SF3B1 Mutations Are Associated with Resistance to Non-Genotoxic MDM2 Inhibition in Chronic Lymphocytic Leukemia
Abstract
1. Introduction
2. Results
2.1. Sanger Sequencing Detects SF3B1 Mutations in CLL Samples
2.2. SF3B1 Mutations in CLL Samples Predict Poor Overall Patient Survival
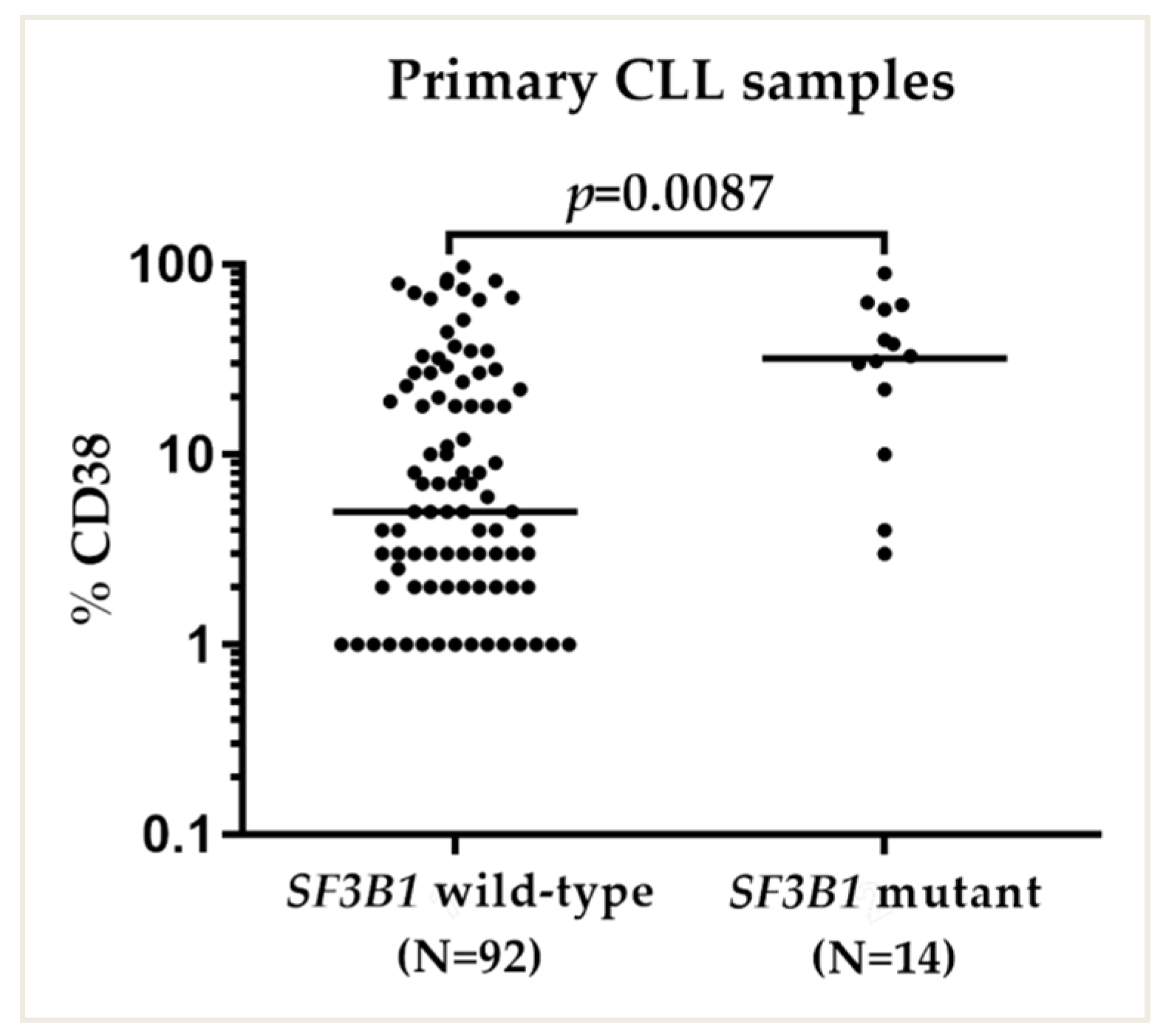
2.3. SF3B1 Mutations Were Associated with High CD38 Expression
2.4. SF3B1 Mutations Negatively Predict for Response to RG7388 in CLL Samples
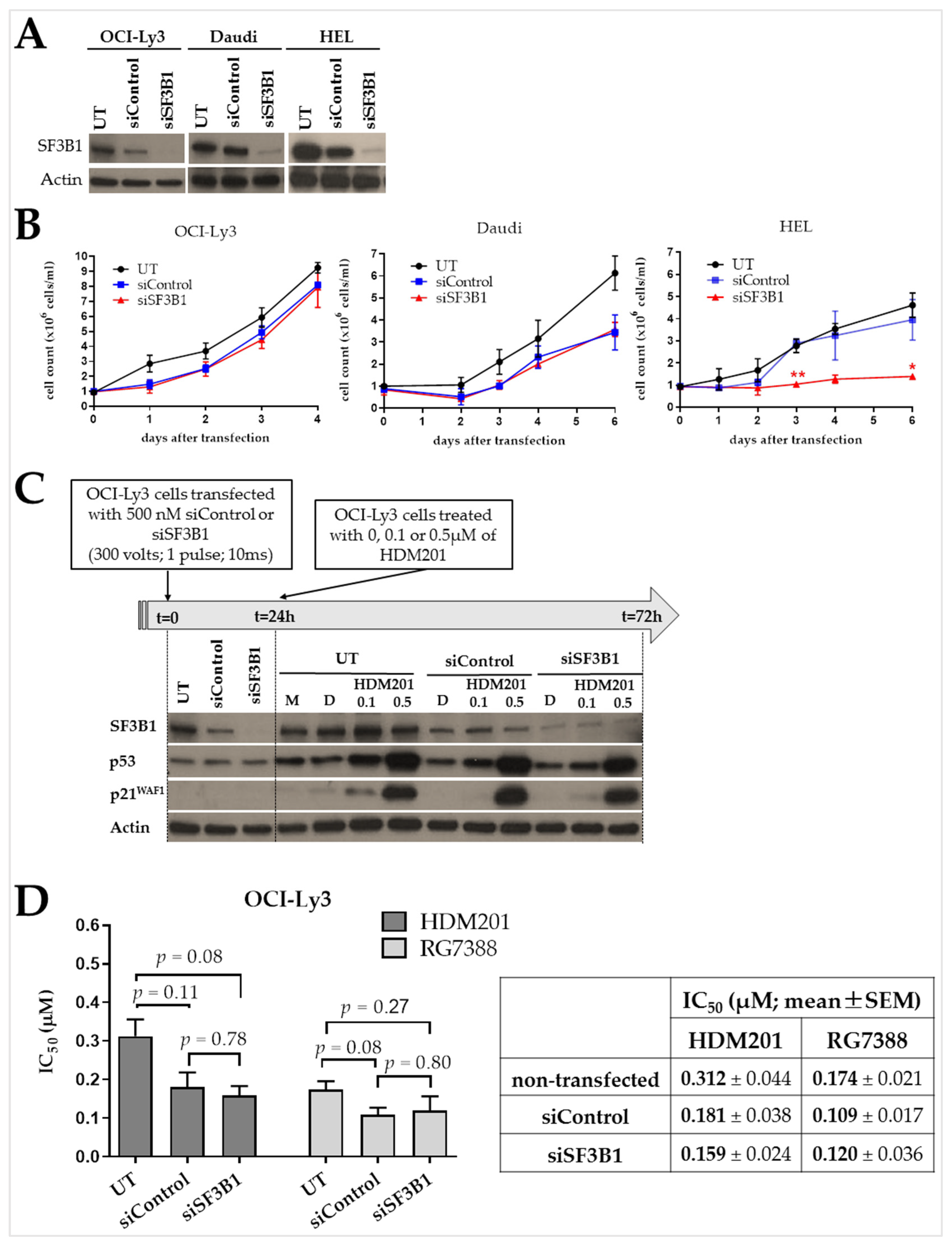
2.5. Nalm-6 SF3B1K700E Cells Are Less Sensitive to Growth Inhibition by RG7388 or HDM201 Compared to Matched SF3B1K700K
2.6. SF3B1 Suppression Has No Effect on the Proliferation of B Cells and Their Response to MDM2 Inhibition
3. Discussion
4. Materials and Methods
4.1. Cell Lines and Compounds
4.2. Patient Samples
4.3. Patient Sample Information
4.4. Cell Viability Assay
4.4.1. Cell Lines
4.4.2. PBMCs
4.5. Design of Synthetic siRNA Molecules
- siSF3B1#1 Sense: CUAGAGAAGUGAUGUUAAU, Antisense: AUUAACAUCACUUCUCUAG
- siSF3B1#2 Sense: GAACACCUAUAUUCGUUAU, Antisense: AUAACGAAUAUAGGUGUUC
- siSF3B1#3 Sense: CAGAGUUCCUGAACUGAAU, Antisense: AUUCAGUUCAGGAACUCUG
- siControl Sense: GCGCGCUUUGUAGGAUUCG, Antisense: CGAAUCCUACAAAGCGCGC
4.6. Transfection Protocol
4.7. Immunoblotting
4.8. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Hallek, M.; Al-Sawaf, O. Chronic lymphocytic leukemia: 2022 update on diagnostic and therapeutic procedures. Am. J. Hematol. 2021, 96, 1679–1705. [Google Scholar] [CrossRef] [PubMed]
- Landau, D.A.; Tausch, E.; Taylor-Weiner, A.N.; Stewart, C.; Reiter, J.G.; Bahlo, J.; Kluth, S.; Bozic, I.; Lawrence, M.; Böttcher, S.; et al. Mutations driving CLL and their evolution in progression and relapse. Nature 2015, 526, 525–530. [Google Scholar] [CrossRef]
- Ahn, I.E.; Tian, X.; Ipe, D.; Cheng, M.; Albitar, M.; Tsao, L.C.; Zhang, L.; Ma, W.; Herman, S.E.M.; Gaglione, E.M.; et al. Prediction of Outcome in Patients With Chronic Lymphocytic Leukemia Treated With Ibrutinib: Development and Validation of a Four-Factor Prognostic Model. J. Clin. Oncol. 2021, 39, 576–585. [Google Scholar] [CrossRef] [PubMed]
- Byrd, J.C.; Wierda, W.G.; Schuh, A.; Devereux, S.; Chaves, J.M.; Brown, J.R.; Hillmen, P.; Martin, P.; Awan, F.T.; Stephens, D.M.; et al. Acalabrutinib monotherapy in patients with relapsed/refractory chronic lymphocytic leukemia: Updated phase 2 results. Blood 2020, 135, 1204–1213. [Google Scholar] [CrossRef]
- Fischer, K.; Al-Sawaf, O.; Bahlo, J.; Fink, A.-M.; Tandon, M.; Dixon, M.; Robrecht, S.; Warburton, S.; Humphrey, K.; Samoylova, O.; et al. Venetoclax and Obinutuzumab in Patients with CLL and Coexisting Conditions. N. Engl. J. Med. 2019, 380, 2225–2236. [Google Scholar] [CrossRef] [PubMed]
- Schulz, H.; Klein, S.K.; Rehwald, U.; Reiser, M.; Hinke, A.; Knauf, W.-U.; Aulitzky, W.-E.; Hensel, M.; Herold, M.; Huhn, D.; et al. Phase 2 study of a combined immunochemotherapy using rituximab and fludarabine in patients with chronic lymphocytic leukemia. Blood 2002, 100, 3115–3120. [Google Scholar] [CrossRef]
- Lane, D.; Levine, A. p53 Research: The Past Thirty Years and the Next Thirty Years. Cold Spring Harb. Perspect. Biol. 2010, 2, a000893. [Google Scholar] [CrossRef]
- Zenz, T.; Eichhorst, B.; Busch, R.; Denzel, T.; Häbe, S.; Winkler, D.; Bühler, A.; Edelmann, J.; Bergmann, M.; Hopfinger, G.; et al. TP53 Mutation and Survival in Chronic Lymphocytic Leukemia. J. Clin. Oncol. 2010, 28, 4473–4479. [Google Scholar] [CrossRef]
- Kojima, K.; Konopleva, M.; McQueen, T.; O’Brien, S.; Plunkett, W.; Andreeff, M. Mdm2 inhibitor Nutlin-3a induces p53-mediated apoptosis by transcription-dependent and transcription-independent mechanisms and may overcome Atm-mediated resistance to fludarabine in chronic lymphocytic leukemia. Blood 2006, 108, 993–1000. [Google Scholar] [CrossRef]
- Wu, C.-E.; Esfandiari, A.; Ho, Y.-H.; Wang, N.; Mahdi, A.; Aptullahoglu, E.; Lovat, P.; Lunec, J. Targeting negative regulation of p53 by MDM2 and WIP1 as a therapeutic strategy in cutaneous melanoma. Br. J. Cancer 2018, 118, 495–508. [Google Scholar] [CrossRef]
- Zanjirband, M.; Edmondson, R.J.; Lunec, J. Pre-clinical efficacy and synergistic potential of the MDM2-p53 antagonists, Nutlin-3 and RG7388, as single agents and in combined treatment with cisplatin in ovarian cancer. Oncotarget 2016, 7, 40115–40134. [Google Scholar] [CrossRef]
- Chamberlain, V.; Drew, Y.; Lunec, J. Tipping Growth Inhibition into Apoptosis by Combining Treatment with MDM2 and WIP1 Inhibitors in p53WT Uterine Leiomyosarcoma. Cancers 2021, 14, 14. [Google Scholar] [CrossRef]
- Ciardullo, C.; Aptullahoglu, E.; Woodhouse, L.; Lin, W.-Y.; Wallis, J.P.; Marr, H.; Marshall, S.; Bown, N.; Willmore, E.; Lunec, J. Non-genotoxic MDM2 inhibition selectively induces a pro-apoptotic p53 gene signature in chronic lymphocytic leukemia cells. Haematologica 2019, 104, 2429–2442. [Google Scholar] [CrossRef] [PubMed]
- Daver, N.G.; Dail, M.; Garcia, J.S.; Jonas, B.A.; Yee, K.W.L.; Kelly, K.R.; Vey, N.; Assouline, S.; Roboz, G.J.; Paolini, S.; et al. Venetoclax and idasanutlin in relapsed/refractory AML: A non-randomized, open-label phase 1b trial. Blood 2022, 141, 1265–1276. [Google Scholar] [CrossRef] [PubMed]
- Gozani, O.; Feld, R.; Reed, R. Evidence that sequence-independent binding of highly conserved U2 snRNP proteins upstream of the branch site is required for assembly of spliceosomal complex A. Genes Dev. 1996, 10, 233–243. [Google Scholar] [CrossRef] [PubMed]
- Kramer, A. The structure and function of proteins involved in mammalian pre-mRNA splicing. Annu. Rev. Biochem. 1996, 65, 367–409. [Google Scholar] [CrossRef]
- Papaemmanuil, E.; Cazzola, M.; Boultwood, J.; Malcovati, L.; Vyas, P.; Bowen, D.; Pellagatti, A.; Wainscoat, J.S.; Hellstrom-Lindberg, E.; Gambacorti-Passerini, C.; et al. SomaticSF3B1Mutation in Myelodysplasia with Ring Sideroblasts. N. Engl. J. Med. 2011, 365, 1384–1395. [Google Scholar] [CrossRef]
- Martin, M.; Maßhöfer, L.; Temming, P.; Rahmann, S.; Metz, C.; Bornfeld, N.; Van De Nes, J.; Klein-Hitpass, L.; Hinnebusch, A.G.; Horsthemke, B.; et al. Exome sequencing identifies recurrent somatic mutations in EIF1AX and SF3B1 in uveal melanoma with disomy 3. Nat. Genet. 2013, 45, 933–936. [Google Scholar] [CrossRef]
- Fabbri, G.; Rasi, S.; Rossi, D.; Trifonov, V.; Khiabanian, H.; Ma, J.; Grunn, A.; Fangazio, M.; Capello, D.; Monti, S.; et al. Analysis of the chronic lymphocytic leukemia coding genome: Role of NOTCH1 mutational activation. J. Exp. Med. 2011, 208, 1389–1401. [Google Scholar] [CrossRef]
- Puente, X.S.; Pinyol, M.; Quesada, V.; Conde, L.; Ordóñez, G.R.; Villamor, N.; Escaramis, G.; Jares, P.; Beà, S.; González-Díaz, M.; et al. Whole-genome sequencing identifies recurrent mutations in chronic lymphocytic leukaemia. Nature 2011, 475, 101–105. [Google Scholar] [CrossRef]
- Obeng, E.A.; Chappell, R.J.; Seiler, M.; Chen, M.C.; Campagna, D.R.; Schmidt, P.J.; Schneider, R.K.; Lord, A.M.; Wang, L.; Gambe, R.G.; et al. Physiologic Expression of Sf3b1(K700E) Causes Impaired Erythropoiesis, Aberrant Splicing, and Sensitivity to Therapeutic Spliceosome Modulation. Cancer Cell 2016, 30, 404–417. [Google Scholar] [CrossRef] [PubMed]
- Mupo, A.; Seiler, M.; Sathiaseelan, V.; Pance, A.; Yang, Y.; Agrawal, A.A.; Iorio, F.; Bautista, R.; Pacharne, S.; Tzelepis, K.; et al. Hemopoietic-specific Sf3b1-K700E knock-in mice display the splicing defect seen in human MDS but develop anemia without ring sideroblasts. Leukemia 2017, 31, 720–727. [Google Scholar] [CrossRef] [PubMed]
- Rossi, D.; Bruscaggin, A.; Spina, V.; Rasi, S.; Khiabanian, H.; Messina, M.; Fangazio, M.; Vaisitti, T.; Monti, S.; Chiaretti, S.; et al. Mutations of the SF3B1 splicing Factor in Chronic Lymphocytic Leukemia: Association with Progression and Fludarabine-Refractoriness. Blood 2011, 118, 215. [Google Scholar] [CrossRef]
- Oscier, D.G.; Rose-Zerilli, M.J.J.; Winkelmann, N.; de Castro, D.G.; Gomez, B.; Forster, J.; Parker, H.; Parker, A.; Gardiner, A.; Collins, A.; et al. The clinical significance of NOTCH1 and SF3B1 mutations in the UK LRF CLL4 trial. Blood 2013, 121, 468–475. [Google Scholar] [CrossRef] [PubMed]
- Stilgenbauer, S.; Schnaiter, A.; Paschka, P.; Zenz, T.; Rossi, M.; Döhner, K.; Bühler, A.; Böttcher, S.; Ritgen, M.; Kneba, M.; et al. Gene mutations and treatment outcome in chronic lymphocytic leukemia: Results from the CLL8 trial. Blood 2014, 123, 3247–3254. [Google Scholar] [CrossRef]
- Landau, D.A.; Carter, S.L.; Stojanov, P.; McKenna, A.; Stevenson, K.; Lawrence, M.S.; Sougnez, C.; Stewart, C.; Sivachenko, A.; Wang, L.; et al. Evolution and Impact of Subclonal Mutations in Chronic Lymphocytic Leukemia. Cell 2013, 152, 714–726. [Google Scholar] [CrossRef]
- Wang, L.; Brooks, A.N.; Fan, J.; Wan, Y.; Gambe, R.; Li, S.; Hergert, S.; Yin, S.; Freeman, S.S.; Levin, J.Z.; et al. Transcriptomic Characterization of SF3B1 Mutation Reveals Its Pleiotropic Effects in Chronic Lymphocytic Leukemia. Cancer Cell 2016, 30, 750–763. [Google Scholar] [CrossRef]
- Raa, G.D.T.; Derks, I.A.M.; Navrkalova, V.; Skowronska, A.; Moerland, P.D.; van Laar, J.; Oldreive, C.; Monsuur, H.; Trbusek, M.; Malcikova, J.; et al. The impact of SF3B1 mutations in CLL on the DNA-damage response. Leukemia 2015, 29, 1133–1142. [Google Scholar] [CrossRef] [PubMed]
- Hallek, M.; Fischer, K.; Fingerle-Rowson, G.; Fink, A.; Busch, R.; Mayer, J.; Hensel, M.; Hopfinger, G.; Hess, G.; von Grünhagen, U.; et al. Addition of rituximab to fludarabine and cyclophosphamide in patients with chronic lymphocytic leukaemia: A randomised, open-label, phase 3 trial. Lancet 2010, 376, 1164–1174. [Google Scholar] [CrossRef]
- Delgado, J.; Espinet, B.; Oliveira, A.C.; Abrisqueta, P.; de la Serna, J.; Collado, R.; Loscertales, J.; Lopez, M.; Hernandez-Rivas, J.A.; Ferra, C.; et al. Chronic lymphocytic leukaemia with 17p deletion: A retrospective analysis of prognostic factors and therapy results. Br. J. Haematol. 2012, 157, 67–74. [Google Scholar] [CrossRef]
- Mian, S.A.; Smith, A.E.; Kulasekararaj, A.G.; Kizilors, A.; Mohamedali, A.M.; Lea, N.C.; Mitsopoulos, K.; Ford, K.; Nasser, E.; Seidl, T.; et al. Spliceosome mutations exhibit specific associations with epigenetic modifiers and proto-oncogenes mutated in myelodysplastic syndrome. Haematologica 2013, 98, 1058–1066. [Google Scholar] [CrossRef]
- te Raa, D.; Derks, I.A.M.; Luijks, D.M.; van Laar, J.; Monsuur, H.; Oldreive, C.; Jethwa, A.; Hüllein, J.; Stankovic, T.; Zenz, T.; et al. SF3B1 Mutations in CLL Are Equivalent to p53/ATM Dysfunction and Cause Defective Puma Upregulation in Response to Chemotherapy. Blood 2012, 120, 711. [Google Scholar] [CrossRef]
- Damle, R.N.; Wasil, T.; Fais, F.; Ghiotto, F.; Valetto, A.; Allen, S.L.; Buchbinder, A.; Budman, D.; Dittmar, K.; Kolitz, J.; et al. Ig V gene mutation status and CD38 expression as novel prognostic indicators in chronic lymphocytic leukemia. Blood 1999, 94, 1840–1847. [Google Scholar] [CrossRef]
- Jeromin, S.; Weissmann, S.; Haferlach, C.; Dicker, F.; Bayer, K.; Grossmann, V.; Alpermann, T.; Roller, A.; Kohlmann, A.; Kern, W.; et al. SF3B1 mutations correlated to cytogenetics and mutations in NOTCH1, FBXW7, MYD88, XPO1 and TP53 in 1160 untreated CLL patients. Leukemia 2014, 28, 108–117. [Google Scholar] [CrossRef]
- Miao, Y.; Zou, Y.-X.; Gu, D.-L.; Zhu, H.-C.; Zhu, H.-Y.; Wang, L.; Liang, J.-H.; Xia, Y.; Wu, J.-Z.; Shao, C.-L.; et al. SF3B1 mutation predicts unfavorable treatment-free survival in Chinese chronic lymphocytic leukemia patients. Ann. Transl. Med. 2019, 7, 176. [Google Scholar] [CrossRef] [PubMed]
- Ghia, P.; Stamatopoulos, K.; Belessi, C.; Moreno, C.; Stilgenbauer, S.; Stevenson, F.; Davi, F.; Rosenquist, R.; European Research Initiative on CLL. ERIC recommendations on IGHV gene mutational status analysis in chronic lymphocytic leukemia. Leukemia 2007, 21, 1–3. [Google Scholar] [CrossRef] [PubMed]
- Nakajima, H.; Takase, S.; Terano, H.; Tanaka, H. New Antitumor Substances, FR901463, FR901464 and FR901465. III. Structures of FR901463, FR901464 and FR901465. J. Antibiot. 1997, 50, 96–99. [Google Scholar] [CrossRef]
- Kaida, D.; Motoyoshi, H.; Tashiro, E.; Nojima, T.; Hagiwara, M.; Ishigami, K.; Watanabe, H.; Kitahara, T.; Yoshida, T.; Nakajima, H.; et al. Spliceostatin A targets SF3b and inhibits both splicing and nuclear retention of pre-mRNA. Nat. Chem. Biol. 2007, 3, 576–583. [Google Scholar] [CrossRef]
- Fan, L.; Lagisetti, C.; Edwards, C.C.; Webb, T.R.; Potter, P.M. Sudemycins, Novel Small Molecule Analogues of FR901464, Induce Alternative Gene Splicing. ACS Chem. Biol. 2011, 6, 582–589. [Google Scholar] [CrossRef] [PubMed]
- Seiler, M.; Yoshimi, A.; Darman, R.; Chan, B.; Keaney, G.; Thomas, M.; Agrawal, A.A.; Caleb, B.; Csibi, A.; Sean, E.; et al. H3B-8800, an orally available small-molecule splicing modulator, induces lethality in spliceosome-mutant cancers. Nat. Med. 2018, 24, 497–504. [Google Scholar] [CrossRef]
- Mizui, Y.; Sakai, T.; Iwata, M.; Uenaka, T.; Okamoto, K.; Shimizu, H.; Yamori, T.; Yoshimatsu, K.; Asada, M. Pladienolides, new substances from culture of Streptomyces platensis Mer-11107. III. In vitro and in vivo antitumor activities. J. Antibiot. 2004, 57, 188–196. [Google Scholar] [CrossRef] [PubMed]
- Aptullahoglu, E.; Ciardullo, C.; Wallis, J.P.; Marr, H.; Marshall, S.; Bown, N.; Willmore, E.; Lunec, J. Splicing Modulation Results in Aberrant Isoforms and Protein Products of p53 Pathway Genes and the Sensitization of B Cells to Non-Genotoxic MDM2 Inhibition. Int. J. Mol. Sci. 2023, 24, 2410. [Google Scholar] [CrossRef] [PubMed]
- Saez, B.; Walter, M.J.; Graubert, T.A. Splicing factor gene mutations in hematologic malignancies. Blood 2017, 129, 1260–1269. [Google Scholar] [CrossRef] [PubMed]
- Leeksma, A.C.; Derks, I.A.M.; Kasem, M.H.; Kilic, E.; de Klein, A.; Jager, M.J.; van de Loosdrecht, A.A.; Jansen, J.H.; Navrkalova, V.; Faber, L.M.; et al. The Effect of SF3B1 Mutation on the DNA Damage Response and Nonsense-Mediated mRNA Decay in Cancer. Front. Oncol. 2020, 10, 609409. [Google Scholar] [CrossRef]
- Dürig, J.; Naschar, M.; Schmücker, U.; Renzing-Köhler, K.; Hölter, T.; Hüttmann, A.; Dührsen, U. CD38 expression is an important prognostic marker in chronic lymphocytic leukaemia. Leukemia 2002, 16, 30–35. [Google Scholar] [CrossRef]
- Walewska, R.; Parry-Jones, N.; Eyre, T.A.; Follows, G.; Martinez-Calle, N.; McCarthy, H.; Parry, H.; Patten, P.E.M.; Riches, J.C.; Hillmen, P.; et al. Guideline for the treatment of chronic lymphocytic leukaemia. Br. J. Haematol. 2022, 197, 544–557. [Google Scholar] [CrossRef]
- Bourdon, J.-C.; Surget, S.; Khoury, M.P. Uncovering the role of p53 splice variants in human malignancy: A clinical perspective. OncoTargets Ther. 2013, 7, 57–68. [Google Scholar] [CrossRef] [PubMed]
- Matsumoto, R.; Tada, M.; Nozaki, M.; Zhang, C.L.; Sawamura, Y.; Abe, H. Short alternative splice transcripts of the mdm2 oncogene correlate to malignancy in human astrocytic neoplasms. Cancer Res. 1998, 58, 609–613. [Google Scholar]
- Thomas, A.L.; Price, C.; Martin, S.G.; Carmichael, J.; Murray, J.C. Identification of two novel mRNA splice variants of bax. Cell Death Differ. 1999, 6, 97–98. [Google Scholar] [CrossRef]
- Wang, Z.; Sun, Y. Identification and characterization of two splicing variants of human Noxa. Anticancer Res. 2008, 28, 1667–1674. [Google Scholar]
- Yin, S.; Gambe, R.G.; Sun, J.; Martinez, A.Z.; Cartun, Z.J.; Regis, F.F.D.; Wan, Y.; Fan, J.; Brooks, A.N.; Herman, S.E.; et al. A Murine Model of Chronic Lymphocytic Leukemia Based on B Cell-Restricted Expression of Sf3b1 Mutation and Atm Deletion. Cancer Cell 2019, 35, 283–296.e5. [Google Scholar] [CrossRef]
- Quesada, V.; Conde, L.; Villamor, N.; Ordóñez, G.R.; Jares, P.; Bassaganyas, L.; Ramsay, A.J.; Beà, S.; Pinyol, M.; Martínez-Trillos, A.; et al. Exome sequencing identifies recurrent mutations of the splicing factor SF3B1 gene in chronic lymphocytic leukemia. Nat. Genet. 2012, 44, 47–52. [Google Scholar] [CrossRef] [PubMed]
- Canbezdi, C.; Tarin, M.; Houy, A.; Bellanger, D.; Popova, T.; Stern, M.-H.; Roman-Roman, S.; Alsafadi, S. Functional and conformational impact of cancer-associated SF3B1 mutations depends on the position and the charge of amino acid substitution. Comput. Struct. Biotechnol. J. 2021, 19, 1361–1370. [Google Scholar] [CrossRef] [PubMed]
- Dolatshad, H.; Pellagatti, A.; Fernandez-Mercado, M.; Yip, B.H.; Malcovati, L.; Attwood, M.; Przychodzen, B.; Sahgal, N.; Kanapin, A.A.; Lockstone, H.; et al. Disruption of SF3B1 results in deregulated expression and splicing of key genes and pathways in myelodysplastic syndrome hematopoietic stem and progenitor cells. Leukemia 2015, 29, 1092–1093. [Google Scholar] [CrossRef] [PubMed]
- Sigmund, A.M.; Kittai, A.S. Richter’s Transformation. Curr. Oncol. Rep. 2022, 24, 1081–1090. [Google Scholar] [CrossRef] [PubMed]
- Nadeu, F.; Royo, R.; Massoni-Badosa, R.; Playa-Albinyana, H.; Garcia-Torre, B.; Duran-Ferrer, M.; Dawson, K.J.; Kulis, M.; Diaz-Navarro, A.; Villamor, N.; et al. Detection of early seeding of Richter transformation in chronic lymphocytic leukemia. Nat. Med. 2022, 28, 1662–1671. [Google Scholar] [CrossRef]
- Ahn, I.E.; Farooqui, M.Z.H.; Tian, X.; Valdez, J.; Sun, C.; Soto, S.; Lotter, J.; Housel, S.; Stetler-Stevenson, M.; Yuan, C.M.; et al. Depth and durability of response to ibrutinib in CLL: 5-year follow-up of a phase 2 study. Blood 2018, 131, 2357–2366. [Google Scholar] [CrossRef]
- Brown, J.R.; Hillmen, P.; O’brien, S.; Barrientos, J.C.; Reddy, N.M.; Coutre, S.E.; Tam, C.S.; Mulligan, S.P.; Jaeger, U.; Barr, P.M.; et al. Extended follow-up and impact of high-risk prognostic factors from the phase 3 RESONATE study in patients with previously treated CLL/SLL. Leukemia 2018, 32, 83–91. [Google Scholar] [CrossRef]
- Jain, P.; Keating, M.; Wierda, W.; Estrov, Z.; Ferrajoli, A.; Jain, N.; George, B.; James, D.; Kantarjian, H.; Burger, J.; et al. Outcomes of patients with chronic lymphocytic leukemia after discontinuing ibrutinib. Blood 2015, 125, 2062–2067. [Google Scholar] [CrossRef]
- Stilgenbauer, S.; Eichhorst, B.; Schetelig, J.; Coutre, S.; Seymour, J.F.; Munir, T.; Puvvada, S.D.; Wendtner, C.-M.; Roberts, A.W.; Jurczak, W.; et al. Venetoclax in relapsed or refractory chronic lymphocytic leukaemia with 17p deletion: A multicentre, open-label, phase 2 study. Lancet Oncol. 2016, 17, 768–778. [Google Scholar] [CrossRef]
- Seymour, J.F.; Kipps, T.J.; Eichhorst, B.; Hillmen, P.; D’rozario, J.; Assouline, S.; Owen, C.; Gerecitano, J.; Robak, T.; De la Serna, J.; et al. Venetoclax–Rituximab in Relapsed or Refractory Chronic Lymphocytic Leukemia. N. Engl. J. Med. 2018, 378, 1107–1120. [Google Scholar] [CrossRef] [PubMed]
- Eyre, T.A.; Clifford, R.; Bloor, A.; Boyle, L.; Roberts, C.; Cabes, M.; Collins, G.P.; Devereux, S.; Follows, G.; Fox, C.P.; et al. NCRI phase II study of CHOP in combination with ofatumumab in induction and maintenance in newly diagnosed Richter syndrome. Br. J. Haematol. 2016, 175, 43–54. [Google Scholar] [CrossRef] [PubMed]
- Rogers, K.A.; Huang, Y.; Dotson, E.; Lundberg, J.; Andritsos, L.A.; Awan, F.T.; Woyach, J.A.; Byrd, J.C. Use ofPD-1 (PDCD1) inhibitors for the treatment of Richter syndrome: Experience at a single academic centre. Br. J. Haematol. 2019, 185, 363–366. [Google Scholar] [CrossRef] [PubMed]
- Hallek, M.; Cheson, B.D.; Catovsky, D.; Caligaris-Cappio, F.; Dighiero, G.; Döhner, H.; Hillmen, P.; Keating, M.J.; Montserrat, E.; Rai, K.R.; et al. Guidelines for the diagnosis and treatment of chronic lymphocytic leukemia: A report from the International Workshop on Chronic Lymphocytic Leukemia updating the National Cancer Institute–Working Group 1996 guidelines. Blood 2008, 111, 5446–5456. [Google Scholar] [CrossRef]
- Dignum, H.M.; Summerfield, G.P.; Proctor, S.J.; Mainou-Fowler, T. Quantification of CD38 expression in B-cell chronic lymphocytic leukemia (B-CLL): A comparison between antibody binding capacity (ABC) and relative median fluorescence (RMF). Leuk. Lymphoma 2004, 45, 1167–1173. [Google Scholar] [CrossRef]
- Elliott, S.L.; Crawford, C.; Mulligan, E.; Summerfield, G.; Newton, P.; Wallis, J.; Mainou-Fowler, T.; Evans, P.; Bedwell, C.; Durkacz, B.W.; et al. Mitoxantrone in combination with an inhibitor of DNA-dependent protein kinase: A potential therapy for high risk B-cell chronic lymphocytic leukaemia. Br. J. Haematol. 2011, 152, 61–71. [Google Scholar] [CrossRef]

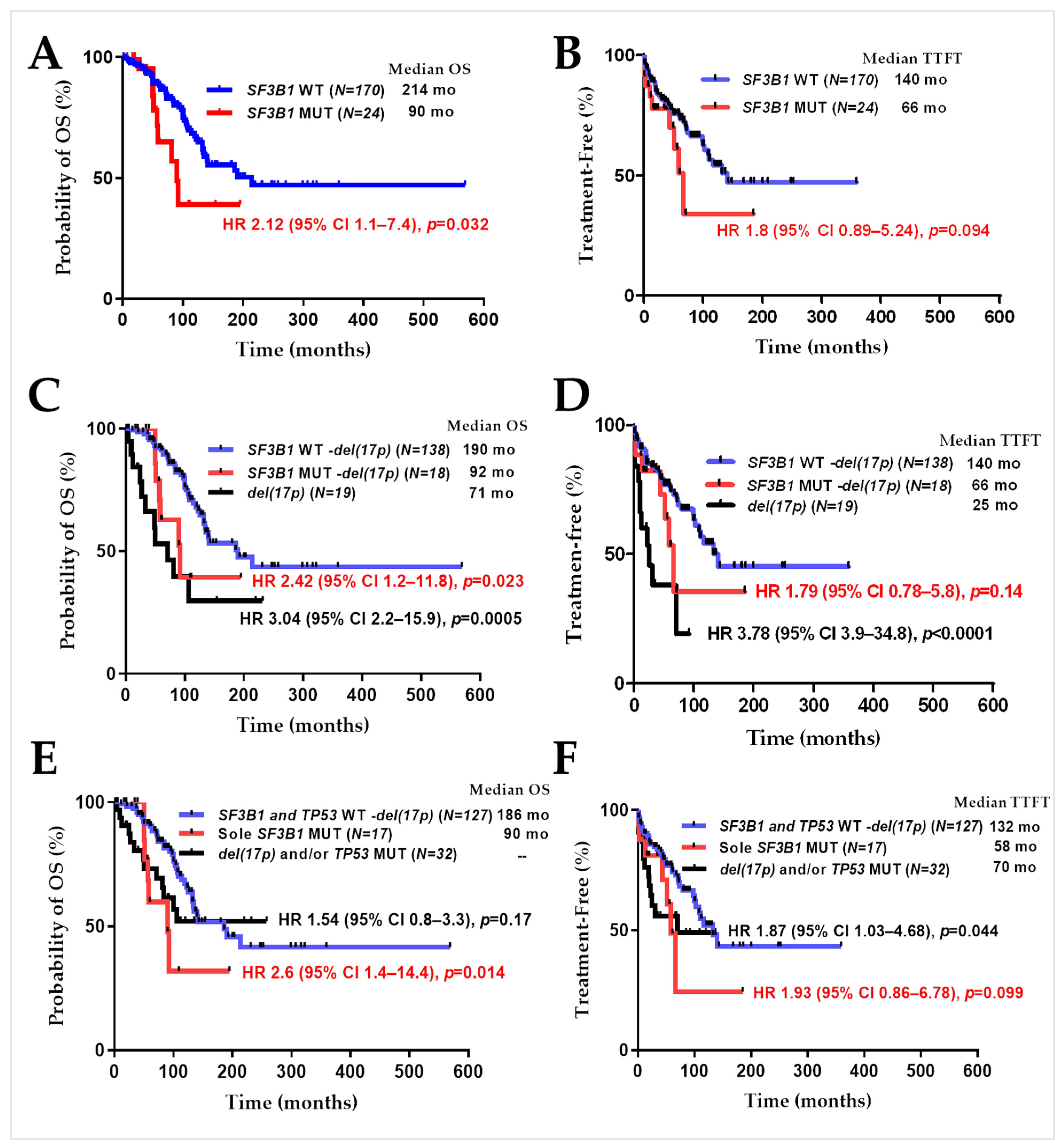


| Features | SF3B1WT No. (%) | SF3B1MUT No. (%) | 1 p Value | |
|---|---|---|---|---|
| AGE (years) | Median (range) | 71 (32–92) | 67.5 (45–86) | |
| OS (months) | Median (range) | 214 (1–569) | 90 (7–195) | 0.032 |
| TTFT (months) | Median (range) | 140 (1–359) | 66 (1–185) | 0.094 |
| SEX | Male | 141 (66%) | 17 (61%) | 0.67 |
| Female | 72 (34%) | 11 (39%) | ||
| BINET STAGE | A | 101 (56%) | 10 (50%) | 0.22 |
| B-C | 78 (44%) | 10 (50%) | ||
| Unknown | 34 | 8 | ||
| 2 IGVH STATUS | Mutated | 106 (61%) | 10 (48%) | 0.25 |
| Unmutated | 68 (39%) | 11 (52%) | ||
| Unknown | 39 | 7 | ||
| CD38 EXPRESSION | CD38 negative (<30%) | 78 (81%) | 5 (36%) | 0.0004 |
| CD38 positive (≥30%) | 18 (19%) | 9 (64%) | ||
| Unknown | 117 | 14 | ||
| TP53 STATUS | Mutated | 30 (16%) | 4 (15%) | 0.93 |
| Unmutated | 163 (84%) | 22 (85%) | ||
| Unknown | 20 | 2 | ||
| RECEIVED ANY TREATMENT | Yes | 90 (44%) | 12 (46%) | 0.68 |
| No | 115 (56%) | 14 (54%) | ||
| Unknown | 8 | 2 | ||
| 3 FISH STRATIFICATION | Low risk | 139 (69%) | 16 (64%) | 0.51 |
| Intermediate/high risk | 62 (31%) | 9 (36%) | ||
| Unknown | 12 | 3 | ||
| Variable | No. (%) | RG7388 LC50 (µM) | Univariate | Multivariate | ||||
|---|---|---|---|---|---|---|---|---|
| 1 η2 | 95% CI | 2 Sig. | 1 ηp2 | 95% CI | 3 Sig. | |||
| SF3B1 Status Wild type Mutant | 65 (83) 13 (17) | ≤1 vs. 1<…<10 ≤1 vs. ≥10 | −0.29/0.19 | 0.863 | −0.27/0.32 | 1 | ||
| −0.71/−0.06 | 0.018 | −0.70/−0.12 | 0.024 | |||||
| 1<…<10 vs. ≤1 1<…<10 vs. ≥10 | 0.095 | −0.19/0.29 | 0.863 | 0.078 | −0.32/0.27 | 1 | ||
| −0.70/0.03 | 0.083 | −0.77/0.13 | 0.257 | |||||
| ≥10 vs. ≤1 ≥10 vs. 1<…<10 | 0.06/0.71 | 0.018 | 0.12/0.70 | 0.024 | ||||
| −0.03/0.70 | 0.083 | −0.13/0.77 | 0.257 | |||||
| TP53 Status Wild type Mutant | 52 (83) 11 (17) | ≤1 vs. 1<…<10 ≤1 vs. ≥10 | −0.74/−0.24 | <0.001 | −0.74/−0.11 | 0.005 | ||
| −0.51/0.11 | 0.263 | −0.57/0.32 | 1 | |||||
| 1<…<10 vs. ≤1 1<…<10 vs. ≥10 | 0.269 | 0.24/0.74 | <0.001 | 0.214 | 0.11/0.74 | 0.005 | ||
| −0.07/0.65 | 0.139 | −0.19/0.79 | 0.396 | |||||
| ≥10 vs. ≤1 ≥10 vs. 1<…<10 | −0.11/0.51 | 0.263 | −0.32/0.57 | 1 | ||||
| −0.65/0.07 | 0.139 | −0.79/0.19 | 0.396 | |||||
| 4 IGVH Status Wild type Mutant | 15 (36) 27 (64) | ≤1 vs. 1<…<10 ≤1 vs. ≥10 | −0.38/0.56 | 0.888 | ||||
| −0.14/0.91 | 0.193 | |||||||
| 1<…<10 vs. ≤1 1<…<10 vs. ≥10 | 0.075 | −0.56/0.38 | 0.888 | |||||
| −0.34/0.92 | 0.502 | |||||||
| ≥10 vs. ≤1 ≥10 vs. 1<…<10 | −0.91/0.14 | 0.193 | ||||||
| −0.92/0.34 | 0.502 | |||||||
| Treatment Received Yes No | 30 (42) 42 (58) | ≤1 vs. 1<…<10 ≤1 vs. ≥10 | −0.14/0.52 | 0.342 | ||||
| −0.41/0.55 | 0.941 | |||||||
| 1<…<10 vs. ≤1 1<…<10 vs. ≥10 | 0.028 | −0.52/0.14 | 0.342 | |||||
| −0.66/0.40 | 0.834 | |||||||
| ≥10 vs. ≤1 ≥10 vs. 1<…<10 | −0.55/0.41 | 0.941 | ||||||
| −0.40/0.66 | 0.834 | |||||||
| CD38 Expression <30% ≥30% | 28 (74) 10 (26) | ≤1 vs. 1<…<10 ≤1 vs. ≥10 | −0.22/0.58 | 0.516 | ||||
| −1.04/0.26 | 0.324 | |||||||
| 1<…<10 vs. ≤1 1<…<10 vs. ≥10 | 0.103 | −0.58/0.22 | 0.516 | |||||
| −1.27/0.13 | 0.132 | |||||||
| ≥10 vs. ≤1 ≥10 vs. 1<…<10 | −0.26/1.04 | 0.324 | ||||||
| −0.13/1.27 | 0.132 | |||||||
| 5 FISH Stratification Low risk Intermediate/ high risk | 53 (76) 17 (24) | ≤1 vs. 1<…<10 ≤1 vs. ≥10 | −0.35/0.27 | 0.943 | ||||
| −0.56/0.34 | 0.833 | |||||||
| 1<…<10 vs. ≤1 1<…<10 vs. ≥10 | 0.006 | −0.27/0.35 | 0.943 | |||||
| −0.57/0.44 | 0.947 | |||||||
| ≥10 vs. ≤1 ≥10 vs. 1<…<10 | −0.34/0.56 | 0.833 | ||||||
| −0.44/0.57 | 0.947 | |||||||
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Aptullahoglu, E.; Wallis, J.P.; Marr, H.; Marshall, S.; Bown, N.; Willmore, E.; Lunec, J. SF3B1 Mutations Are Associated with Resistance to Non-Genotoxic MDM2 Inhibition in Chronic Lymphocytic Leukemia. Int. J. Mol. Sci. 2023, 24, 11335. https://doi.org/10.3390/ijms241411335
Aptullahoglu E, Wallis JP, Marr H, Marshall S, Bown N, Willmore E, Lunec J. SF3B1 Mutations Are Associated with Resistance to Non-Genotoxic MDM2 Inhibition in Chronic Lymphocytic Leukemia. International Journal of Molecular Sciences. 2023; 24(14):11335. https://doi.org/10.3390/ijms241411335
Chicago/Turabian StyleAptullahoglu, Erhan, Jonathan P. Wallis, Helen Marr, Scott Marshall, Nick Bown, Elaine Willmore, and John Lunec. 2023. "SF3B1 Mutations Are Associated with Resistance to Non-Genotoxic MDM2 Inhibition in Chronic Lymphocytic Leukemia" International Journal of Molecular Sciences 24, no. 14: 11335. https://doi.org/10.3390/ijms241411335
APA StyleAptullahoglu, E., Wallis, J. P., Marr, H., Marshall, S., Bown, N., Willmore, E., & Lunec, J. (2023). SF3B1 Mutations Are Associated with Resistance to Non-Genotoxic MDM2 Inhibition in Chronic Lymphocytic Leukemia. International Journal of Molecular Sciences, 24(14), 11335. https://doi.org/10.3390/ijms241411335






