Thermochemical Study of the Interaction of Cytosine and Uracil with Peptides in a Buffered Saline: Complex Formation with beta-Endorphin 30-31 (Human), L-Glutathion (Reduced) and α-L-Alanyl-L-Tyrosine
Abstract
1. Introduction
- (a)
- To obtain thermochemical characteristics of the interaction of uracil and cytosine with a number of peptides of various structures by calorimetry of dissolution;
- (b)
- To calculate the values of the complexation constant, and the change in Gibbs energy, enthalpy, and entropy from calorimetric data;
- (c)
- To study the effect of the charge and the number of donors and acceptors of the H-bond in the structure of peptide on the patterns of binding to nucleic bases.
2. Results and Discussion
2.1. Distribution of Ionic Forms of Peptides
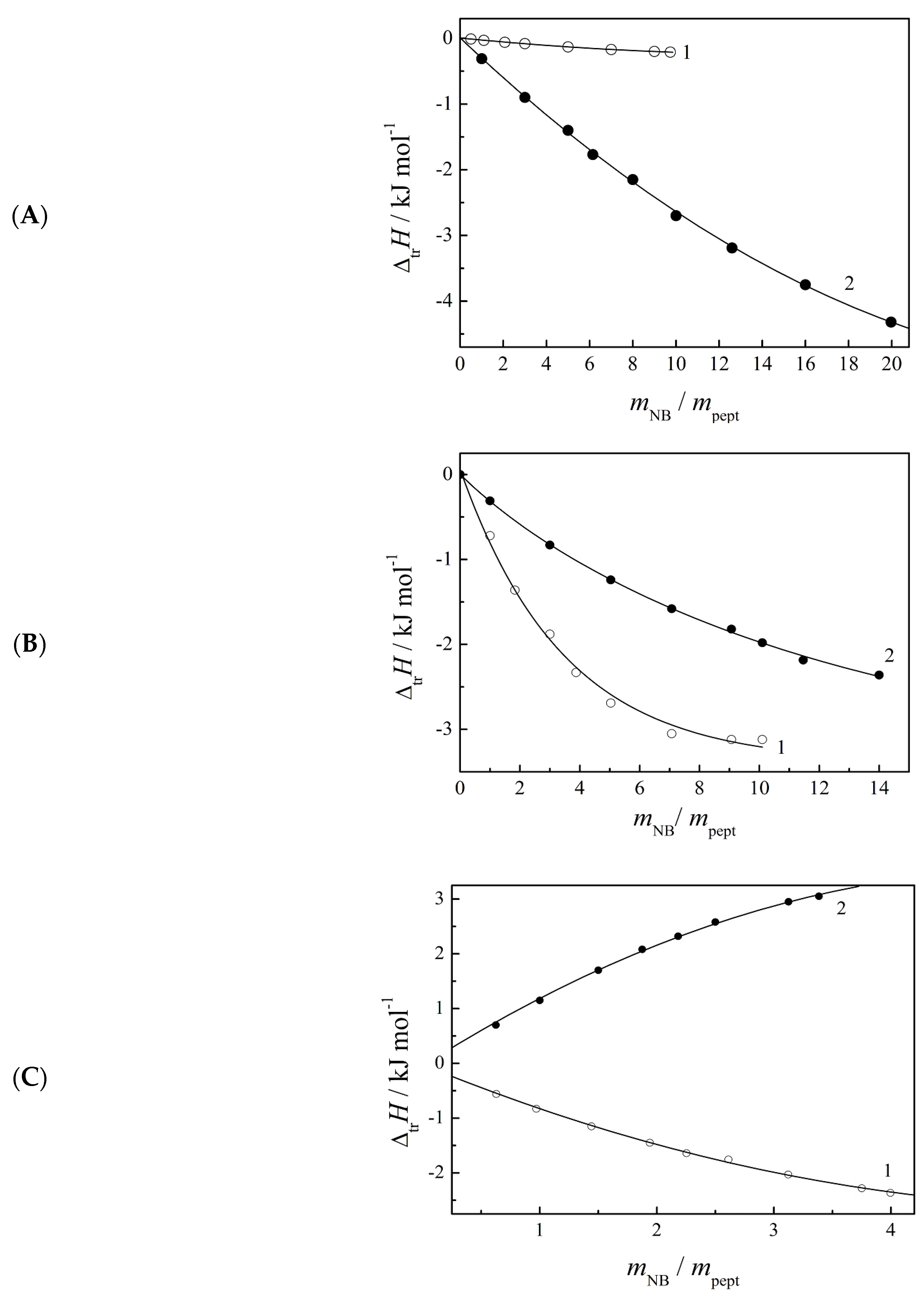
2.2. Enthalpies of Dissolution of Peptides
2.3. Thermodynamic Parameters of Complexation of Peptides with Nucleic Bases
3. Materials and Methods
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Kruse, M.; Möser, C.; Smith, D.M.; Müller-Landau, H.; Rant, U.; Hölzel, R.; Bier, F.F. Measuring Influenza A Virus and Peptide Interaction Using Electrically Controllable DNA Nanolevers. Adv. Mater. Technol. 2022, 7, 2101141. [Google Scholar] [CrossRef]
- Kruse, M.; Altattan, B.; Laux, E.-M.; Grasse, N.; Heinig, L.; Möser, C.; Smith, D.M.; Hölzel, R. Characterization of binding interactions of SARS-CoV–2 spike protein and DNA—Peptide nanostructures. Sci. Rep. 2022, 12, 12828. [Google Scholar] [CrossRef] [PubMed]
- Kolchina, N.; Khavinson, V.; Linkova, N.; Yakimov, A.; Baitin, D.; Afanasyeva, A.; Petukhov, M. Systematic search for structural motifs of peptide binding to double-stranded DNA. Nucleic Acids Res. 2019, 47, 10553–10563. [Google Scholar] [CrossRef] [PubMed]
- Costa, D.; Albuquerque, T.; Queiroz, J.A.; Valente, A.J.M. A co-delivery platform based on plasmid DNA peptide-surfactant complexes: Formation, characterization and release behavior. Colloids Surf. B Biointerfaces 2019, 178, 430–438. [Google Scholar] [CrossRef] [PubMed]
- Uittenbogaard, J.P.; Zomer, B.; Hoogerhout, P.; Metz, B. Reactions of Propiolactone with Nucleobase Analogues, Nucleosides, and Peptides. J. Biol. Chem. 2011, 286, 36198–36214. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.-C.; Huang, J.T.B.; Jeng, K.-C.G.; Yang, R.C.K.; Liao, M.-K.; Chen, C.-S.; Chien, W.-J.; Wey, M.-T.; Kan, L.-S.; Sheh, L. Determination of Allosteric Effects and Interstrand Bidentate Interactions in DNA-peptide Molecular Recognition. J. Chin. Chem. Soc. 2010, 57, 266–274. [Google Scholar] [CrossRef]
- Bajpayee, N.S.; McGrath, W.J.; Mangel, W.F. Interaction of the adenovirus proteinase with protein cofactors with high negative charge densities. Biochemistry 2005, 44, 8721–8729. [Google Scholar] [CrossRef] [PubMed]
- Balodis, E.; Madekufamba, M.; Trevani, L.N.; Tremaine, P.R. Ionization constants and thermal stabilities of uracil and adenine under hydrothermal conditions as measured by in situ UV-visible spectroscopy. Geochim. Cosmochim. Acta 2012, 93, 182–204. [Google Scholar] [CrossRef]
- Vasiliev, V.P.; Borodin, V.A.; Kozlovsky, E.V. The Use of Computers in Chemical-Analytical Calculations; Higher School: Moscow, Russia, 1993; p. 110. Available online: https://chemequ.ru/online-progs/rrsu-online/ (accessed on 1 January 2023).
- Badelin, V.G.; Barannikov, V.P.; Katrovtseva, A.V.; Tarasova, G.N. Dissociation Constants of Protolytic Dissociation of Glutamyl-Glutamic and Glycyl-Glutamic Acids in Aqueous Solution at 298 K. Russ. J. Gen. Chem. 2013, 83, 945–948. [Google Scholar] [CrossRef]
- Lytkin, A.I.; Krutova, O.N.; Chernikov, V.V.; Mokhova, Y.V.; Krutova, E.D.; Tyunina, E.Y. Thermochemical study of reactions of acid–base interaction in an L-glutathione aqueous solution. Russ. J. Phys. Chem. A 2021, 95, 2051–2054. [Google Scholar] [CrossRef]
- Aslan, N.; Erden, P.E.; Doğan, A.; Canel, E.; Kılıç, E. Protonation Constants of Some Alanyl Dipeptides in Mixed Aqueous Organic Solvents. J. Solut. Chem. 2016, 45, 299–312. [Google Scholar] [CrossRef]
- Lytkin, I.; Chernikov, V.V.; Krutova, O.N.; Golubev, A.A.; Romanov, R.A. Standard Enthalpies of Formation of L-Glutathion and Products of Its Dissociation in Aqueous Solutions. Russ. J. Phys. Chem. A 2021, 95, 1831–1834. [Google Scholar] [CrossRef]
- Solovyev, A.Y.; Tarnovskaya, S.I.; Chernova, I.A.; Shataeva Larisa, K.; Skorik, Y.A. The interaction of amino acids, peptides, and proteins with DNA. Int. J. Biol. Macromol. 2015, 78, 39–45. [Google Scholar] [CrossRef] [PubMed]
- Alkorta, I.; Elguero, J. Interaction of Protein Backbone with Nucleic Acid Bases. J. Phys. Chem. B 2003, 107, 5306–5310. [Google Scholar] [CrossRef]
- Hunter, K.C.; Millen, A.L.; Wetmore, S.D. Effects of Hydrogen-Bonding and Stacking Interactions with Amino Acids on the Acidity of Uracil. J. Phys. Chem. B 2007, 111, 1858–1871. [Google Scholar] [CrossRef] [PubMed]
- Rutledge, L.R.; Campbell-Verduyn, L.S.; Hunter, K.C.; Wetmore, S.D. Characterization of Nucleobase-Amino Acid Stacking Interactions Utilized by a DNA Repair Enzyme. J. Phys. Chem. B 2006, 110, 19652–19663. [Google Scholar] [CrossRef] [PubMed]
- Borodin, V.A.; Vasil’ev, V.P.; Kozlovsky, E.V. Universal program package for treatment experimental data on the complex equilibria in solutions. In Mathematical Problems of Chemical Thermodynamics; Nauka: Novosibirsk, Russia, 1985; pp. 219–226. (In Russian) [Google Scholar]
- Borodin, V.A.; Kozlovsky, E.V.; Vasil’ev, V.P. Treatment of the results of calorimetric measurements on digital computer in the study of complex equilibriums in solutions. Russ. J. Inorg. Chem. 1982, 27, 2169–2172. [Google Scholar]
- Gurney, R.W. Ionic Processes in Solution; McGraw Hill: New York, NY, USA, 1953. [Google Scholar]
- Kaur, N.; Banipal, P.K.; Banipal, T.S. Physico-chemical effects of caffeine on aqueous solutions of pyrimidine based model compounda of nucleic acids. J. Mol. Liq. 2016, 221, 721–732. [Google Scholar] [CrossRef]
- Barannikov, V.P.; Kurbatova, M.S.; Mezhevoi, I.N. The influence of structure of isomolecular dipeptides of α-Ala-α-Ala and β-Ala-β-Ala on their behavior in aqueous micellar solution of SDS. Thermochim. Acta 2020, 689, 178647. [Google Scholar] [CrossRef]
- Gunther, C.; Pfestorf, R.; Rother, M.; Seidel, J.; Zimmermann, R.; Wolf, G.; Schreder, V. An interlaboratory test for certification of potassium chloride as a certified reference material (CRM) for solution calorimetry. J. Therm. Anal. 1988, 33, 359–363. [Google Scholar] [CrossRef]

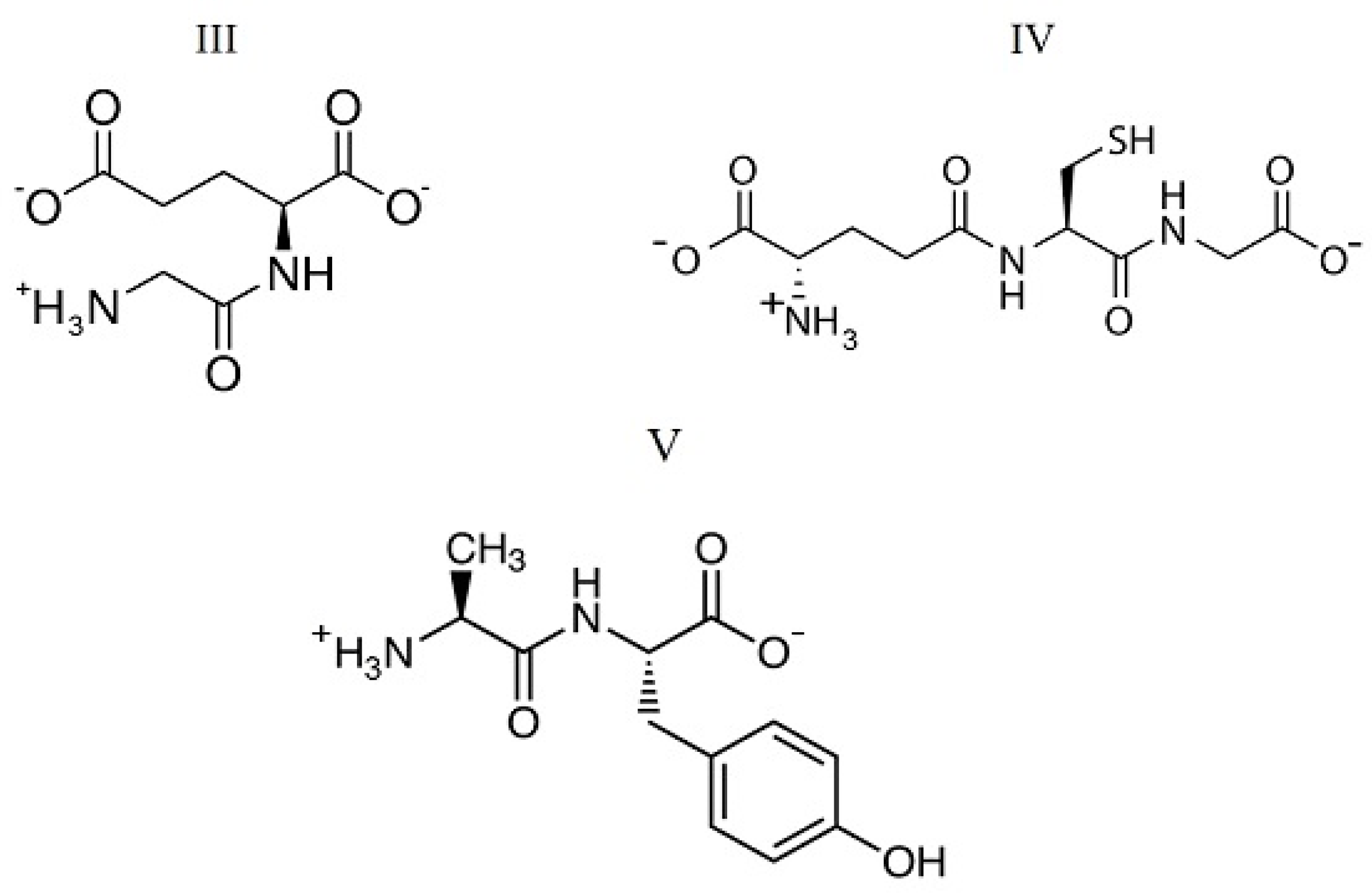

| mNB | ΔsolHm | mNB | ΔsolHm |
|---|---|---|---|
| GlyGlu + uracil | GlyGlu + cytosine | ||
| 0 | 11.43 | 0.0030 | 11.12 |
| 0.0015 | 11.42 | 0.0091 | 10.53 |
| 0.0033 | 11.39 | 0.0153 | 10.03 |
| 0.0062 | 11.37 | 0.0185 | 9.66 |
| 0.0090 | 11.35 | 0.0242 | 9.28 |
| 0.0151 | 11.30 | 0.0303 | 8.73 |
| 0.0212 | 11.25 | 0.0378 | 8.24 |
| 0.0272 | 11.20 | 0.0480 | 7.68 |
| 0.0292 | 11.21 | 0.0599 | 7.11 |
| γGluCysGly + uracil | γGluCysGly + cytosine | ||
| 0 | 20.36 | 0.0030 | 20.05 |
| 0.0030 | 19.64 | 0.0090 | 19.53 |
| 0.0055 | 19.00 | 0.0150 | 19.12 |
| 0.0090 | 18.48 | 0.0210 | 18.78 |
| 0.0116 | 18.03 | 0.0240 | 18.64 |
| 0.0151 | 17.67 | 0.0300 | 18.38 |
| 0.0212 | 17.24 | 0.0344 | 18.18 |
| 0.0272 | 17.21 | 0.0420 | 17.98 |
| 0.0303 | 17.28 | ||
| AlaTyr + uracil | AlaTyr + cytosine | ||
| 0 | 10.89 | 0.0050 | 11.59 |
| 0.0050 | 10.33 | 0.0080 | 12.04 |
| 0.0078 | 10.06 | 0.0115 | 12.59 |
| 0.0115 | 9.74 | 0.0155 | 12.97 |
| 0.0155 | 9.44 | 0.0175 | 13.21 |
| 0.0180 | 9.27 | 0.0201 | 13.47 |
| 0.0201 | 9.13 | 0.0250 | 13.84 |
| 0.0250 | 8.86 | 0.0271 | 13.94 |
| 0.0300 | 8.61 | ||
| 0.0320 | 8.53 | ||
| AlaAla + uracil | |||
| 0 | −7.46 | ||
| 0.0050 | −7.44 | ||
| 0.0078 | −7.41 | ||
| 0.0115 | −7.36 | ||
| 0.0155 | −7.32 | ||
| 0.0201 | −7.27 | ||
| 0.0250 | −7.25 | ||
| 0.0300 | −7.24 | ||
| 0.0320 | −7.23 | ||
| lgKr * | ΔrG | ΔrH | T ΔrS | |
|---|---|---|---|---|
| kJ·mol−1 | ||||
| Ur + GlyGlu | 1.16 ± 0.001 | −6.64 ± 0.01 | −0.7 ± 0.1 | 5.9 ± 0.1 |
| Ur + γ-GluCysGly | 2.01 ± 0.04 | −11.4 ± 0.2 | −4.4 ± 0.1 | 7.0 ± 0.3 |
| Ur + AlaTyr | 1.45 ± 0.001 | −8.3 ± 0.01 | −5.30 ± 0.05 | 3.0 ± 0.06 |
| Ur + AlaAla | 1.03 ± 0.05 | −5.9 ± 0.3 | 1.0 ± 0.04 | 6.9 ± 0.3 |
| Cyt + GlyGlu | 0.95 ± 0.001 | −5.40 ± 0.01 | −12.7 ± 0.08 | −7.3 ± 0.1 |
| Cyt + γ-GluCysGly | 1.39 ± 0.002 | −7.89 ± 0.01 | −4.79 ± 0.07 | 3.1 ± 0.1 |
| Cyt + AlaTyr | 1.29 ± 0.002 | −7.35 ± 0.01 | 9.8 ± 0.1 | 17.15 ± 0.1 |
| Chemical a | M b | CAS No. c | Origin | Purity d |
|---|---|---|---|---|
| Uracil | 112.09 | 66-22-8 | Termo Ficher Scientific | 0.996 |
| Cytosine | 111.10 | 70-30-7 | Appolo Scientific | 0.996 |
| Glycyl-L-glutamic acid (beta-endorphin 30-31 human) | 204.18 | 7412-78-4 | Bachem | >0.99 |
| L-γ-Glutamyl-L-cysteinyl-glycine (glutathion reduced) | 307.32 | 108457-42-7 | Tokyo Chemical Industry | >0.99 |
| L-α-Alanyl-L-tyrosine | 252.27 | 3061-88-9 | Abcr GmbH | >0.98 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Barannikov, V.P.; Smirnov, V.I.; Mezhevoi, I.N.; Koltyshev, D.R. Thermochemical Study of the Interaction of Cytosine and Uracil with Peptides in a Buffered Saline: Complex Formation with beta-Endorphin 30-31 (Human), L-Glutathion (Reduced) and α-L-Alanyl-L-Tyrosine. Int. J. Mol. Sci. 2023, 24, 9764. https://doi.org/10.3390/ijms24119764
Barannikov VP, Smirnov VI, Mezhevoi IN, Koltyshev DR. Thermochemical Study of the Interaction of Cytosine and Uracil with Peptides in a Buffered Saline: Complex Formation with beta-Endorphin 30-31 (Human), L-Glutathion (Reduced) and α-L-Alanyl-L-Tyrosine. International Journal of Molecular Sciences. 2023; 24(11):9764. https://doi.org/10.3390/ijms24119764
Chicago/Turabian StyleBarannikov, Vladimir P., Valeriy I. Smirnov, Igor N. Mezhevoi, and Damir R. Koltyshev. 2023. "Thermochemical Study of the Interaction of Cytosine and Uracil with Peptides in a Buffered Saline: Complex Formation with beta-Endorphin 30-31 (Human), L-Glutathion (Reduced) and α-L-Alanyl-L-Tyrosine" International Journal of Molecular Sciences 24, no. 11: 9764. https://doi.org/10.3390/ijms24119764
APA StyleBarannikov, V. P., Smirnov, V. I., Mezhevoi, I. N., & Koltyshev, D. R. (2023). Thermochemical Study of the Interaction of Cytosine and Uracil with Peptides in a Buffered Saline: Complex Formation with beta-Endorphin 30-31 (Human), L-Glutathion (Reduced) and α-L-Alanyl-L-Tyrosine. International Journal of Molecular Sciences, 24(11), 9764. https://doi.org/10.3390/ijms24119764





