The Many Faces of MLKL, the Executor of Necroptosis
Abstract
1. Introduction
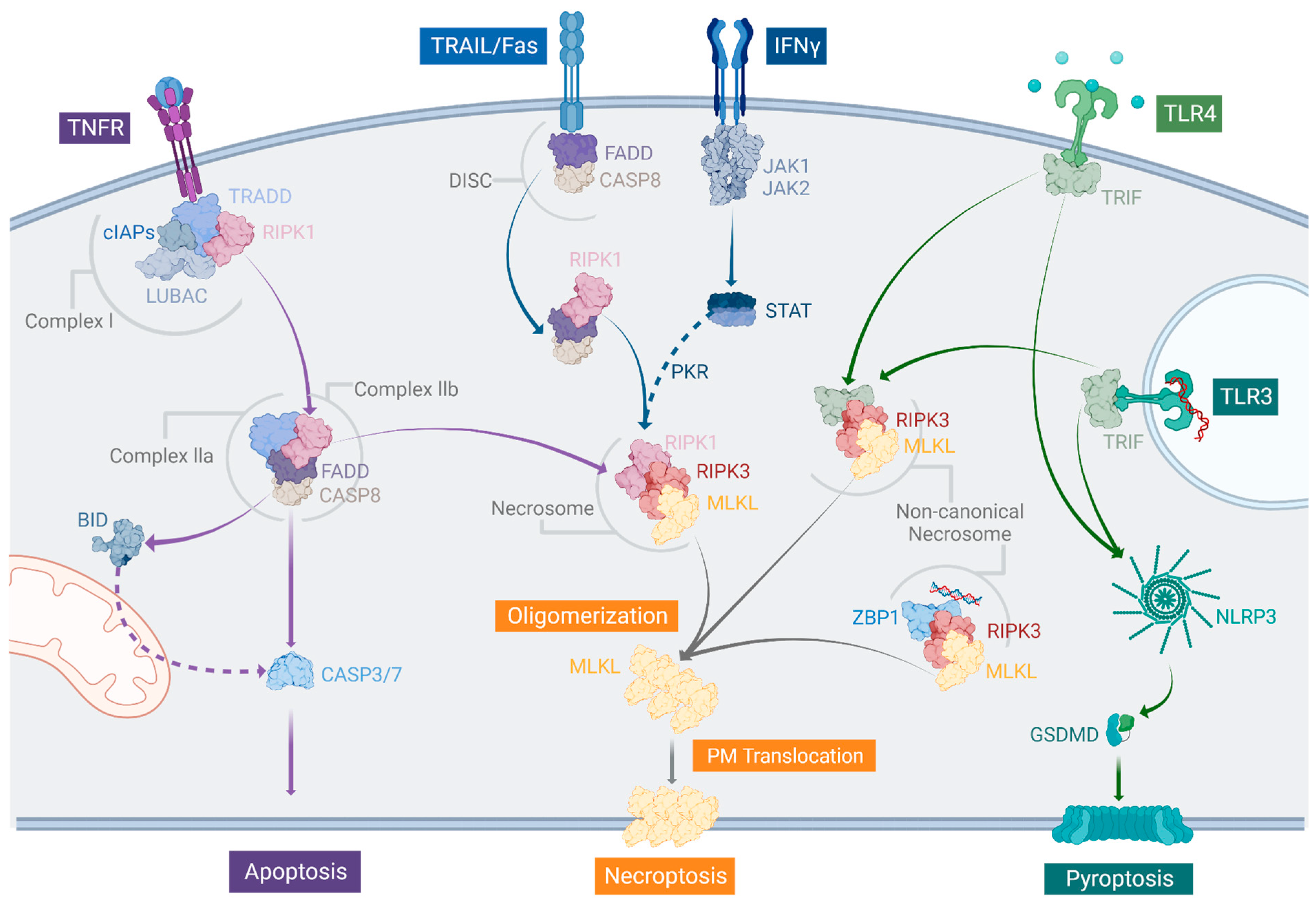
2. The Necroptotic Pathway and Its Relation to Other Types of Regulated Cell Death
2.1. The Canonical TNF Pathway
2.2. TRAIL-, Fas/CD95-, and IFN-Dependent Manners to Induce Necroptosis
2.3. The Non-Canonical Necrosome
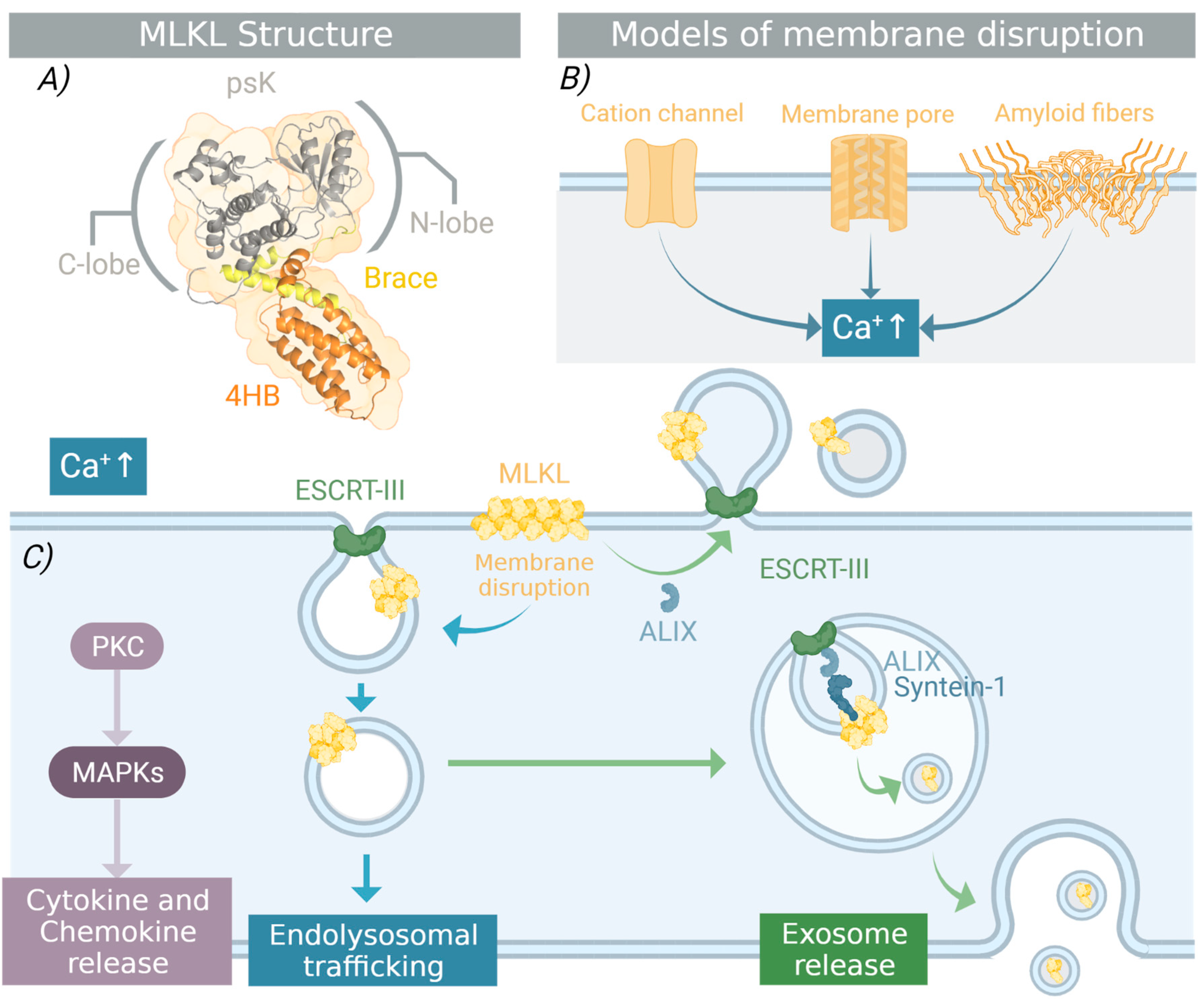
3. Structural Requirements for MLKL Necroptotic Function
3.1. MLKL and RIPK3: Till Death Do Them Part
3.2. Post-Translational Modifications Governing MLKL Function
3.3. Oligomerization and Membrane Translocation
4. The Necroptotic and Non-Necroptotic Functions of MLKL
4.1. Punching Holes in the Plasma Membrane
4.2. One Protein with Many Faces
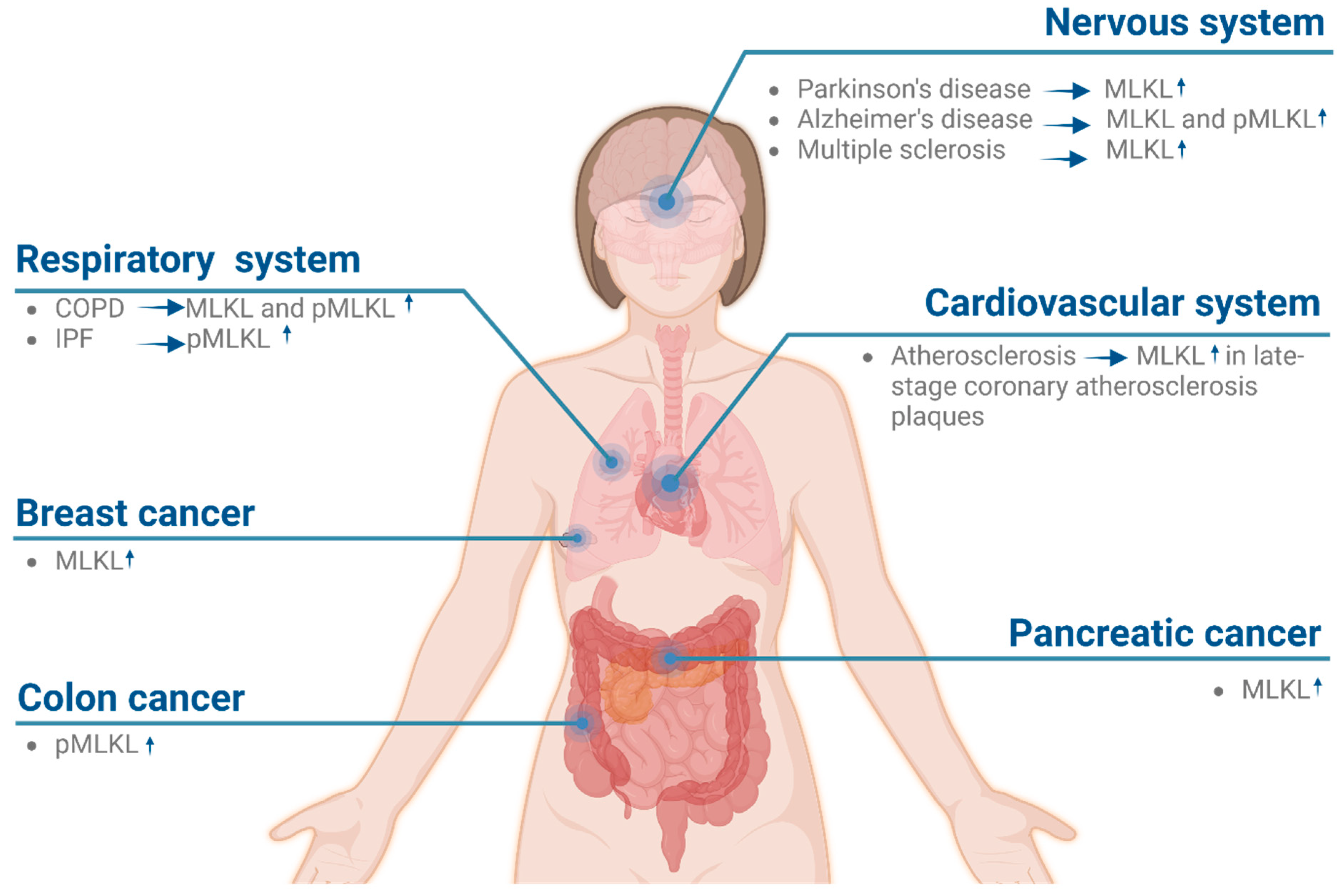
5. MLKL Is a Potential Target for Treating Human Diseases
6. Inhibition of MLKL with Small Molecules
6.1. Irreversible Inhibitors
6.2. Inhibitors That Target the psK Domain
6.3. Fragment-Based Allosteric and Reversible Binders
7. Conclusions and Future Perspectives
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Henriquez, M.; Armisen, R.; Stutzin, A.; Quest, A. Cell Death by Necrosis, a Regulated Way to Go. Curr. Mol. Med. 2008, 8, 187–206. [Google Scholar] [CrossRef]
- Rodriguez, D.A.; Weinlich, R.; Brown, S.; Guy, C.; Fitzgerald, P.; Dillon, C.P.; Oberst, A.; Quarato, G.; Low, J.; Cripps, J.G.; et al. Characterization of RIPK3-mediated phosphorylation of the activation loop of MLKL during necroptosis. Cell Death Differ. 2016, 23, 76–88. [Google Scholar] [CrossRef]
- Elmore, S. Apoptosis: A Review of Programmed Cell Death. Toxicol. Pathol. 2007, 35, 495–516. [Google Scholar] [CrossRef]
- Galluzzi, L.; Vitale, I.; Aaronson, S.A.; Abrams, J.M.; Adam, D.; Agostinis, P.; Alnemri, E.S.; Altucci, L.; Amelio, I.; Andrews, D.W.; et al. Molecular mechanisms of cell death: Recommendations of the Nomenclature Committee on Cell Death 2018. Cell Death Differ. 2018, 25, 486–541. [Google Scholar] [CrossRef]
- Murphy, J.M. The killer pseudokinase mixed lineage kinase domain-like protein (MLKL). Cold Spring Harb. Perspect. Biol. 2020, 12, a036376. [Google Scholar] [CrossRef]
- Samson, A.L.; Zhang, Y.; Geoghegan, N.D.; Gavin, X.J.; Davies, K.A.; Mlodzianoski, M.J.; Whitehead, L.W.; Frank, D.; Garnish, S.E.; Fitzgibbon, C.; et al. MLKL trafficking and accumulation at the plasma membrane control the kinetics and threshold for necroptosis. Nat. Commun. 2020, 11, 3151. [Google Scholar] [CrossRef] [PubMed]
- Liccardi, G.; Annibaldi, A. MLKL post-translational modifications: Road signs to infection, inflammation and unknown destinations. Cell Death Differ. 2022, 30, 269–278. [Google Scholar] [CrossRef] [PubMed]
- Khoury, M.K.; Gupta, K.; Franco, S.R.; Liu, B. Necroptosis in the Pathophysiology of Disease. Am. J. Pathol. 2020, 190, 272–285. [Google Scholar] [CrossRef]
- Laster, S.M.; Wood, J.G.; Gooding, L.R. Tumor necrosis factor can induce both apoptic and necrotic forms of cell lysis. J. Immunol. 1988, 141, 2629–2634. [Google Scholar] [CrossRef]
- Vercammen, D.; Beyaert, R.; Denecker, G.; Goossens, V.; Van Loo, G.; Declercq, W.; Grooten, J.; Fiers, W.; Vandenabeele, P. Inhibition of caspases increases the sensitivity of L929 cells to necrosis mediated by tumor necrosis factor. J. Exp. Med. 1998, 187, 1477–1485. [Google Scholar] [CrossRef] [PubMed]
- Taraborrelli, L.; Peltzer, N.; Montinaro, A.; Kupka, S.; Rieser, E.; Hartwig, T.; Sarr, A.; Darding, M.; Draber, P.; Haas, T.L.; et al. LUBAC prevents lethal dermatitis by inhibiting cell death induced by TNF, TRAIL and CD95L. Nat. Commun. 2018, 9, 3910. [Google Scholar] [CrossRef] [PubMed]
- He, S.; Liang, Y.; Shao, F.; Wang, X. Toll-like receptors activate programmed necrosis in macrophages through a receptor-interacting kinase-3-mediated pathway. Proc. Natl. Acad. Sci. USA 2011, 108, 20054–20059. [Google Scholar] [CrossRef]
- Cho, Y.S.; Challa, S.; Moquin, D.; Genga, R.; Ray, T.D.; Guildford, M.; Chan, F.K.M. Phosphorylation-Driven Assembly of the RIP1-RIP3 Complex Regulates Programmed Necrosis and Virus-Induced Inflammation. Cell 2009, 137, 1112–1123. [Google Scholar] [CrossRef] [PubMed]
- Aglietti, R.A.; Dueber, E.C. Recent Insights into the Molecular Mechanisms Underlying Pyroptosis and Gasdermin Family Functions. Trends Immunol. 2017, 38, 261–271. [Google Scholar] [CrossRef]
- Feoktistova, M.; Geserick, P.; Kellert, B.; Dimitrova, D.P.; Langlais, C.; Hupe, M.; Cain, K.; MacFarlane, M.; Häcker, G.; Leverkus, M. CIAPs Block Ripoptosome Formation, a RIP1/Caspase-8 Containing Intracellular Cell Death Complex Differentially Regulated by cFLIP Isoforms. Mol. Cell 2011, 43, 449–463. [Google Scholar] [CrossRef]
- McComb, S.; Cheung, H.H.; Korneluk, R.G.; Wang, S.; Krishnan, L.; Sad, S. CIAP1 and cIAP2 limit macrophage necroptosis by inhibiting Rip1 and Rip3 activation. Cell Death Differ. 2012, 19, 1791–1801. [Google Scholar] [CrossRef]
- Zhou, Z.; Han, V.; Han, J. New components of the necroptotic pathway. Protein Cell 2012, 3, 811–817. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.; Chen, Z.J. Regulation of NF-κB by ubiquitination. Curr. Opin. Immunol. 2013, 25. [Google Scholar] [CrossRef]
- Tenev, T.; Bianchi, K.; Darding, M.; Broemer, M.; Langlais, C.; Wallberg, F.; Zachariou, A.; Lopez, J.; MacFarlane, M.; Cain, K.; et al. The Ripoptosome, a Signaling Platform that Assembles in Response to Genotoxic Stress and Loss of IAPs. Mol. Cell 2011, 43, 432–448. [Google Scholar] [CrossRef]
- Li, H.; Zhu, H.; Xu, C.J.; Yuan, J. Cleavage of BID by caspase 8 mediates the mitochondrial damage in the Fas pathway of apoptosis. Cell 1998, 94, 491–501. [Google Scholar] [CrossRef]
- Kantari, C.; Walczak, H. Caspase-8 and Bid: Caught in the act between death receptors and mitochondria. Biochim. Biophys. Acta—Mol. Cell Res. 2011, 1813, 558–563. [Google Scholar] [CrossRef] [PubMed]
- Demarco, B.; Grayczyk, J.P.; Bjanes, E.; Le Roy, D.; Tonnus, W.; Assenmacher, C.A.; Radaelli, E.; Fettrelet, T.; Mack, V.; Linkermann, A.; et al. Caspase-8-dependent gasdermin D cleavage promotes antimicrobial defense but confers susceptibility to TNF-induced lethality. Sci. Adv. 2020, 6, eabc3465. [Google Scholar] [CrossRef] [PubMed]
- Newton, K.; Wickliffe, K.E.; Dugger, D.L.; Maltzman, A.; Roose-Girma, M.; Dohse, M.; Kőműves, L.; Webster, J.D.; Dixit, V.M. Cleavage of RIPK1 by caspase-8 is crucial for limiting apoptosis and necroptosis. Nature 2019, 574, 428–431. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.; Lu, H.C.; Steelman, A.J.; Li, J. Myeloid caspase-8 restricts RIPK3-dependent proinflammatory IL-1β production and CD4 T cell activation in autoimmune demyelination. Proc. Natl. Acad. Sci. USA 2022, 119, e2117636119. [Google Scholar] [CrossRef] [PubMed]
- Ju, E.; Park, K.A.; Shen, H.M.; Hur, G.M. The resurrection of RIP kinase 1 as an early cell death checkpoint regulator—A potential target for therapy in the necroptosis era. Exp. Mol. Med. 2022, 54, 1401–1411. [Google Scholar] [CrossRef]
- Mocarski, E.S.; Upton, J.W.; Kaiser, W.J. Viral infection and the evolution of caspase 8-regulated apoptotic and necrotic death pathways. Nat. Rev. Immunol. 2012, 12, 79–88. [Google Scholar] [CrossRef]
- Mompeán, M.; Li, W.; Li, J.; Laage, S.; Siemer, A.B.; Bozkurt, G.; Wu, H.; McDermott, A.E. The Structure of the Necrosome RIPK1-RIPK3 Core, a Human Hetero-Amyloid Signaling Complex. Cell 2018, 173, 1244–1253. [Google Scholar] [CrossRef]
- Galluzzi, L.; Kepp, O.; Kroemer, G. MLKL regulates necrotic plasma membrane permeabilization. Cell Res. 2014, 24, 139–140. [Google Scholar] [CrossRef]
- Meurette, O.; Rebillard, A.; Huc, L.; Le Moigne, G.; Merino, D.; Micheau, O.; Lagadic-Gossmann, D.; Dimanche-Boitrel, M.T. TRAIL induces receptor-interacting protein 1-dependent and caspase-dependent necrosis-like cell death under acidic extracellular conditions. Cancer Res. 2007, 67, 218–226. [Google Scholar] [CrossRef]
- Jouan-Lanhouet, S.; Arshad, M.I.; Piquet-Pellorce, C.; Martin-Chouly, C.; Le Moigne-Muller, G.; Van Herreweghe, F.; Takahashi, N.; Sergent, O.; Lagadic-Gossmann, D.; Vandenabeele, P.; et al. TRAIL induces necroptosis involving RIPK1/RIPK3-dependent PARP-1 activation. Cell Death Differ. 2012, 19, 2003–2014. [Google Scholar] [CrossRef]
- Füllsack, S.; Rosenthal, A.; Wajant, H.; Siegmund, D. Redundant and receptor-specific activities of TRADD, RIPK1 and FADD in death receptor signaling. Cell Death Dis. 2019, 10, 122. [Google Scholar] [CrossRef]
- Peter, M.E.; Krammer, P.H. The CD95(APO-1/Fas) DISC and beyond. Cell Death Differ. 2003, 10, 26–35. [Google Scholar] [CrossRef]
- Vandenabeele, P.; Galluzzi, L.; Vanden Berghe, T.; Kroemer, G. Molecular mechanisms of necroptosis: An ordered cellular explosion. Nat. Rev. Mol. Cell Biol. 2010, 11, 700–714. [Google Scholar] [CrossRef]
- Thapa, R.J.; Nogusa, S.; Chen, P.; Maki, J.L.; Lerro, A.; Andrake, M.; Rall, G.F.; Degterev, A.; Balachandran, S. Interferon-induced RIP1/RIP3-mediated necrosis requires PKR and is licensed by FADD and caspases. Proc. Natl. Acad. Sci. USA 2013, 110, 3109–3118. [Google Scholar] [CrossRef]
- Yang, D.; Liang, Y.; Zhao, S.; Ding, Y.; Zhuang, Q.; Shi, Q.; Ai, T.; Wu, S.Q.; Han, J. ZBP1 mediates interferon-induced necroptosis. Cell. Mol. Immunol. 2020, 17, 356–368. [Google Scholar] [CrossRef]
- Jiao, H.; Wachsmuth, L.; Kumari, S.; Schwarzer, R.; Lin, J.; Eren, R.O.; Fisher, A.; Lane, R.; Young, G.R.; Kassiotis, G.; et al. Z-nucleic-acid sensing triggers ZBP1-dependent necroptosis and inflammation. Nature 2020, 580, 391–395. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.Y.; Dai, Y.H.; Wan, X.X.; Hu, X.M.; Zhao, W.J.; Ban, X.X.; Wan, H.; Huang, K.; Zhang, Q.; Xiong, K. ZBP1-Mediated Necroptosis: Mechanisms and Therapeutic Implications. Molecules 2023, 28, 52. [Google Scholar] [CrossRef] [PubMed]
- Simpson, D.S.; Pang, J.; Weir, A.; Kong, I.Y.; Fritsch, M.; Rashidi, M.; Cooney, J.P.; Davidson, K.C.; Speir, M.; Djajawi, T.M.; et al. Interferon-γ primes macrophages for pathogen ligand-induced killing via a caspase-8 and mitochondrial cell death pathway. Immunity 2022, 55, 423–441.e9. [Google Scholar] [CrossRef]
- Muendlein, H.I.; Connolly, W.M.; Magri, Z.; Smirnova, I.; Ilyukha, V.; Gautam, A.; Degterev, A.; Poltorak, A. ZBP1 promotes LPS-induced cell death and IL-1β release via RHIM-mediated interactions with RIPK1. Nat. Commun. 2021, 12, 86. [Google Scholar] [CrossRef] [PubMed]
- Lawlor, K.E.; Khan, N.; Mildenhall, A.; Gerlic, M.; Croker, B.A.; D’Cruz, A.A.; Hall, C.; Kaur Spall, S.; Anderton, H.; Masters, S.L.; et al. RIPK3 promotes cell death and NLRP3 inflammasome activation in the absence of MLKL. Nat. Commun. 2015, 6, 6282. [Google Scholar] [CrossRef]
- Kang, S.; Fernandes-Alnemri, T.; Rogers, C.; Mayes, L.; Wang, Y.; Dillon, C.; Roback, L.; Kaiser, W.; Oberst, A.; Sagara, J.; et al. Caspase-8 scaffolding function and MLKL regulate NLRP3 inflammasome activation downstream of TLR3. Nat. Commun. 2015, 6, 7515. [Google Scholar] [CrossRef] [PubMed]
- Conos, S.A.; Chen, K.W.; De Nardo, D.; Hara, H.; Whitehead, L.; Núñez, G.; Masters, S.L.; Murphy, J.M.; Schroder, K.; Vaux, D.L.; et al. Active MLKL triggers the NLRP3 inflammasome in a cell-intrinsic manner. Proc. Natl. Acad. Sci. USA 2017, 114, E961–E969. [Google Scholar] [CrossRef] [PubMed]
- Davies, K.A.; Tanzer, M.C.; Griffin, M.D.W.; Mok, Y.F.; Young, S.N.; Qin, R.; Petrie, E.J.; Czabotar, P.E.; Silke, J.; Murphy, J.M. The brace helices of MLKL mediate interdomain communication and oligomerisation to regulate cell death by necroptosis. Cell Death Differ. 2018, 25, 1567–1580. [Google Scholar] [CrossRef] [PubMed]
- Berger, S.B.; Bertin, J.; Gough, P.J. Life after death: RIP1 and RIP3 move beyond necroptosis. Cell Death Discov. 2016, 2, 16056. [Google Scholar] [CrossRef][Green Version]
- Meng, Y.; Davies, K.A.; Fitzgibbon, C.; Young, S.N.; Garnish, S.E.; Horne, C.R.; Luo, C.; Garnier, J.M.; Liang, L.Y.; Cowan, A.D.; et al. Human RIPK3 maintains MLKL in an inactive conformation prior to cell death by necroptosis. Nat. Commun. 2021, 12, 6783. [Google Scholar] [CrossRef]
- Garnish, S.E.; Meng, Y.; Koide, A.; Sandow, J.J.; Denbaum, E.; Jacobsen, A.V.; Yeung, W.; Samson, A.L.; Horne, C.R.; Fitzgibbon, C.; et al. Conformational interconversion of MLKL and disengagement from RIPK3 precede cell death by necroptosis. Nat. Commun. 2021, 12, 2211. [Google Scholar] [CrossRef]
- Samson, A.L.; Fitzgibbon, C.; Patel, K.M.; Hildebrand, J.M.; Whitehead, L.W.; Rimes, J.S.; Jacobsen, A.V.; Horne, C.R.; Gavin, X.J.; Young, S.N.; et al. A toolbox for imaging RIPK1, RIPK3, and MLKL in mouse and human cells. Cell Death Differ. 2021, 28, 2126–2144. [Google Scholar] [CrossRef]
- Wang, H.; Sun, L.; Su, L.; Rizo, J.; Liu, L.; Wang, L.F.; Wang, F.S.; Wang, X. Mixed Lineage Kinase Domain-like Protein MLKL Causes Necrotic Membrane Disruption upon Phosphorylation by RIP3. Mol. Cell 2014, 54, 133–146. [Google Scholar] [CrossRef]
- Najafov, A.; Mookhtiar, A.K.; Luu, H.S.; Ordureau, A.; Pan, H.; Amin, P.P.; Li, Y.; Lu, Q.; Yuan, J. TAM Kinases Promote Necroptosis by Regulating Oligomerization of MLKL. Mol. Cell 2019, 75, 457–468.e4. [Google Scholar] [CrossRef]
- Garcia, L.R.; Tenev, T.; Newman, R.; Haich, R.O.; Liccardi, G.; John, S.W.; Annibaldi, A.; Yu, L.; Pardo, M.; Young, S.N.; et al. Ubiquitylation of MLKL at lysine 219 positively regulates necroptosis-induced tissue injury and pathogen clearance. Nat. Commun. 2021, 12, 3364. [Google Scholar] [CrossRef]
- Liu, Z.; Dagley, L.F.; Shield-Artin, K.; Young, S.N.; Bankovacki, A.; Wang, X.; Tang, M.; Howitt, J.; Stafford, C.A.; Nachbur, U.; et al. Oligomerization-driven MLKL ubiquitylation antagonizes necroptosis. EMBO J. 2021, 40, e103718. [Google Scholar] [CrossRef]
- Yoon, S.; Bogdanov, K.; Wallach, D. Site-specific ubiquitination of MLKL targets it to endosomes and targets Listeria and Yersinia to the lysosomes. Cell Death Differ. 2022, 29, 306–322. [Google Scholar] [CrossRef]
- Petrie, E.J.; Hildebrand, J.M.; Murphy, J.M. Insane in the membrane: A structural perspective of MLKL function in necroptosis. Immunol. Cell Biol. 2017, 95, 152–159. [Google Scholar] [CrossRef]
- Espiritu, R.A.; Pedrera, L.; Ros, U. Tuning the way to die: Implications of membrane perturbations in necroptosis. In Advances in Biomembranes and Lipid Self-Assembly; Academic Press: Cambridge, MA, USA, 2019; Volume 29. [Google Scholar] [CrossRef]
- Cai, Z.; Jitkaew, S.; Zhao, J.; Chiang, H.C.; Choksi, S.; Liu, J.; Ward, Y.; Wu, L.G.; Liu, Z.G. Plasma membrane translocation of trimerized MLKL protein is required for TNF-induced necroptosis. Nat. Cell Biol. 2014, 16, 55–65. [Google Scholar] [CrossRef] [PubMed]
- Yoon, S.; Kovalenko, A.; Bogdanov, K.; Wallach, D. MLKL, the Protein that Mediates Necroptosis, Also Regulates Endosomal Trafficking and Extracellular Vesicle Generation. Immunity 2017, 47, 51–65. [Google Scholar] [CrossRef]
- Dondelinger, Y.; Declercq, W.; Montessuit, S.; Roelandt, R.; Goncalves, A.; Bruggeman, I.; Hulpiau, P.; Weber, K.; Sehon, C.A.; Marquis, R.W.; et al. MLKL Compromises Plasma Membrane Integrity by Binding to Phosphatidylinositol Phosphates. Cell Rep. 2014, 7, 971–981. [Google Scholar] [CrossRef]
- Petrie, E.J.; Birkinshaw, R.W.; Koide, A.; Denbaum, E.; Hildebrand, J.M.; Garnish, S.E.; Davies, K.A.; Sandow, J.J.; Samson, A.L.; Gavin, X.; et al. Identification of MLKL membrane translocation as a checkpoint in necroptotic cell death using Monobodies. Proc. Natl. Acad. Sci. USA 2020, 117, 8468–8475. [Google Scholar] [CrossRef] [PubMed]
- Dovey, C.M.; Diep, J.; Clarke, B.P.; Hale, A.T.; McNamara, D.E.; Guo, H.; Brown, N.W.; Cao, J.Y.; Grace, C.R.; Gough, P.J.; et al. MLKL Requires the Inositol Phosphate Code to Execute Necroptosis. Mol. Cell 2018, 70, 936–948.e7. [Google Scholar] [CrossRef] [PubMed]
- Xia, B.; Fang, S.; Chen, X.; Hu, H.; Chen, P.; Wang, H.; Gao, Z. MLKL forms cation channels. Cell Res. 2016, 26, 517–528. [Google Scholar] [CrossRef] [PubMed]
- Ros Quincoces, U.L.; Garcia-Saez, A.J. Necroptosis Execution is Mediated by Plasma Membrane Nanopores that are Independent of Calcium. Biophys. J. 2017, 112, 400a. [Google Scholar] [CrossRef][Green Version]
- Flores-romero, H.; Ros, U.; Garcia-saez, A.J. Pore formation in regulated cell death. EMBO J. 2020, 39, e105753. [Google Scholar] [CrossRef]
- Liu, S.; Liu, H.; Johnston, A.; Hanna-Addams, S.; Reynoso, E.; Xiang, Y.; Wang, Z. MLKL forms disulfide bond-dependent amyloid-like polymers to induce necroptosis. Proc. Natl. Acad. Sci. USA 2017, 114, E7450–E7459. [Google Scholar] [CrossRef] [PubMed]
- Gong, Y.N.; Guy, C.; Olauson, H.; Becker, J.U.; Yang, M.; Fitzgerald, P.; Linkermann, A.; Green, D.R. ESCRT-III Acts Downstream of MLKL to Regulate Necroptotic Cell Death and Its Consequences. Cell 2017, 169, 286–300. [Google Scholar] [CrossRef] [PubMed]
- Fan, W.; Guo, J.; Gao, B.; Zhang, W.; Ling, L.; Xu, T.; Pan, C.; Li, L.; Chen, S.; Wang, H.; et al. Flotillin-mediated endocytosis and ALIX–syntenin-1–mediated exocytosis protect the cell membrane from damage caused by necroptosis. Sci. Signal. 2019, 12, eaaw3423. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.; Prokopec, J.S.; Zhang, Y.; Sukhoplyasova, M.; Shinglot, H.; Wang, M.T.; Linkermann, A.; Stewart-Ornstein, J.; Gong, Y.N. Sensing plasma membrane pore formation induces chemokine production in survivors of regulated necrosis. Dev. Cell 2022, 57, 228–245. [Google Scholar] [CrossRef]
- Tang, Y.; Chen, Y.; Nursalim, Y.; Groom, K.; Hickey, A.; Chamley, L.; Chen, Q. Endoplasmic reticulum stress occurs in association with the extrusion of toxic extracellular vesicles from human placentae treated with antiphospholipid antibodies. Clin. Sci. 2020, 134, 459–472. [Google Scholar] [CrossRef]
- Sai, K.; Parsons, C.; House, J.S.; Kathariou, S.; Ninomiya-Tsuji, J. Necroptosis mediators RIPK3 and MLKL suppress intracellular Listeria replication independently of host cell killing. J. Cell Biol. 2019, 218, 1994–2005. [Google Scholar] [CrossRef]
- Valenti, M.; Molina, M.; Cid, V.J. Human gasdermin D and MLKL disrupt mitochondria, endocytic traffic and TORC1 signalling in budding yeast. Open Biol. 2023, 13, 220366. [Google Scholar] [CrossRef]
- Douanne, T.; André-Grégoire, G.; Trillet, K.; Thys, A.; Papin, A.; Feyeux, M.; Hulin, P.; Chiron, D.; Gavard, J.; Bidère, N. Pan-nexin-1 limits the production of proinflammatory cytokines during necroptosis. EMBO Rep. 2019, 20, e47840. [Google Scholar] [CrossRef]
- Chen, J.; Kuroki, S.; Someda, M.; Yonehara, S. Interferon-γ induces the cell surface exposure of phosphatidylserine by activating the protein MLKL in the absence of caspase-8 activity. J. Biol. Chem. 2019, 294, 11994–12006. [Google Scholar] [CrossRef]
- Zhang, S.; Su, Y.; Ying, Z.; Guo, D.; Pan, C.; Guo, J.; Zou, Z.; Wang, L.; Zhang, Z.; Jiang, Z.; et al. RIP1 kinase inhibitor halts the progression of an immune-induced demyelination disease at the stage of monocyte elevation. Proc. Natl. Acad. Sci. USA 2019, 116, 5675–5680. [Google Scholar] [CrossRef]
- Ying, Z.; Pan, C.; Shao, T.; Liu, L.; Li, L.; Guo, D.; Zhang, S.; Yuan, T.; Cao, R.; Jiang, Z.; et al. Mixed Lineage Kinase Domain-like Protein MLKL Breaks Down Myelin following Nerve Injury. Mol. Cell 2018, 72, 457–468.e5. [Google Scholar] [CrossRef] [PubMed]
- Yoon, S.; Bogdanov, K.; Kovalenko, A.; Wallach, D. Necroptosis is preceded by nuclear translocation of the signaling proteins that induce it. Cell Death Differ. 2016, 23, 253–260. [Google Scholar] [CrossRef]
- Weber, K.; Roelandt, R.; Bruggeman, I.; Estornes, Y.; Vandenabeele, P. Nuclear RIPK3 and MLKL contribute to cytosolic necrosome formation and necroptosis. Commun. Biol. 2018, 1, 6. [Google Scholar] [CrossRef] [PubMed]
- Dai, J.; Zhang, C.; Guo, L.; He, H.; Jiang, K.; Huang, Y.; Zhang, X.; Zhang, H.; Wei, W.; Zhang, Y.; et al. A necroptotic-independent function of MLKL in regulating endothelial cell adhesion molecule expression. Cell Death Dis. 2020, 11, 282. [Google Scholar] [CrossRef]
- Liu, Y.; Liu, T.; Lei, T.; Zhang, D.; Du, S.; Girani, L.; Qi, D.; Lin, C.; Tong, R.; Wang, Y. RIP1/RIP3-regulated necroptosis as a target for multifaceted disease therapy (Review). Int. J. Mol. Med. 2019, 44, 771–786. [Google Scholar] [CrossRef]
- Zhang, G.; Wang, J.; Zhao, Z.; Xin, T.; Fan, X.; Shen, Q.; Raheem, A.; Lee, C.R.; Jiang, H.; Ding, J. Regulated necrosis, a proinflammatory cell death, potentially counteracts pathogenic infections. Cell Death Dis. 2022, 13, 637. [Google Scholar] [CrossRef] [PubMed]
- Martens, S.; Bridelance, J.; Roelandt, R.; Vandenabeele, P.; Takahashi, N. MLKL in cancer: More than a necroptosis regulator. Cell Death Differ. 2021, 28, 1757–1772. [Google Scholar] [CrossRef]
- Griffiths, C.E.; Barker, J.N. Pathogenesis and clinical features of psoriasis. Lancet 2007, 370, 263–271. [Google Scholar] [CrossRef]
- Hao, Q.; Shetty, S.; Tucker, T.A.; Idell, S. Interferon-γ Preferentially Promotes Necroptosis of Lung Epithelial Cells by Upregulating MLKL. Cells 2022, 11, 563. [Google Scholar] [CrossRef]
- Sarhan, J.; Liu, B.C.; Muendlein, H.I.; Weindel, C.G.; Smirnova, I.; Tang, A.Y.; Ilyukha, V.; Sorokin, M.; Buzdin, A.; Fitzgerald, K.A.; et al. Constitutive interferon signaling maintains critical threshold of MLKL expression to license necroptosis. Cell Death Differ. 2019, 26, 332–347. [Google Scholar] [CrossRef]
- Caccamo, A.; Branca, C.; Piras, I.S.; Ferreira, E.; Huentelman, M.J.; Liang, W.S.; Readhead, B.; Dudley, J.T.; Spangenberg, E.E.; Green, K.N.; et al. Necroptosis activation in Alzheimer’s disease. Nat. Neurosci. 2017, 20, 1236–1246. [Google Scholar] [CrossRef]
- Richard, R.; Mousa, S. Necroptosis in Alzheimer’s disease: Potential therapeutic target. Biomed. Pharmacother. 2022, 152, 113203. [Google Scholar] [CrossRef]
- Iannielli, A.; Bido, S.; Folladori, L.; Segnali, A.; Cancellieri, C.; Maresca, A.; Massimino, L.; Rubio, A.; Morabito, G.; Caporali, L.; et al. Pharmacological Inhibition of Necroptosis Protects from Dopaminergic Neuronal Cell Death in Parkinson’s Disease Models. Cell Rep. 2018, 22, 2066–2079. [Google Scholar] [CrossRef]
- Picon, C.; Jayaraman, A.; James, R.; Beck, C.; Gallego, P.; Witte, M.E.; van Horssen, J.; Mazarakis, N.D.; Reynolds, R. Neuron-specific activation of necroptosis signaling in multiple sclerosis cortical grey matter. Acta Neuropathol. 2021, 141, 585–604. [Google Scholar] [CrossRef]
- Karunakaran, D.; Geoffrion, M.; Wei, L.; Gan, W.; Richards, L.; Shangari, P.; DeKemp, E.M.; Beanlands, R.A.; Perisic, L.; Maegdefessel, L.; et al. Targeting macrophage necroptosis for therapeutic and diagnostic interventions in atherosclerosis. Sci. Adv. 2016, 2, e1600224. [Google Scholar] [CrossRef]
- Yang, J.; Zhao, Y.; Zhang, L.; Fan, H.; Qi, C.; Zhang, K.; Liu, X.; Fei, L.; Chen, S.; Wang, M.; et al. RIPK3/MLKL-Mediated Neuronal Necroptosis Modulates the M1/M2 Polarization of Microglia/Macrophages in the Ischemic Cortex. Cereb. Cortex 2018, 28, 2622–2635. [Google Scholar] [CrossRef]
- Lu, Z.; Van Eeckhoutte, H.P.; Liu, G.; Nair, P.M.; Jones, B.; Gillis, C.M.; Nalkurthi, B.C.; Verhamme, F.; Buyle-Huybrecht, T.; Vandenabeele, P.; et al. Necroptosis Signaling Promotes Inflammation, Airway Remodeling, and Emphysema in Chronic Obstructive Pulmonary Disease. Am. J. Respir. Crit. Care Med. 2021, 204, 667–681. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.-M.; Yoshida, M.; Kim, M.-S.; Lee, J.-H.; Baek, A.-R.; Jang, A.S.; Kim, D.J.; Minagawa, S.; Chin, S.S.; Park, C.-S.; et al. Involvement of Alveolar Epithelial Cell Necroptosis in Idiopathic Pulmonary Fibrosis Pathogenesis. Am. J. Respir. Cell Mol. Biol. 2018, 59, 215–224. [Google Scholar] [CrossRef] [PubMed]
- Seifert, L.; Werba, G.; Tiwari, S.; Giao Ly, N.N.; Alothman, S.; Alqunaibit, D.; Avanzi, A.; Barilla, R.; Daley, D.; Greco, S.H.; et al. The necrosome promotes pancreatic oncogenesis via CXCL1 and Mincle-induced immune suppression. Nature 2016, 532, 245–249. [Google Scholar] [CrossRef] [PubMed]
- Ando, Y.; Ohuchida, K.; Otsubo, Y.; Kibe, S.; Takesue, S.; Abe, T.; Iwamoto, C.; Shindo, K.; Moriyama, T.; Nakata, K.; et al. Necroptosis in pancreatic cancer promotes cancer cell migration and invasion by release of CXCL5. PLoS ONE 2020, 15, e0228015. [Google Scholar] [CrossRef] [PubMed]
- Colbert, L.E.; Fisher, S.B.; Hardy, C.W.; Hall, W.A.; Saka, B.; Shelton, J.W.; Petrova, A.V.; Warren, M.D.; Pantazides, B.G.; Gandhi, K.; et al. Pronecrotic mixed lineage kinase domain-like protein expression is a prognostic biomarker in patients with early-stage resected pancreatic adenocarcinoma. Cancer 2013, 119, 3148–3155. [Google Scholar] [CrossRef] [PubMed]
- Stoll, G.; Ma, Y.; Yang, H.; Kepp, O.; Zitvogel, L.; Kroemer, G. Pro-necrotic molecules impact local immunosurveillance in human breast cancer. Oncoimmunology 2017, 6, e1299302. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Huang, S.; Zeng, L.; Li, K.; Yang, L.; Gao, S.; Guan, C.; Zhang, S.; Lao, X.; Liao, G.; et al. Necroptosis in head and neck squamous cell carcinoma: Characterization of clinicopathological relevance and in vitro cell model. Cell Death Dis. 2020, 11, 391. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Ros, U.; Agrawal, D.; Keller, E.C.; Slotta-Huspenina, J.; Dill, V.; Shen, B.; Shi, R.; Herold, T.; Belka, C.; et al. MLKL promotes cellular differentiation in myeloid leukemia by facilitating the release of G-CSF. Cell Death Differ. 2021, 28, 3235–3250. [Google Scholar] [CrossRef]
- Ruan, J.; Mei, L.; Zhu, Q.; Shi, G.; Wang, H. Mixed lineage kinase domain-like protein is a prognostic biomarker for cervical squamous cell cancer. Int. J. Clin. Exp. Pathol. 2015, 8, 15035–15038. [Google Scholar]
- Zhao, Q.; Cheng, X.; Guo, J.; Bi, Y.; Kuang, L.; Ren, J.; Zhong, J.; Pan, L.; Zhang, X.; Guo, Y.; et al. MLKL inhibits intestinal tumorigenesis by suppressing STAT3 signaling pathway. Int. J. Biol. Sci. 2021, 17, 869–881. [Google Scholar] [CrossRef]
- Seifert, L.; Werba, G.; Tiwari, S.; Giao Ly, N.N.; Alothman, S.; Alqunaibit, D.; Avanzi, A.; Barilla, R.; Daley, D.; Greco, S.H.; et al. Necrosulfonamide inhibits necroptosis by selectively targeting the mixed lineage kinase domain-like protein. Cell Death Dis. 2020, 5, 333–337. [Google Scholar] [CrossRef]
- Yan, B.; Liu, L.; Huang, S.; Ren, Y.; Wang, H.; Yao, Z.; Li, L.; Chen, S.; Wang, X.; Zhang, Z. Discovery of a new class of highly potent necroptosis inhibitors targeting the mixed lineage kinase domain-like protein. Chem. Commun. 2017, 53, 3637–3640. [Google Scholar] [CrossRef]
- Rathkey, J.K.; Zhao, J.; Liu, Z.; Chen, Y.; Yang, J.; Kondolf, H.C.; Benson, B.L.; Chirieleison, S.M.; Huang, A.Y.; Dubyak, G.R.; et al. Chemical disruption of the pyroptotic pore-forming protein gasdermin D inhibits inflammatory cell death and sepsis. Sci. Immunol. 2018, 3, eaat2738. [Google Scholar] [CrossRef]
- Cui, B.; Yan, B.; Wang, K.; Li, L.; Chen, S.; Zhang, Z. Discovery of a New Class of Uracil Derivatives as Potential Mixed Lineage Kinase Domain-like Protein (MLKL) Inhibitors. J. Med. Chem. 2022, 65, 12747–12780. [Google Scholar] [CrossRef] [PubMed]
- Rübbelke, M.; Fiegen, D.; Bauer, M.; Binder, F.; Hamilton, J.; King, J.; Thamm, S.; Nar, H.; Zeeb, M. Locking mixed-lineage kinase domain-like protein in its auto-inhibited state prevents necroptosis. Proc. Natl. Acad. Sci. USA 2020, 117, 33272–33281. [Google Scholar] [CrossRef] [PubMed]
- Hildebrand, J.M.; Tanzer, M.C.; Lucet, I.S.; Young, S.N.; Spall, S.K.; Sharma, P.; Pierotti, C.; Garnier, J.M.; Dobson, R.C.; Webb, A.I.; et al. Activation of the pseudokinase MLKL unleashes the four-helix bundle domain to induce membrane localization and necroptotic cell death. Proc. Natl. Acad. Sci. USA 2014, 111, 15072–15077. [Google Scholar] [CrossRef] [PubMed]
- Pierotti, C.L.; Tanzer, M.C.; Jacobsen, A.V.; Hildebrand, J.M.; Garnier, J.M.; Sharma, P.; Lucet, I.S.; Cowan, A.D.; Kersten, W.J.; Luo, M.X.; et al. Potent Inhibition of Necroptosis by Simultaneously Targeting Multiple Effectors of the Pathway. ACS Chem. Biol. 2020, 15, 2702–2713. [Google Scholar] [CrossRef]
- Sammond, D.M.; Nailor, K.E.; Veal, J.M.; Nolte, R.T.; Wang, L.; Knick, V.B.; Rudolph, S.K.; Truesdale, A.T.; Nartey, E.N.; Stafford, J.A.; et al. Discovery of a novel and potent series of dianilinopyrimidineurea and urea isostere inhibitors of VEGFR2 tyrosine kinase. Bioorg. Med. Chem. Lett. 2005, 15, 3519–3523. [Google Scholar] [CrossRef]
- Rübbelke, M.; Hamilton, J.; Binder, F.; Bauer, M.; King, J.; Nar, H.; Zeeb, M. Discovery and Structure-Based Optimization of Fragments Binding the Mixed Lineage Kinase Domain-like Protein Executioner Domain. J. Med. Chem. 2021, 64, 15629–15638. [Google Scholar] [CrossRef]
- Ji, Y.; Ward, L.A.; Hawkins, C.J. Reconstitution of Human Necrosome Interactions in Saccharomyces cerevisiae. Biomolecules 2021, 11, 153. [Google Scholar] [CrossRef]
| Class | Mechanism | Inhibitor | Structure | Potency | Effect on Hallmarks of MLKL Activation | Limitations | Ref. |
|---|---|---|---|---|---|---|---|
| Irreversible inhibitors | Covalent binding to the 4HB via C86 | NSA | 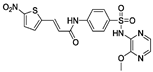 | IC50 = 1.4 nM |
|
| [99,100,101,102,103] |
| Xanthine-derivatives ex:TC13172 |  | IC50 = 2 nM |
| ||||
| Uracil-derivatives | 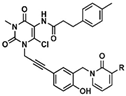 Cpd 56 R = H Cpd 66 R = OCH3 | Cpd 56 IC50 = 82 nM Cpd 66 IC50 = 31 nM |
| ||||
| Targeting the ATP binding site | Non-covalent binding to the psK domain | Compound 1 or GW806742X |  | IC50 < 50 nM Kd = 0.53 µM |
|
| [104,105,106] |
| Compound 2 |  | IC50 < 50 nM Kd = 1.8 µM | |||||
| Allosteric and reversible binders | Non-covalent binding to the 4HB domain | Compound 5 | 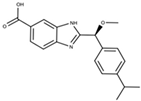 | Kd = 50 µM |
|
| [107] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Martinez-Osorio, V.; Abdelwahab, Y.; Ros, U. The Many Faces of MLKL, the Executor of Necroptosis. Int. J. Mol. Sci. 2023, 24, 10108. https://doi.org/10.3390/ijms241210108
Martinez-Osorio V, Abdelwahab Y, Ros U. The Many Faces of MLKL, the Executor of Necroptosis. International Journal of Molecular Sciences. 2023; 24(12):10108. https://doi.org/10.3390/ijms241210108
Chicago/Turabian StyleMartinez-Osorio, Veronica, Yasmin Abdelwahab, and Uris Ros. 2023. "The Many Faces of MLKL, the Executor of Necroptosis" International Journal of Molecular Sciences 24, no. 12: 10108. https://doi.org/10.3390/ijms241210108
APA StyleMartinez-Osorio, V., Abdelwahab, Y., & Ros, U. (2023). The Many Faces of MLKL, the Executor of Necroptosis. International Journal of Molecular Sciences, 24(12), 10108. https://doi.org/10.3390/ijms241210108





