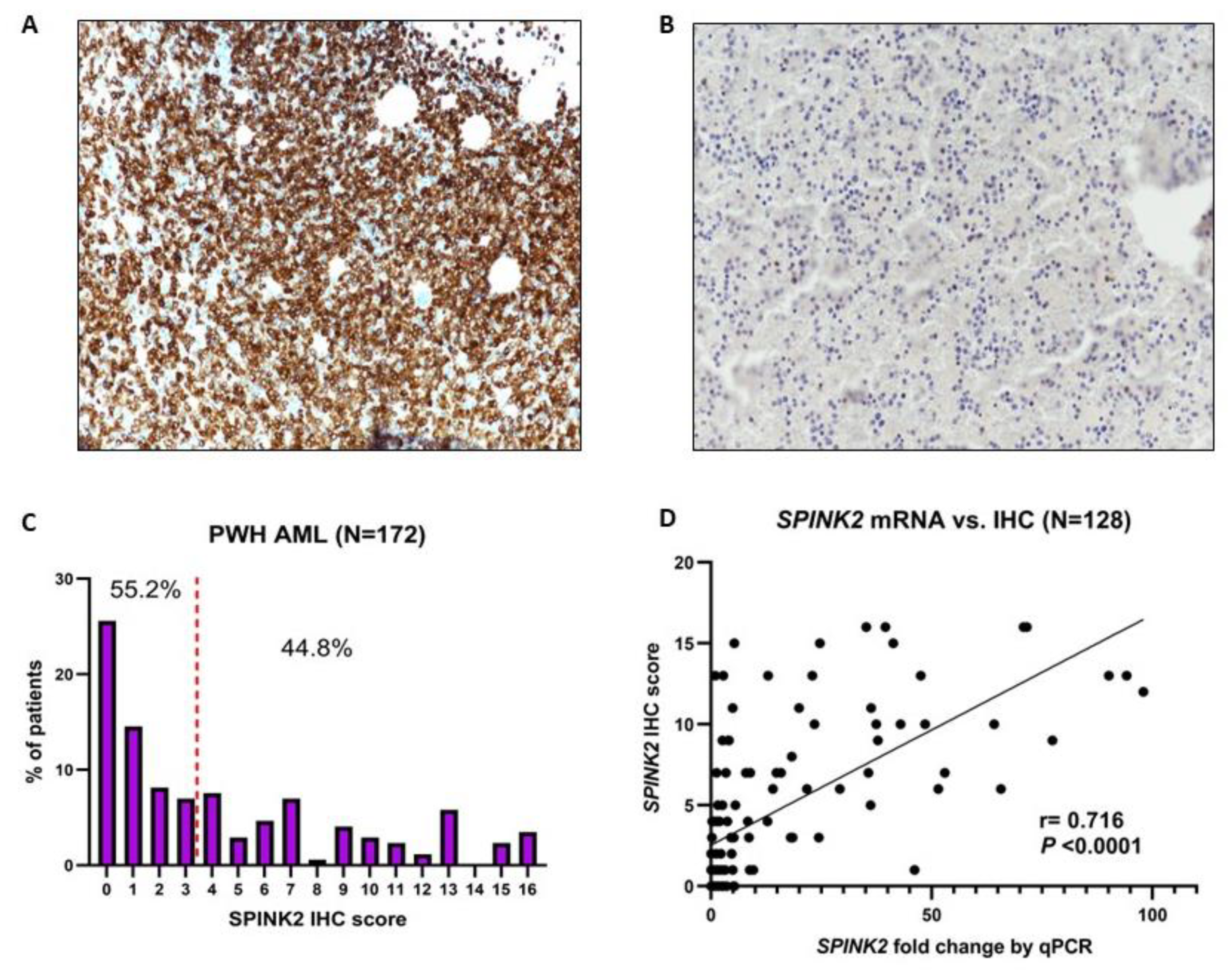
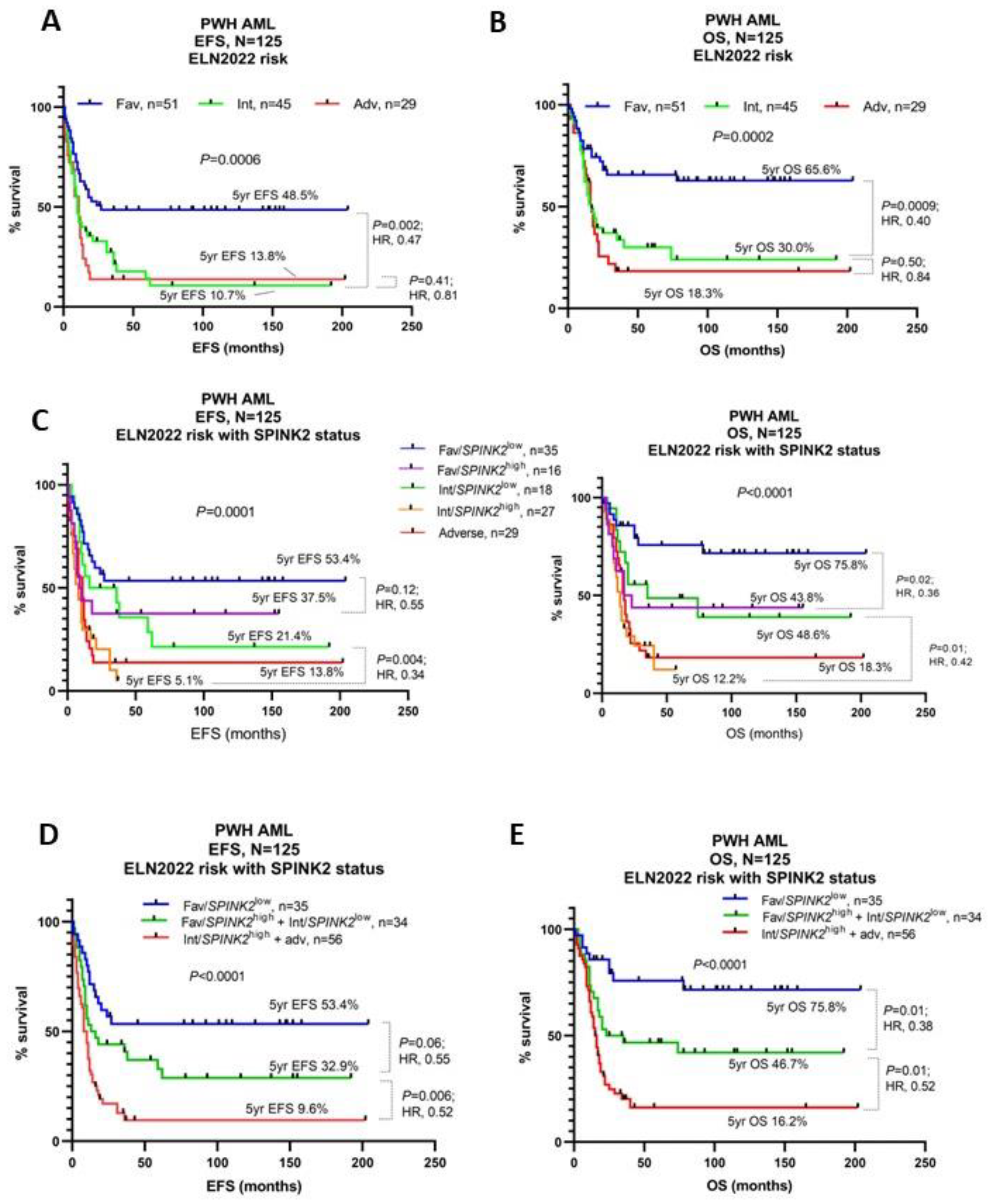
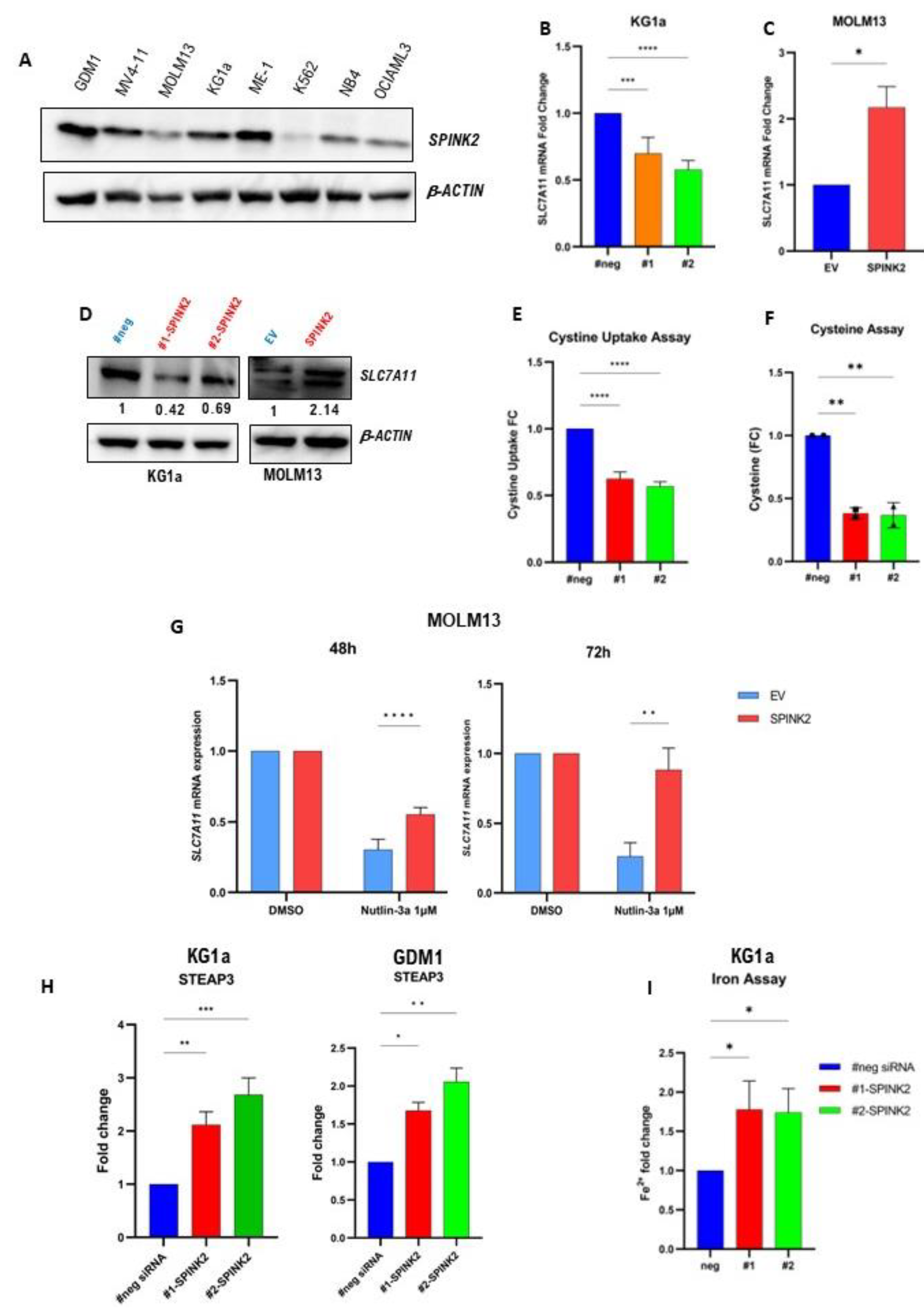

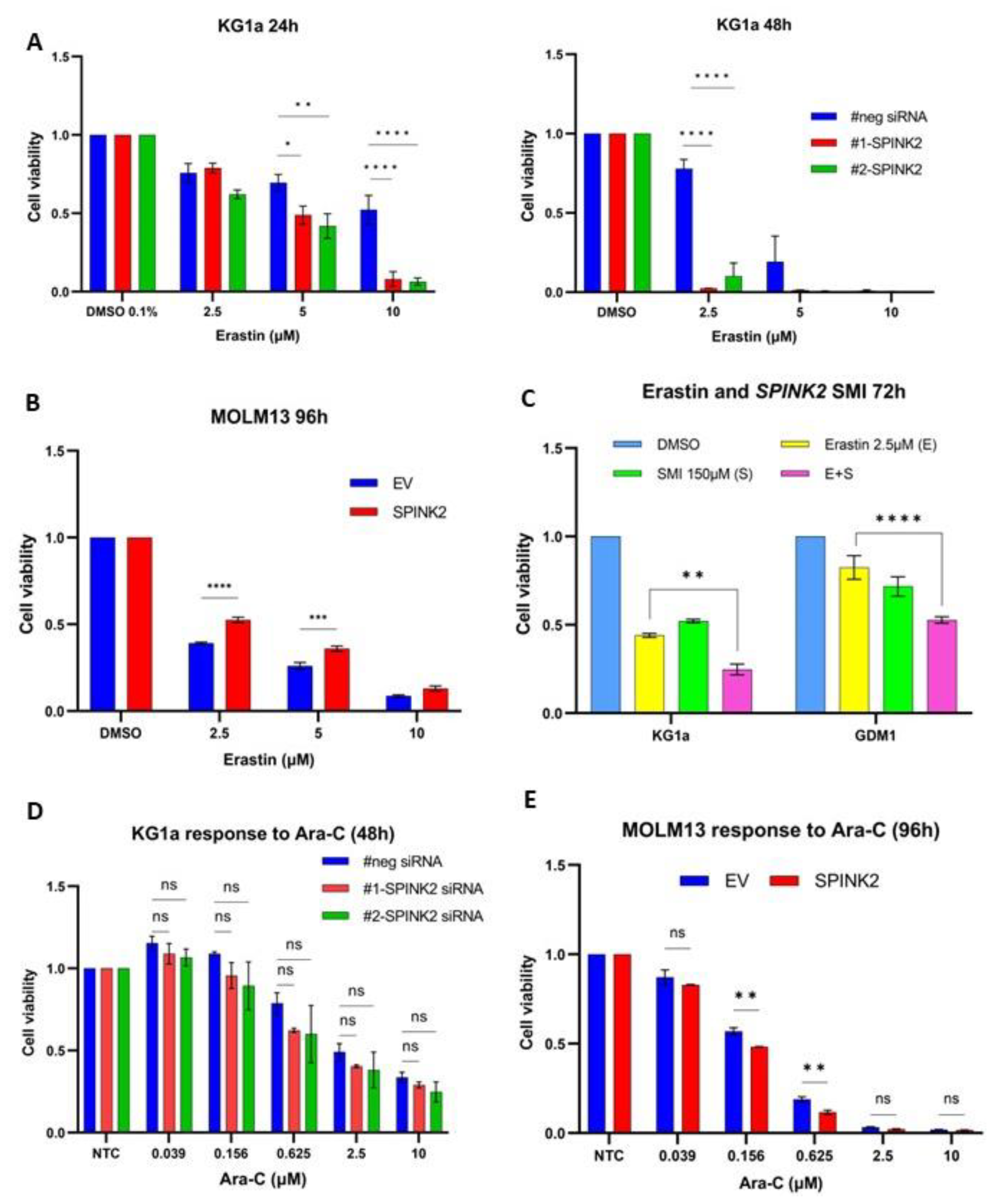
| Characteristic | High SPINK2 (n = 77) | Low SPINK2 (n = 95) | p-Value |
|---|---|---|---|
| Sex | |||
| Male | 41 (53.3%) | 48 (50.5%) | 0.76 |
| Female | 36 (46.7%) | 47 (49.5%) | |
| Age, years | |||
| Median (range) | 54 (20–75) | 51 (18–86) | 0.23 |
| Hb level, g/dL | |||
| Median (range) | 8.5 (3–13.6) | 7.8 (2.9–12.9) | 0.16 |
| Bone Marrow blast, % | |||
| Median (range) | 69 (11–98) | 69 (12–98) | 0.65 |
| WBC level, ×109/L | |||
| Median (range) | 28.1 (1.3–517) | 19.8 (0.9–330.4) | 0.18 |
| Platelets, ×109/L | |||
| Median (range) | 67 (4–748) | 43 (2–247) | <0.001 |
| FAB classification | |||
| M0 | 1/48 (2.1%) | 4/68 (5.9%) | 0.40 |
| M1 | 11/48 (22.9%) | 21/68 (30.9%) | 0.40 |
| M2 | 8/48 (16.7%) | 17/68 (25.0%) | 0.36 |
| M4 (incl. M4Eo) | 11/48 (22.9%) | 13/68 (19.1%) | 0.65 |
| M5 | 16/48 (33.3%) | 11/68 (16.2%) | 0.04 |
| M6 | 1/48 (2.1%) | 2/68 (2.9%) | 0.99 |
| Unclassified | 29/77 (37.6%) | 27/95 (28.4%) | - |
| AML type | |||
| De novo | 71 (92.2%) | 85 (89.5%) | 0.61 |
| Secondary/t-AML | 6 (7.8%) | 10 (10.5%) | |
| MRC Cytogenetic Risk | |||
| Favorable | 5 (6.5%) | 19 (20.7%) | 0.014 |
| Intermediate | 64 (83.1%) | 61 (66.3%) | 0.014 |
| Adverse | 8 (10.4%) | 12 (13.0%) | 0.64 |
| Unclassified | - | 3 | - |
| ELN 2022 risk | |||
| Favorable | 19/73 (26.0%) | 39/92 (42.4%) | 0.033 |
| Intermediate | 34/73 (46.6%) | 24/92 (26.1%) | 0.009 |
| Adverse | 20/73 (27.4%) | 29/92 (31.5%) | 0.61 |
| Cytogenetics | |||
| Normal | 51 (66.2%) | 44 (47.8%) | 0.019 |
| t(8;21) | 0 (0.0%) | 14 (15.2%) | <0.001 |
| inv(16) | 5 (6.5%) | 4 (4.4%) | 0.73 |
| Complex | 5 (6.5%) | 5 (5.4%) | 0.99 |
| Others | 12 (15.6%) | 20 (21.7%) | 0.33 |
| Unknown | - | 3 (3.2%) | - |
| Mutations | |||
| FLT3-ITD | 26/77 (33.8%) | 21/95 (22.1%) | 0.12 |
| NPM1 | 33/77 (42.9%) | 14/95 (14.7%) | <0.0001 |
| CEBPA bZIP | 2/74 (2.7%) | 18/95 (19.0%) | 0.001 |
| DNMT3A | 26/74 (35.1%) | 18/95 (19.0%) | 0.022 |
| NPM1+/DNMT3A+ | 17/74 (23.0%) | 7/95 (7.4%) | 0.007 |
| NPM1+/FLT3-ITD+ | 20/77 (26.0%) | 11/95 (11.6%) | 0.017 |
| NPM1+/FLT3-ITD+/DNMT3A+ | 9/74 (12.2%) | 6/95 (6.3%) | 0.275 |
| TP53 | 1/69 (1.5%) | 2/86 (2.3%) | 0.99 |
| RUNX1 | 8/69 (11.6%) | 12/86 (14.0%) | 0.81 |
| ASXL1 | 4/69 (5.8%) | 4/86 (4.7%) | 0.99 |
| BCOR | 2/69 (2.9%) | 2/86 (2.3%) | 0.99 |
| EZH2 | 1/69 (1.5%) | 3/86 (3.5%) | 0.63 |
| SF3B1 | 0/69 (0.0%) | 1/86 (1.2%) | 0.99 |
| SRSF2 | 4/69 (5.8%) | 4/86 (4.7%) | 0.99 |
| STAG2 | 1/69 (1.5%) | 6/86 (7.0%) | 0.13 |
| U2AF1 | 0/69 (0.0%) | 2/86 (2.3%) | 0.50 |
| ZRSR2 | 2/69 (2.9%) | 0/86 (0.0%) | 0.20 |
| Factor | High SPINK2 | Low SPINK2 | p-Value |
|---|---|---|---|
| Whole cohort (N = 137) | N = 60 | N = 77 | |
| Response to induction | |||
| CR | 73.3% | 88.3% | 0.028 |
| NR1 | 51.7% | 33.8% | 0.038 |
| Relapse ‡ after CR | |||
| Median RFS | 9 months | 37 months | 0.004 |
| 6-month relapse rate | 31.8% | 9.1% | |
| 5 year RFS | 25.8% | 46.8% | |
| Intermediate cytogenetic risk (N = 101) | N = 51 | N = 50 | |
| Response to induction | |||
| CR | 68.6% | 90.0% | 0.01 |
| NR1 | 66.7% | 37.5% | 0.005 |
| Relapse ‡ after CR | |||
| Median RFS | 12 months | 37 months | 0.018 |
| 6-month relapse rate | 31.4% | 6.9% | |
| 5 year RFS | 27.0% | 44.6% | |
| Intermediate risk ELN2022 (N = 47) | N = 28 | N = 19 | |
| Response to induction | |||
| CR | 67.9% | 84.2% | 0.31 |
| NR1 | 67.9% | 21.1% | 0.003 |
| Relapse ‡ after CR | |||
| Median RFS | 14 months | 37 months | 0.034 |
| 6-month relapse rate | 26.3% | 6.7% | |
| 5 year RFS | 17.9% | 34.3% | |
| Normal karyotype (N = 76) | N = 40 | N = 36 | |
| Response to induction | |||
| CR | 72.5% | 91.7% | 0.040 |
| NR1 | 55.0% | 27.8% | 0.021 |
| Relapse‡after CR | |||
| Median RFS | 12 months | 35 months | 0.07 |
| 6-month relapse rate | 31.0% | 6.2% | |
| 5 year RFS | 30.2% | 41.9% | |
| NPM1mut (N = 46) | N = 33 | N = 13 | |
| Response to induction | |||
| CR | 75.8% | 100% | 0.08 |
| NR1 | 48.5% | 15.4% | 0.049 |
| Relapse‡after CR | |||
| Median RFS | 14 months | Unreached | 0.095 |
| 6-month relapse rate | 28.0% | 0.0% | |
| 5 year RFS | 35.5% | 50.5% |
| Covariates | OS | EFS | RFS § | ||||||
|---|---|---|---|---|---|---|---|---|---|
| Whole Cohort ‡ N = 125 ¶ | HR | 95%C.I. | p-value | HR | 95%C.I. | p-value | HR | 95%C.I. | p-value |
| Age ≥ 60 years | 1.29 | 0.68–2.36 | 0.416 | / | / | / | / | / | / |
| SPINK2high | 2.45 | 1.48–4.07 | <0.001 | 2.08 | 1.31–3.32 | 0.002 | 1.89 | 1.12–3.15 | 0.015 |
| CR1 | 0.40 | 0.24–0.67 | <0.001 | 0.33 | 0.21–0.52 | <0.001 | / | / | / |
| SCT ¥ in CR | 0.11 | 0.02–0.37 | 0.0023 | 0.15 | 0.04–0.36 | <0.001 | 0.11 | 0.02–0.37 | 0.003 |
| DNMT3A | 1.20 | 0.72–1.96 | 0.479 | 1.18 | 0.73–1.87 | 0.490 | 1.602 | 0.91–2.73 | 0.090 |
| ELN2022 adv | 1.78 | 1.02–3.02 | 0.037 | 1.86 | 0.94–3.43 | 0.060 | 2.16 | 1.21–3.73 | 0.007 |
| IDH2 | 2.33 | 1.18–4.31 | 0.010 | 1.58 | 0.93–2.59 | 0.080 | / | / | / |
| NPM1mut † N = 42 ¶ | HR | 95%C.I. | p-value | HR | 95%C.I. | p-value | HR | 95%C.I. | p-value |
| Age ≥ 60 years | 9.10 | 2.36–34.39 | 0.001 | 7.53 | 2.01–27.45 | 0.002 | 3.58 | 0.92–12.13 | 0.046 |
| SPINK2high | 5.55 | 1.89–21.32 | 0.005 | 5.11 | 1.91–16.65 | 0.003 | 3.52 | 1.23–11.72 | 0.027 |
| FLT3-ITD | 2.54 | 0.94–8.18 | 0.085 | 3.9 | 1.37–11.94 | 0.017 | 2.47 | 0.88–7.84 | 0.100 |
| DNMT3A | 0.81 | 0.34–1.99 | 0.635 | 1.10 | 0.49–2.57 | 0.824 | 3.12 | 1.20–9.65 | 0.029 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Pitts, H.A.; Cheng, C.-K.; Cheung, J.S.; Sun, M.K.-H.; Yung, Y.-L.; Chan, H.-Y.; Wong, R.S.M.; Yip, S.-F.; Lau, K.-N.; Wong, W.S.; et al. SPINK2 Protein Expression Is an Independent Adverse Prognostic Marker in AML and Is Potentially Implicated in the Regulation of Ferroptosis and Immune Response. Int. J. Mol. Sci. 2023, 24, 9696. https://doi.org/10.3390/ijms24119696
Pitts HA, Cheng C-K, Cheung JS, Sun MK-H, Yung Y-L, Chan H-Y, Wong RSM, Yip S-F, Lau K-N, Wong WS, et al. SPINK2 Protein Expression Is an Independent Adverse Prognostic Marker in AML and Is Potentially Implicated in the Regulation of Ferroptosis and Immune Response. International Journal of Molecular Sciences. 2023; 24(11):9696. https://doi.org/10.3390/ijms24119696
Chicago/Turabian StylePitts, Herbert Augustus, Chi-Keung Cheng, Joyce Sin Cheung, Murphy Ka-Hei Sun, Yuk-Lin Yung, Hoi-Yun Chan, Raymond S. M. Wong, Sze-Fai Yip, Ka-Ngai Lau, Wai Shan Wong, and et al. 2023. "SPINK2 Protein Expression Is an Independent Adverse Prognostic Marker in AML and Is Potentially Implicated in the Regulation of Ferroptosis and Immune Response" International Journal of Molecular Sciences 24, no. 11: 9696. https://doi.org/10.3390/ijms24119696
APA StylePitts, H. A., Cheng, C.-K., Cheung, J. S., Sun, M. K.-H., Yung, Y.-L., Chan, H.-Y., Wong, R. S. M., Yip, S.-F., Lau, K.-N., Wong, W. S., Raghupathy, R., Chan, N. P. H., & Ng, M. H. L. (2023). SPINK2 Protein Expression Is an Independent Adverse Prognostic Marker in AML and Is Potentially Implicated in the Regulation of Ferroptosis and Immune Response. International Journal of Molecular Sciences, 24(11), 9696. https://doi.org/10.3390/ijms24119696







