Genome-Wide Analysis of Strictosidine Synthase-like Gene Family Revealed Their Response to Biotic/Abiotic Stress in Poplar
Abstract
:1. Introduction
2. Result
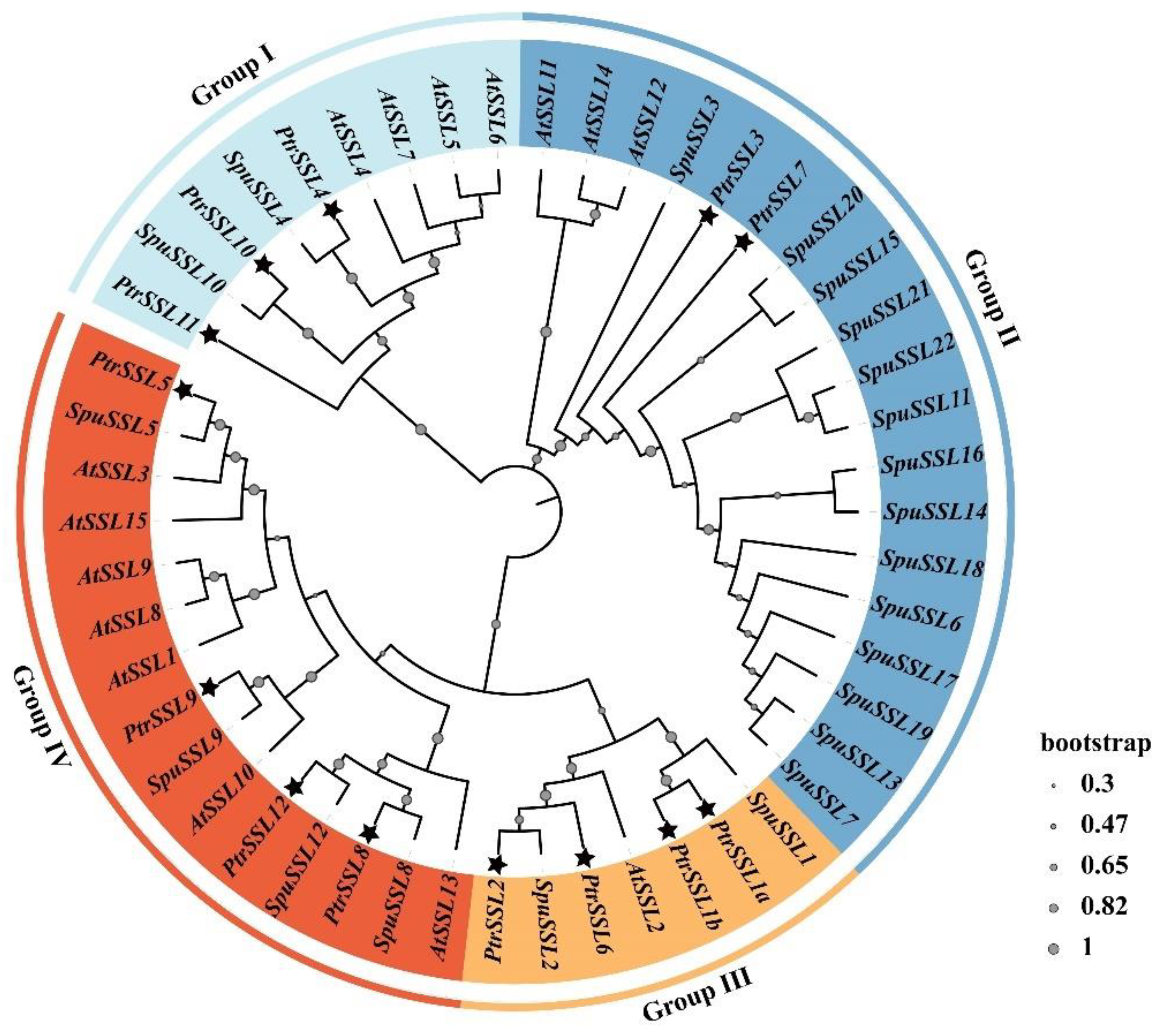
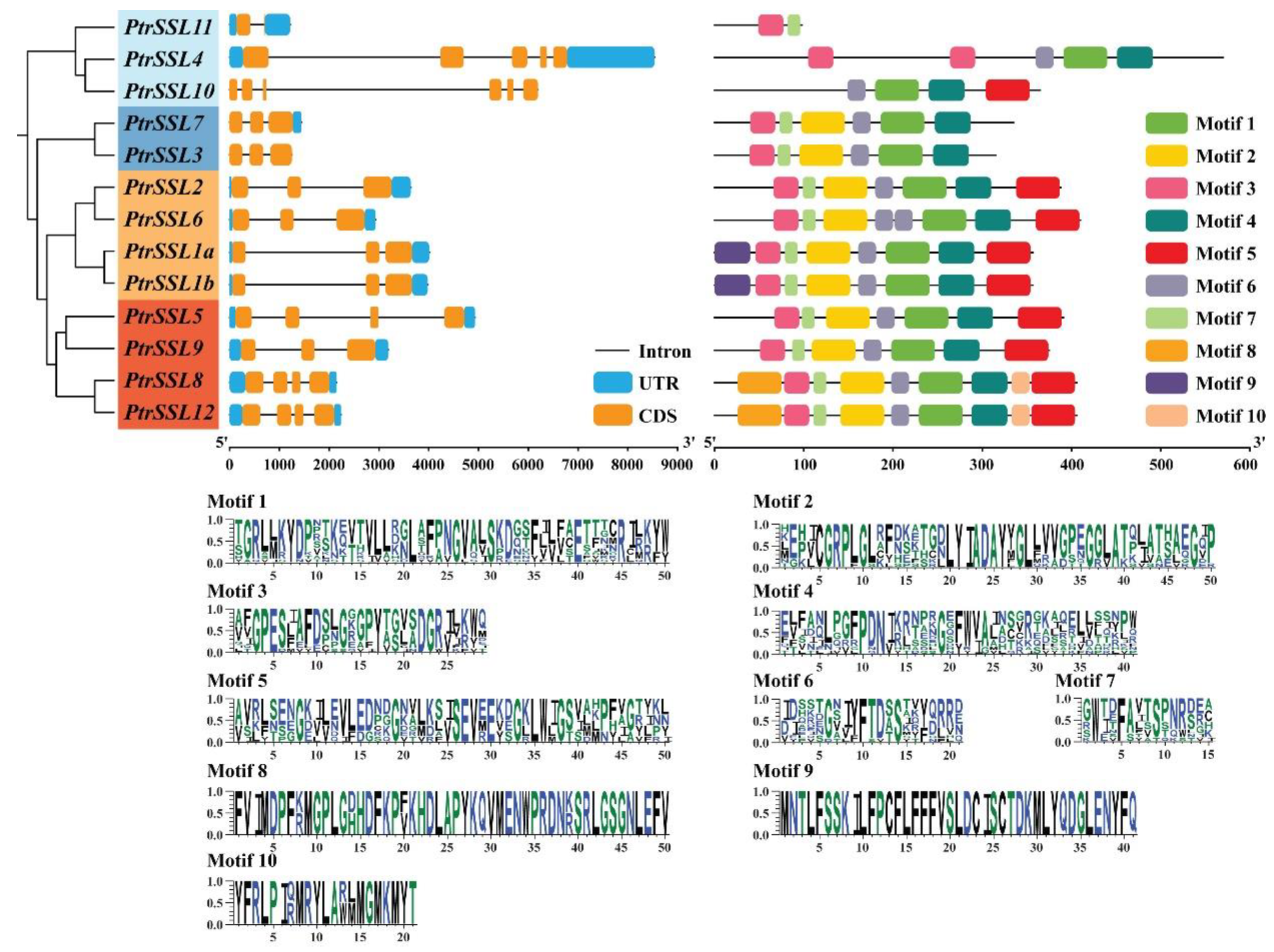
2.1. Phylogenetic and Structural Analysis of PtrSSL Family
2.2. Chromosomal Distribution and Collinearity Analysis of PtrSSLs
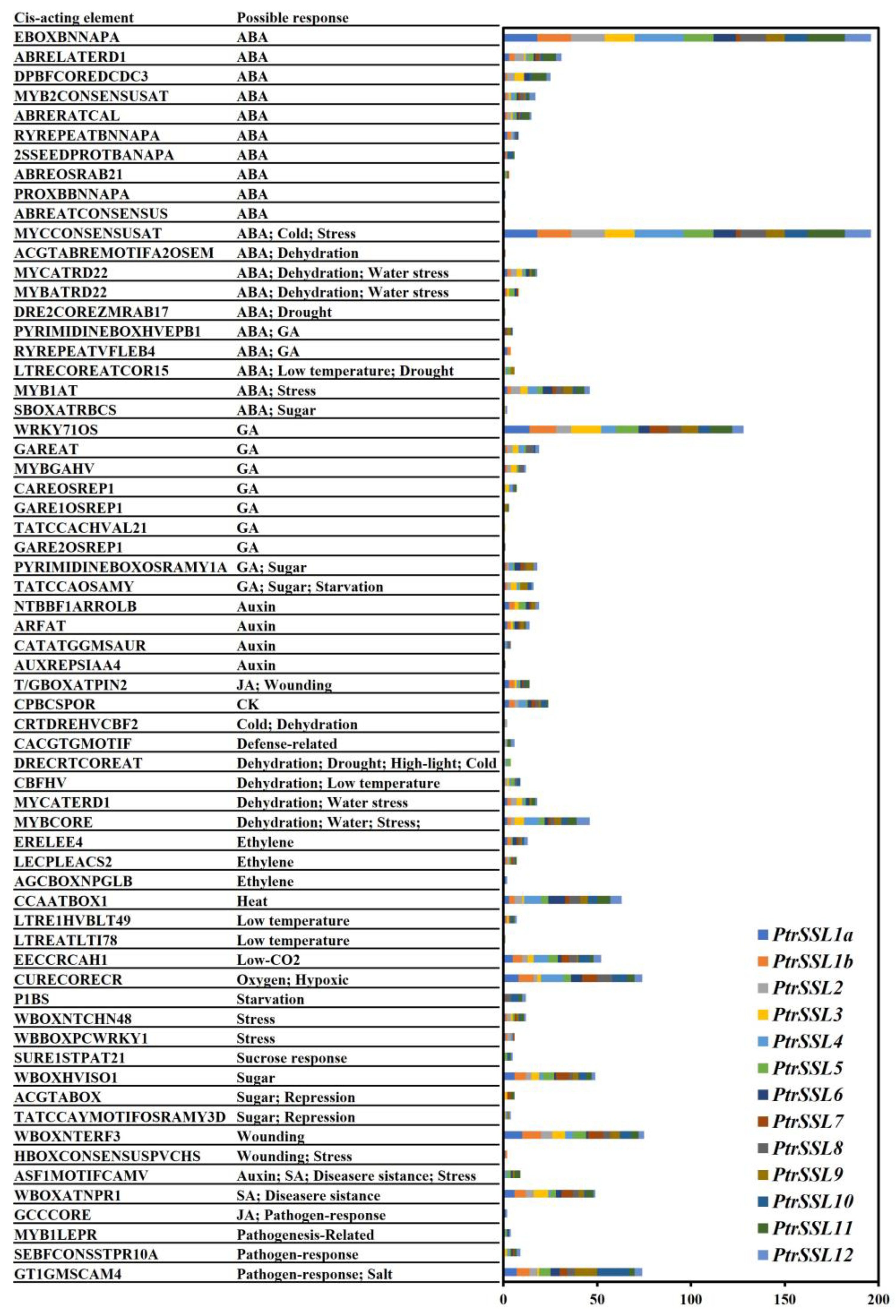
2.3. Cis-Elements Analysis of PtrSSLs Promoters
2.4. Expression Patterns of PtrSSLs in Roots, Stems, and Leaves
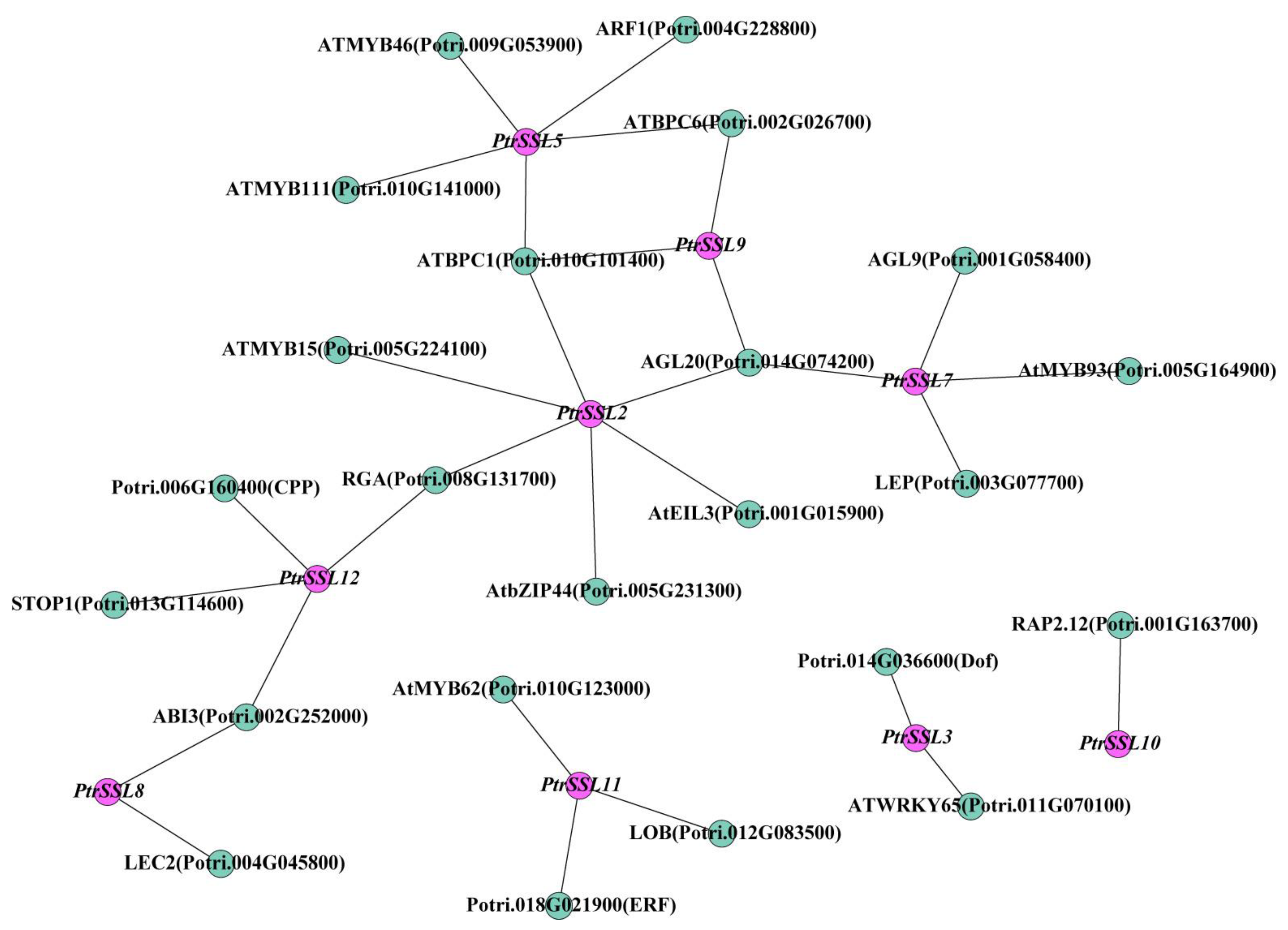
2.5. Analysis of Upstream TF Regulation Network
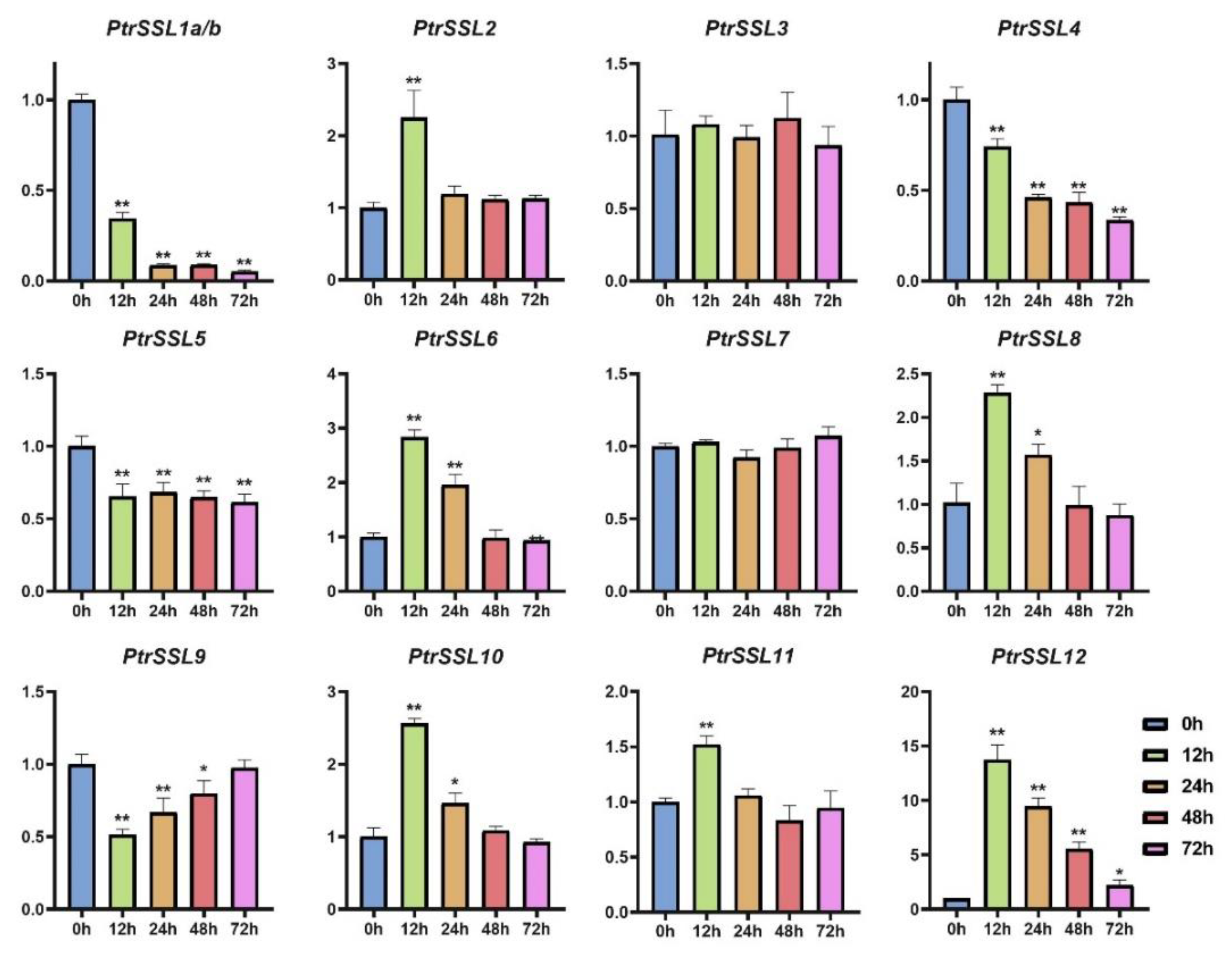
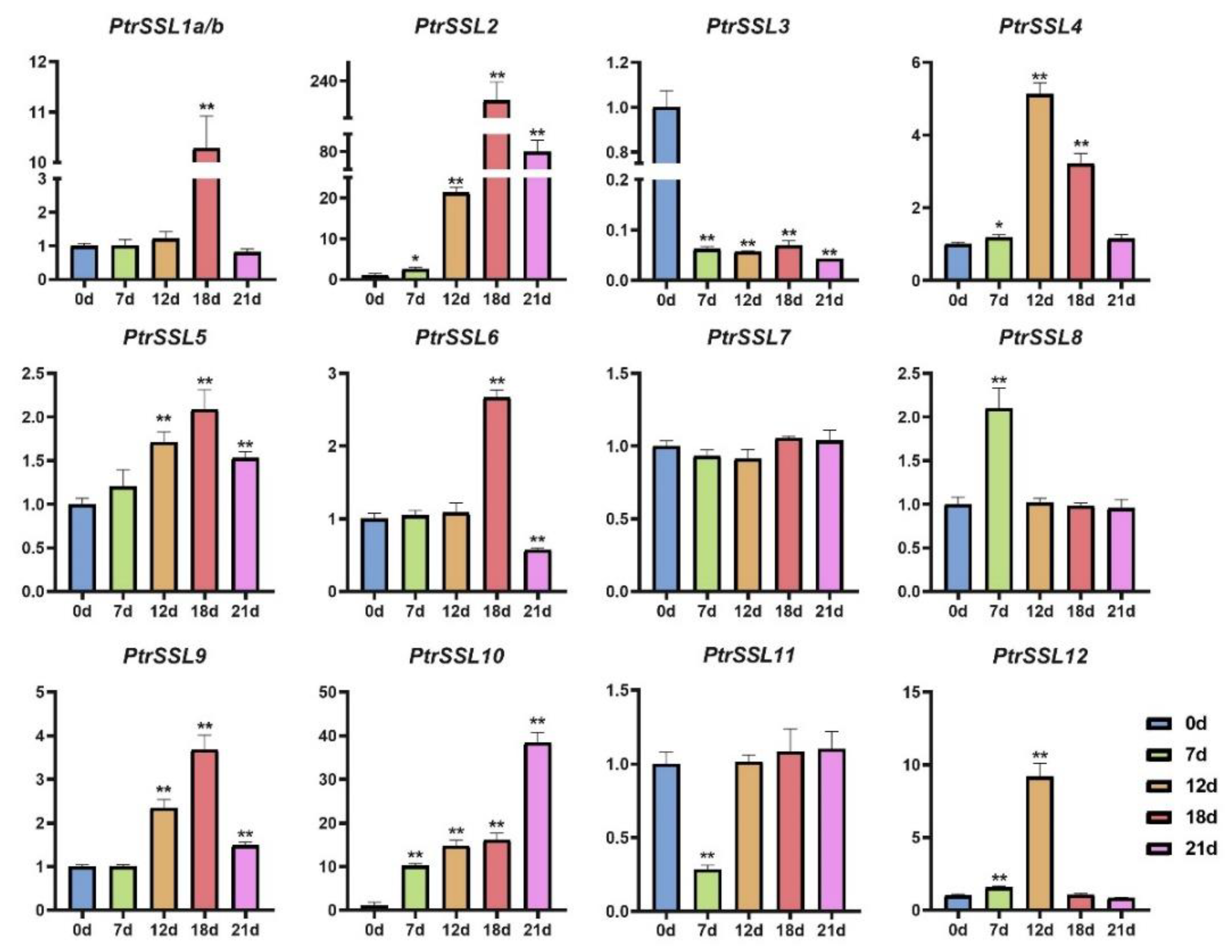
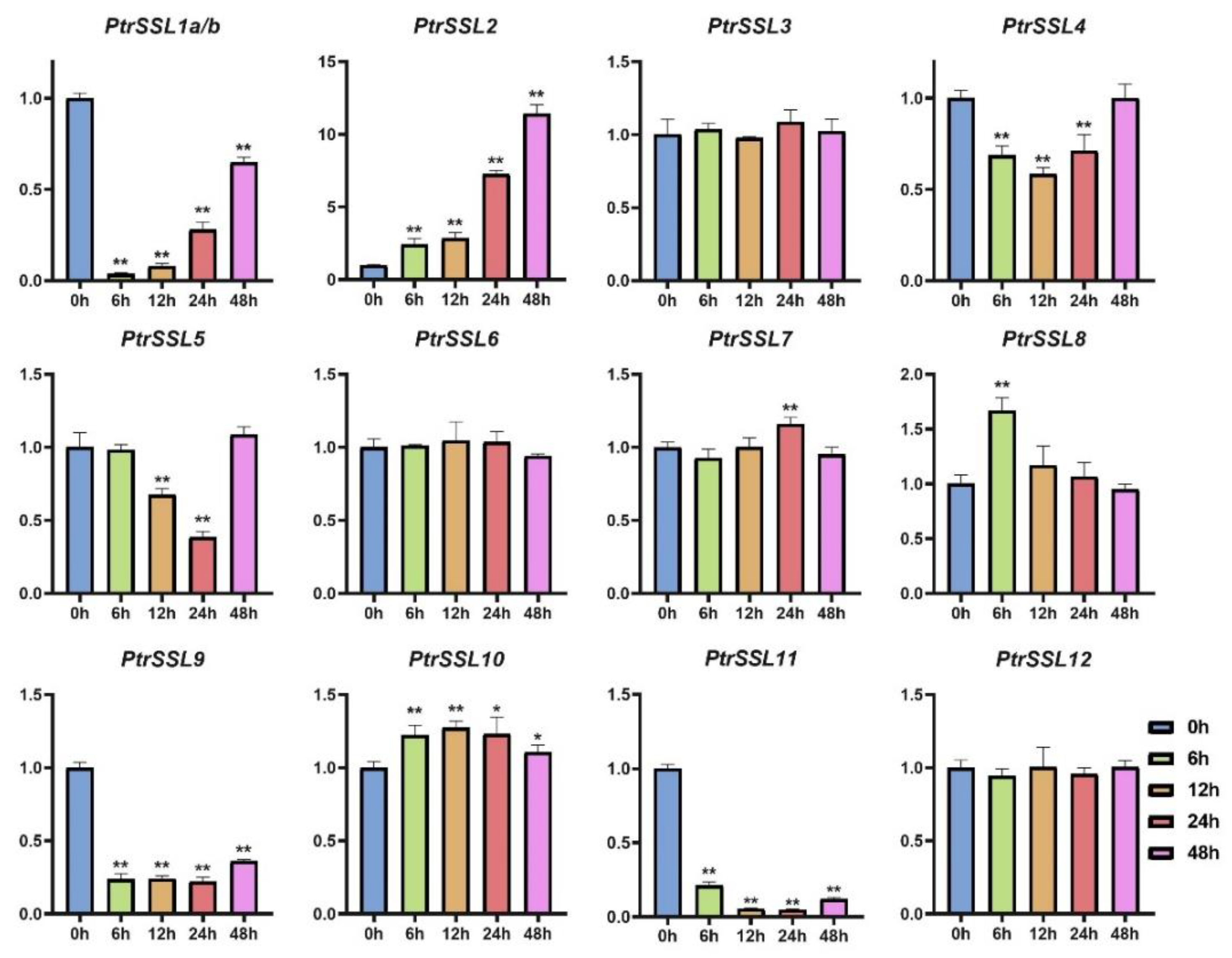
2.6. RT-qPCR Validation of PtrSSLs under Different Stresses
3. Discussion
4. Materials and Methods
4.1. Identification of SSLs in Poplar
4.2. Phylogenetic and Structural Analysis of PtrSSLs
4.3. Chromosomal Localization and Collinearity Analysis
4.4. Cis-Acting Element Analysis
4.5. Upstream TF Regulation Network Analysis
4.6. Plant Treatments
4.7. qRT-PCR Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Sohani, M.M.; Schenk, P.M.; Schultz, C.J.; Schmidt, O. Phylogenetic and transcriptional analysis of a strictosidine synthase-like gene family in Arabidopsis thaliana reveals involvement in plant defence responses. Plant Biol. 2009, 11, 105–117. [Google Scholar] [CrossRef] [PubMed]
- Takahashi, F.; Kuromori, T.; Sato, H.; Shinozaki, K. Regulatory Gene Networks in Drought Stress Responses and Resistance in Plants. Adv. Exp. Med. Biol. 2018, 1081, 189–214. [Google Scholar] [CrossRef] [PubMed]
- Deinlein, U.; Stephan, A.B.; Horie, T.; Luo, W.; Xu, G.; Schroeder, J.I. Plant salt-tolerance mechanisms. Trends Plant Sci. 2014, 19, 371–379. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bashir, K.; Matsui, A.; Rasheed, S.; Seki, M. Recent advances in the characterization of plant transcriptomes in response to drought, salinity, heat, and cold stress. F1000Research 2019, 8, 658. [Google Scholar] [CrossRef] [Green Version]
- Wei, H.; Movahedi, A.; Liu, G.; Li, Y.; Liu, S.; Yu, C.; Chen, Y.; Zhong, F.; Zhang, J. Comprehensive Analysis of Carotenoid Cleavage Dioxygenases Gene Family and Its Expression in Response to Abiotic Stress in Poplar. Int. J. Mol. Sci. 2022, 23, 1418. [Google Scholar] [CrossRef]
- Chai, W.; Si, W.; Ji, W.; Qin, Q.; Zhao, M.; Jiang, H. Genome-Wide Investigation and Expression Profiling of HD-Zip Transcription Factors in Foxtail Millet (Setaria italica L.). BioMed Res. Int. 2018, 2018, 8457614. [Google Scholar] [CrossRef] [Green Version]
- Tounsi, S.; Jemli, S.; Feki, K.; Brini, F.; Najib Saïdi, M. Superoxide dismutase (SOD) family in durum wheat: Promising candidates for improving crop resilience. Protoplasma 2023, 260, 145–158. [Google Scholar] [CrossRef]
- Zeng, D.; Dai, L.J.; Li, X.; Li, W.; Qu, G.Z.; Li, S. Genome-Wide Identification of the ERF Transcription Factor Family for Structure Analysis, Expression Pattern, and Response to Drought Stress in Populus alba × Populus glandulosa. Int. J. Mol. Sci. 2023, 24, 3697. [Google Scholar] [CrossRef]
- Liu, R.; Wu, M.; Liu, H.L.; Gao, Y.M.; Chen, J.; Yan, H.W.; Xiang, Y. Genome-wide identification and expression analysis of the NF-Y transcription factor family in Populus. Physiol. Plant. 2021, 171, 309–327. [Google Scholar] [CrossRef]
- Wang, Y.; Wang, R.; Yu, Y.; Gu, Y.; Wang, S.; Liao, S.; Xu, X.; Jiang, T.; Yao, W. Genome-Wide Analysis of SIMILAR TO RCD ONE (SRO) Family Revealed Their Roles in Abiotic Stress in Poplar. Int. J. Mol. Sci. 2023, 24, 4146. [Google Scholar] [CrossRef]
- Wang, R.; Wang, Y.; Gu, Y.; Yan, P.; Zhao, W.; Jiang, T. Genome-Wide Identification of miR169 Family in Response to ABA and Salt Stress in Poplar. Forests 2023, 14, 961. [Google Scholar] [CrossRef]
- Fabbri, M.; Delp, G.; Schmidt, O.; Theopold, U. Animal and plant members of a gene family with similarity to alkaloid-synthesizing enzymes. Biochem. Biophys. Res. Commun. 2000, 271, 191–196. [Google Scholar] [CrossRef]
- Kibble, N.A.J.; Sohani, M.M.; Shirley, N.; Byrt, C.; Roessner, U.; Bacic, A.; Schmidt, O.; Schultz, C.J. Phylogenetic analysis and functional characterisation of strictosidine synthase-like genes in Arabidopsis thaliana. Funct. Plant Biol. FPB 2010, 36, 1098–1109. [Google Scholar] [CrossRef]
- Ali, E.F.; El-Shehawi, A.M.; Ibrahim, O.H.M.; Abdul-Hafeez, E.Y.; Moussa, M.M.; Hassan, F.A.S. A vital role of chitosan nanoparticles in improvisation the drought stress tolerance in Catharanthus roseus (L.) through biochemical and gene expression modulation. Plant Physiol. Biochem. PPB 2021, 161, 166–175. [Google Scholar] [CrossRef]
- Chen, L.; Meng, J.; Luan, Y. miR1916 plays a role as a negative regulator in drought stress resistance in tomato and tobacco. Biochem. Biophys. Res. Commun 2019, 508, 597–602. [Google Scholar] [CrossRef]
- Liu, Y.; Yang, H.; Wen, F.; Bao, L.; Zhao, Z.; Zhong, Z. Chitooligosaccharide-induced plant stress resistance. Carbohydr. Polym. 2023, 302, 120344. [Google Scholar] [CrossRef]
- Smith, D.L.; Praslickova, D.; Ilangumaran, G. Inter-organismal signaling and management of the phytomicrobiome. Front. Plant Sci. 2015, 6, 722. [Google Scholar] [CrossRef] [Green Version]
- Tang, W.; Liu, X.; He, Y.; Yang, F. Enhancement of Vindoline and Catharanthine Accumulation, Antioxidant Enzymes Activities, and Gene Expression Levels in Catharanthus roseus Leaves by Chitooligosaccharides Elicitation. Mar. Drugs 2022, 20, 188. [Google Scholar] [CrossRef]
- Chen, L.; Meng, J.; He, X.L.; Zhang, M.; Luan, Y.S. Solanum lycopersicum microRNA1916 targets multiple target genes and negatively regulates the immune response in tomato. Plant Cell Environ. 2019, 42, 1393–1407. [Google Scholar] [CrossRef]
- Tuskan, G.A.; Difazio, S.; Jansson, S.; Bohlmann, J.; Grigoriev, I.; Hellsten, U.; Putnam, N.; Ralph, S.; Rombauts, S.; Salamov, A.; et al. The genome of black cottonwood, Populus trichocarpa (Torr. & Gray). Science 2006, 313, 1596–1604. [Google Scholar] [CrossRef] [Green Version]
- Higo, K.; Ugawa, Y.; Iwamoto, M.; Korenaga, T. Plant cis-acting regulatory DNA elements (PLACE) database: 1999. Nucleic Acids Res. 1999, 27, 297–300. [Google Scholar] [CrossRef] [Green Version]
- Bedi, S.; Sengupta, S.; Ray, A.; Nag Chaudhuri, R. ABI3 mediates dehydration stress recovery response in Arabidopsis thaliana by regulating expression of downstream genes. Plant Sci. 2016, 250, 125–140. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.; Guo, T.; Shen, Y.; Wang, Z.; Kang, J.; Zhang, J.; Yi, F.; Yang, Q.; Long, R. Overexpression of MtRAV3 enhances osmotic and salt tolerance and inhibits growth of Medicago truncatula. Plant Physiol. Biochem. 2021, 163, 154–165. [Google Scholar] [CrossRef] [PubMed]
- Barrero-Gil, J.; Mouriz, A.; Piqueras, R.; Salinas, J.; Jarillo, J.A.; Piñeiro, M. A MRG-operated chromatin switch at SOC1 attenuates abiotic stress responses during the floral transition. Plant Physiol. 2021, 187, 462–471. [Google Scholar] [CrossRef] [PubMed]
- Yan, J.; Liu, Y.; Yang, L.; He, H.; Huang, Y.; Fang, L.; Scheller, H.V.; Jiang, M.; Zhang, A. Cell wall β-1,4-galactan regulated by the BPC1/BPC2-GALS1 module aggravates salt sensitivity in Arabidopsis thaliana. Mol. Plant 2021, 14, 411–425. [Google Scholar] [CrossRef]
- Luo, Y.; Bai, R.; Li, J.; Yang, W.; Li, R.; Wang, Q.; Zhao, G.; Duan, D. The transcription factor MYB15 is essential for basal immunity (PTI) in Chinese wild grape. Planta 2019, 249, 1889–1902. [Google Scholar] [CrossRef]
- Agarwal, M.; Hao, Y.; Kapoor, A.; Dong, C.H.; Fujii, H.; Zheng, X.; Zhu, J.K. A R2R3 type MYB transcription factor is involved in the cold regulation of CBF genes and in acquired freezing tolerance. J. Biol. Chem. 2006, 281, 37636–37645. [Google Scholar] [CrossRef] [Green Version]
- Ramírez, V.; Agorio, A.; Coego, A.; García-Andrade, J.; Hernández, M.J.; Balaguer, B.; Ouwerkerk, P.B.; Zarra, I.; Vera, P. MYB46 modulates disease susceptibility to Botrytis cinerea in Arabidopsis. Plant Physiol. 2011, 155, 1920–1935. [Google Scholar] [CrossRef] [Green Version]
- Chen, K.; Song, M.; Guo, Y.; Liu, L.; Xue, H.; Dai, H.; Zhang, Z. MdMYB46 could enhance salt and osmotic stress tolerance in apple by directly activating stress-responsive signals. Plant Biotechnol. J. 2019, 17, 2341–2355. [Google Scholar] [CrossRef] [Green Version]
- Guerrero-Sánchez, V.M.; López-Hidalgo, C.; Rey, M.D.; Castillejo, M.; Jorrín-Novo, J.V.; Escandón, M. Multiomic Data Integration in the Analysis of Drought-Responsive Mechanisms in Quercus ilex Seedlings. Plants 2022, 11, 3067. [Google Scholar] [CrossRef]
- Jaiswal, V.; Kakkar, M.; Kumari, P.; Zinta, G.; Gahlaut, V.; Kumar, S. Multifaceted roles of GRAS transcription factors in growth and stress responses in plants. iScience 2022, 25, 105026. [Google Scholar] [CrossRef]
- Khan, Y.; Xiong, Z.; Zhang, H.; Liu, S.; Yaseen, T.; Hui, T. Expression and roles of GRAS gene family in plant growth, signal transduction, biotic and abiotic stress resistance and symbiosis formation—A review. Plant Biol. 2022, 24, 404–416. [Google Scholar] [CrossRef]
- Sadhukhan, A.; Kobayashi, Y.; Iuchi, S.; Koyama, H. Synergistic and antagonistic pleiotropy of STOP1 in stress tolerance. Trends Plant Sci 2021, 26, 1014–1022. [Google Scholar] [CrossRef]
- Qin, L.; Sun, L.; Wei, L.; Yuan, J.; Kong, F.; Zhang, Y.; Miao, X.; Xia, G.; Liu, S. Maize SRO1e represses anthocyanin synthesis through regulating the MBW complex in response to abiotic stress. Plant J. Cell Mol. Biol. 2021, 105, 1010–1025. [Google Scholar] [CrossRef]
- Cannon, S.B.; Mitra, A.; Baumgarten, A.; Young, N.D.; May, G. The roles of segmental and tandem gene duplication in the evolution of large gene families in Arabidopsis thaliana. BMC Plant Biol. 2004, 4, 10. [Google Scholar] [CrossRef] [Green Version]
- Hamel, L.P.; Sheen, J.; Séguin, A. Ancient signals: Comparative genomics of green plant CDPKs. Trends Plant Sci. 2014, 19, 79–89. [Google Scholar] [CrossRef] [Green Version]
- Hu, W.; Criscione, F.; Liang, S.; Tu, Z. MicroRNAs of two medically important mosquito species: Aedes aegypti and Anopheles stephensi. Insect Mol. Biol. 2015, 24, 240–252. [Google Scholar] [CrossRef] [Green Version]
- Potter, S.C.; Luciani, A.; Eddy, S.R.; Park, Y.; Lopez, R.; Finn, R.D. HMMER web server: 2018 update. Nucleic Acids Res. 2018, 46, W200–W204. [Google Scholar] [CrossRef] [Green Version]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular Evolutionary Genetics Analysis across Computing Platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Tamura, K. MEGA7: Molecular Evolutionary Genetics Analysis Version 7.0 for Bigger Datasets. Mol. Biol. Evol. 2016, 33, 1870–1874. [Google Scholar] [CrossRef] [Green Version]
- Bailey, T.L.; Boden, M.; Buske, F.A.; Frith, M.; Grant, C.E.; Clementi, L.; Ren, J.; Li, W.W.; Noble, W.S. MEME SUITE: Tools for motif discovery and searching. Nucleic Acids Res. 2009, 37, W202–W208. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.; Chen, H.; Zhang, Y.; Thomas, H.R.; Frank, M.H.; He, Y.; Xia, R. TBtools: An Integrative Toolkit Developed for Interactive Analyses of Big Biological Data. Mol. Plant 2020, 13, 1194–1202. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Tang, H.; Debarry, J.D.; Tan, X.; Li, J.; Wang, X.; Lee, T.H.; Jin, H.; Marler, B.; Guo, H.; et al. MCScanX: A toolkit for detection and evolutionary analysis of gene synteny and collinearity. Nucleic Acids Res. 2012, 40, e49. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shannon, P.; Markiel, A.; Ozier, O.; Baliga, N.S.; Wang, J.T.; Ramage, D.; Amin, N.; Schwikowski, B.; Ideker, T. Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Res. 2003, 13, 2498–2504. [Google Scholar] [CrossRef]
- Li, S.; Zhen, C.; Xu, W.; Wang, C.; Cheng, Y. Simple, rapid and efficient transformation of genotype Nisqually-1: A basic tool for the first sequenced model tree. Sci. Rep. 2017, 7, 2638. [Google Scholar] [CrossRef] [Green Version]
- Nemsa, I.; Hernández, M.A.; Lacasa, A.; Porras, I.; García-Lidón, A.; Cifuentes, D.; Bouzid, S.; Ortuño, A.; Del Río, J.A. Pathogenicity of Alternaria alternata on fruits and leaves of ‘Fortune’ mandarin (Citrus clementina × Citrus tangerina). Can. J. Plant Pathol. 2012, 34, 195–202. [Google Scholar] [CrossRef]
- Zhao, H.; Wang, S.; Chen, S.; Jiang, J.; Liu, G. Phylogenetic and stress-responsive expression analysis of 20 WRKY genes in Populus simonii × Populus nigra. Gene 2015, 565, 130–139. [Google Scholar] [CrossRef]
- Egusa, M.; Miwa, T.; Kaminaka, H.; Takano, Y.; Kodama, M. Nonhost resistance of Arabidopsis thaliana against Alternaria alternata involves both pre- and postinvasive defenses but is collapsed by AAL-toxin in the absence of LOH2. Phytopathology 2013, 103, 733–740. [Google Scholar] [CrossRef] [Green Version]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]



| Gene ID | Symbol | Length | Molecular Weight | Theoretical pI | Aliphatic Index | GRAVY | |
|---|---|---|---|---|---|---|---|
| Populus trichocarpa | Potri.T015518 | PtrSSL1b | 357 | 39,445.26 | 7.49 | 89.52 | −0.015 |
| Potri.017G027600 | PtrSSL12 | 409 | 45,895.72 | 6.15 | 86.63 | −0.238 | |
| Potri.016G037900 | PtrSSL9 | 377 | 41,011.22 | 7.09 | 96.4 | −0.032 | |
| Potri.016G037800 | PtrSSL11 | 98 | 10,878.46 | 5.5 | 102.65 | 0.019 | |
| Potri.016G037700 | PtrSSL6 | 410 | 45,566.97 | 7.12 | 87.29 | −0.162 | |
| Potri.015G037700 | PtrSSL7 | 335 | 36,270.25 | 9.14 | 91.37 | 0.018 | |
| Potri.012G046200 | PtrSSL3 | 315 | 34,126.71 | 9.4 | 86.1 | 0.037 | |
| Potri.008G109966 | PtrSSL1a | 357 | 39,445.26 | 7.49 | 89.52 | −0.015 | |
| Potri.007G130700 | PtrSSL8 | 406 | 45,861.89 | 6.68 | 89.26 | −0.193 | |
| Potri.006G140500 | PtrSSL10 | 365 | 40,733.89 | 5.73 | 96.93 | −0.044 | |
| Potri.006G040900 | PtrSSL2 | 388 | 42,747.89 | 9.38 | 90.49 | −0.172 | |
| Potri.005G099400 | PtrSSL4 | 570 | 62,550.88 | 6.11 | 97.3 | −0.015 | |
| Potri.001G214500 | PtrSSL5 | 391 | 43,983.39 | 6.48 | 84.5 | −0.267 | |
| Arabidopsis thaliana | AT5G22020 | AtSSL15 | 395 | 44,527.85 | 6.52 | 86.08 | −0.159 |
| AT3G59530 | AtSSL13 | 403 | 45,628.54 | 6.41 | 84.86 | −0.227 | |
| AT3G57030 | AtSSL10 | 374 | 41,001.16 | 7.71 | 95.67 | 0.009 | |
| AT3G57020 | AtSSL9 | 370 | 41,457.8 | 6.56 | 88.19 | −0.189 | |
| AT3G57010 | AtSSL8 | 376 | 41,980.07 | 5.78 | 83.62 | −0.164 | |
| AT3G51450 | AtSSL7 | 371 | 41,124.39 | 5.4 | 104.29 | 0.07 | |
| AT3G51440 | AtSSL6 | 371 | 41,383.49 | 5.83 | 96.68 | 0.011 | |
| AT3G51430 | AtSSL5 | 371 | 41,641.9 | 6.2 | 95.36 | −0.02 | |
| AT3G51420 | AtSSL4 | 370 | 41,595.67 | 5.65 | 96.16 | 0.054 | |
| AT2G41300 | AtSSL1 | 394 | 44,391.42 | 6.21 | 75.2 | −0.403 | |
| AT2G41290 | AtSSL2 | 376 | 41,475.26 | 6.2 | 83.96 | −0.145 | |
| AT1G74020 | AtSSL12 | 335 | 35,293.13 | 5.6 | 86.12 | 0.07 | |
| AT1G74010 | AtSSL14 | 325 | 34,184.1 | 8.27 | 86.98 | 0.155 | |
| AT1G74000 | AtSSL11 | 329 | 34,666.8 | 9.66 | 83.22 | 0.024 | |
| AT1G08470 | AtSSL3 | 390 | 44,036.55 | 8.11 | 84.95 | −0.239 | |
| Salix purpurea | Sapur.T191900 | SpuSSL6 | 304 | 33,213.88 | 8.98 | 93.98 | 0.118 |
| Sapur.T191200 | SpuSSL7 | 324 | 34,854.57 | 7.58 | 91.2 | 0.151 | |
| Sapur.T190300 | SpuSSL11 | 262 | 28,228.92 | 6.58 | 94.16 | 0.029 | |
| Sapur.15ZG033100 | SpuSSL13 | 324 | 34,902.61 | 7.58 | 90 | 0.148 | |
| Sapur.15WG054500 | SpuSSL14 | 324 | 35,060.81 | 6.34 | 90.59 | 0.141 | |
| Sapur.15WG054100 | SpuSSL15 | 286 | 31,575.3 | 9.16 | 96.43 | 0.173 | |
| Sapur.15WG052000 | SpuSSL16 | 324 | 34,932.64 | 7.58 | 90 | 0.135 | |
| Sapur.15WG051300 | SpuSSL17 | 288 | 30,907.06 | 7.58 | 87.05 | 0.115 | |
| Sapur.15WG051000 | SpuSSL18 | 335 | 36,203.24 | 8.62 | 91.43 | 0.139 | |
| Sapur.15WG050600 | SpuSSL3 | 162 | 17,593.19 | 4.69 | 88.02 | 0.349 | |
| Sapur.15WG050400 | SpuSSL19 | 324 | 34,978.71 | 7.58 | 90 | 0.158 | |
| Sapur.15WG050300 | SpuSSL20 | 200 | 22,144.36 | 9.8 | 94.65 | −0.148 | |
| Sapur.15WG049100 | SpuSSL21 | 262 | 28,296.15 | 8.76 | 96.76 | 0.061 | |
| Sapur.15WG048500 | SpuSSL22 | 271 | 29,352.2 | 6.58 | 93.54 | 0.029 | |
| Sapur.017G016600 | SpuSSL12 | 406 | 46,029.86 | 6.33 | 86.13 | −0.234 | |
| Sapur.016G034000 | SpuSSL9 | 374 | 40,947.06 | 6.52 | 97.46 | −0.034 | |
| Sapur.008G088100 | SpuSSL1 | 357 | 39,231.86 | 5.63 | 93.89 | −0.023 | |
| Sapur.007G116200 | SpuSSL8 | 406 | 45,795.78 | 6.65 | 90.44 | −0.166 | |
| Sapur.006G115900 | SpuSSL10 | 239 | 26,500.28 | 5.05 | 91.38 | −0.055 | |
| Sapur.006G030000 | SpuSSL2 | 340 | 37,236.23 | 8.16 | 80.65 | −0.222 | |
| Sapur.005G078900 | SpuSSL4 | 409 | 45,329.69 | 5.72 | 94.38 | −0.128 | |
| Sapur.004G165500 | SpuSSL5 | 391 | 43,758.23 | 6.56 | 85.5 | −0.204 |
| Duplicated Gene Pairs | Ka | Ks | Ka/Ks | The Length of Homologous Fragment (bp) | Homology/% | Duplication Date (MYA) |
|---|---|---|---|---|---|---|
| PtrSSL1a-PtrSSL1b | 0 | 0 | 0 | 1074 | 100 | 0 |
| PtrSSL2-PtrSSL6 | 0.09842 | 0.346721 | 0.283861 | 1233 | 86.93 | 11.55735 |
| PtrSSL3-PtrSSL7 | 0.187272 | 0.481373 | 0.389037 | 1008 | 80.65 | 16.04577 |
| PtrSSL8-PtrSSL12 | 0.032732 | 0.265984 | 0.123062 | 1221 | 92.38 | 8.866147 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, R.; Zhao, W.; Yao, W.; Wang, Y.; Jiang, T.; Liu, H. Genome-Wide Analysis of Strictosidine Synthase-like Gene Family Revealed Their Response to Biotic/Abiotic Stress in Poplar. Int. J. Mol. Sci. 2023, 24, 10117. https://doi.org/10.3390/ijms241210117
Wang R, Zhao W, Yao W, Wang Y, Jiang T, Liu H. Genome-Wide Analysis of Strictosidine Synthase-like Gene Family Revealed Their Response to Biotic/Abiotic Stress in Poplar. International Journal of Molecular Sciences. 2023; 24(12):10117. https://doi.org/10.3390/ijms241210117
Chicago/Turabian StyleWang, Ruiqi, Wenna Zhao, Wenjing Yao, Yuting Wang, Tingbo Jiang, and Huanzhen Liu. 2023. "Genome-Wide Analysis of Strictosidine Synthase-like Gene Family Revealed Their Response to Biotic/Abiotic Stress in Poplar" International Journal of Molecular Sciences 24, no. 12: 10117. https://doi.org/10.3390/ijms241210117
APA StyleWang, R., Zhao, W., Yao, W., Wang, Y., Jiang, T., & Liu, H. (2023). Genome-Wide Analysis of Strictosidine Synthase-like Gene Family Revealed Their Response to Biotic/Abiotic Stress in Poplar. International Journal of Molecular Sciences, 24(12), 10117. https://doi.org/10.3390/ijms241210117








