BSR1, a Rice Receptor-like Cytoplasmic Kinase, Positively Regulates Defense Responses to Herbivory
Abstract
1. Introduction
2. Results
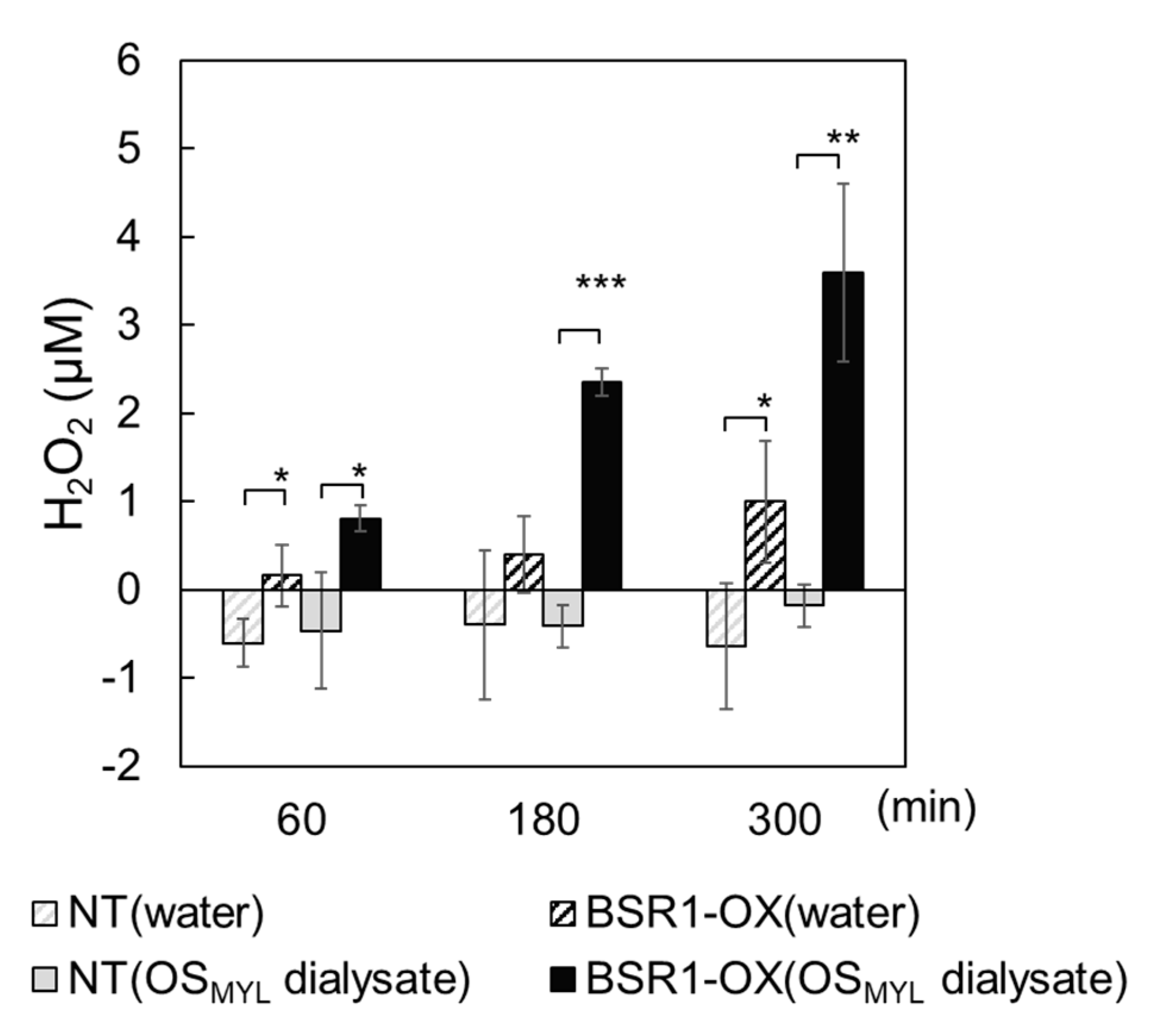
2.1. BSR1 Contributes to Defense Responses Triggered by Larval OS
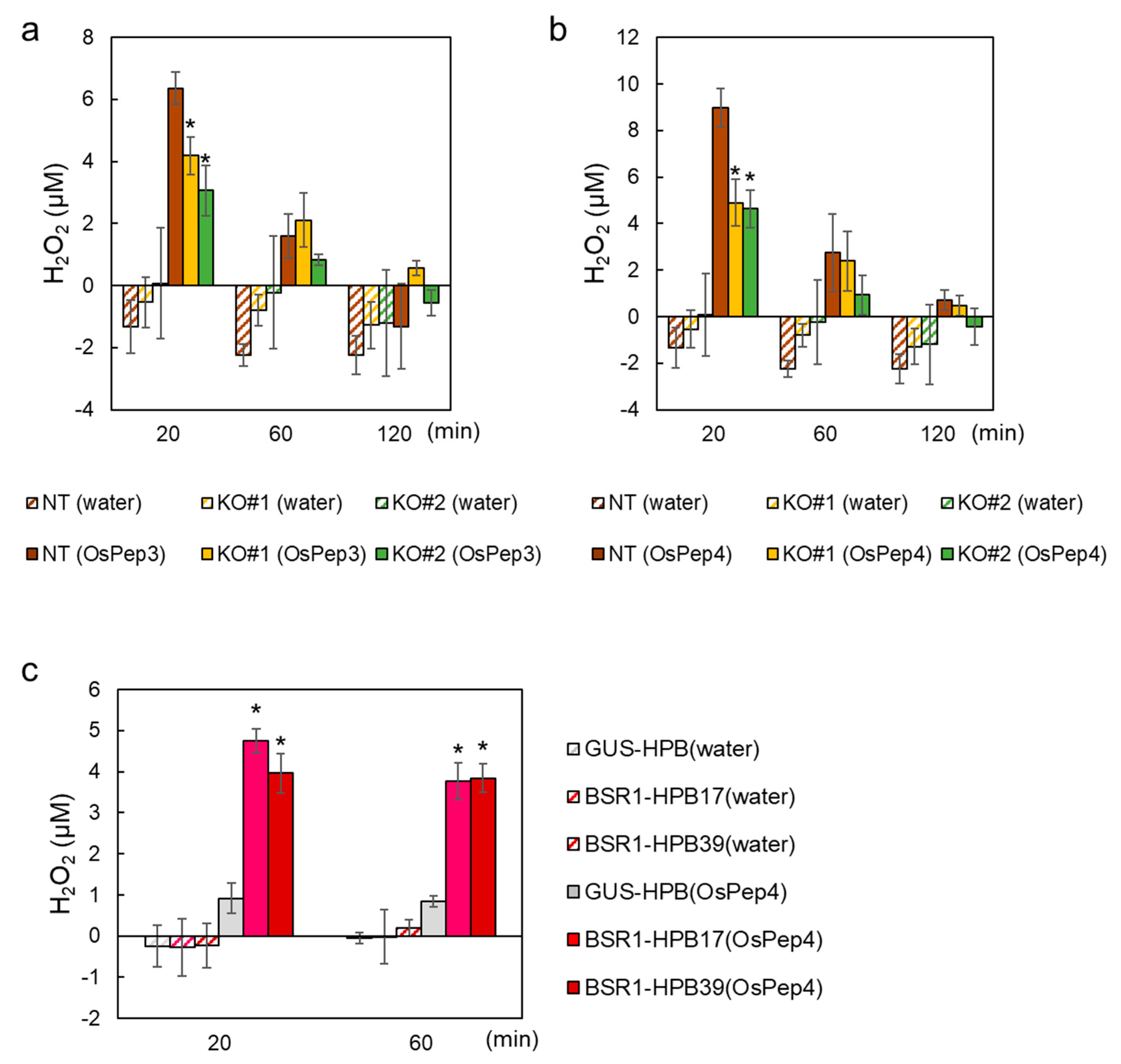
2.2. BSR1 Mediates the OsPep-Triggered Defense Response in Rice
2.3. Overexpression of BSR1 Enhances Phytoalexin Production
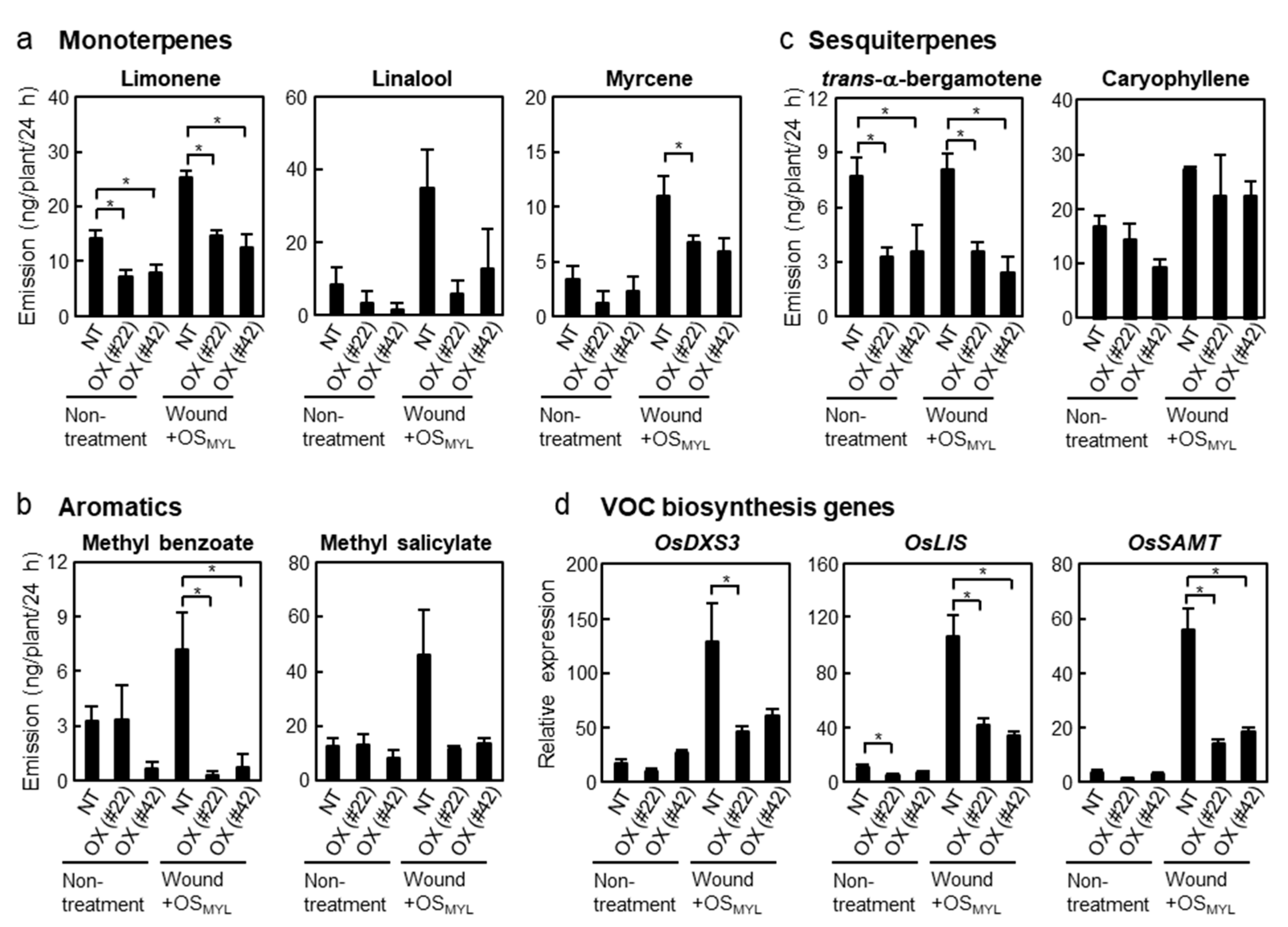
2.4. Overexpression of BSR1 Alters the Level of Volatile Organic Compounds
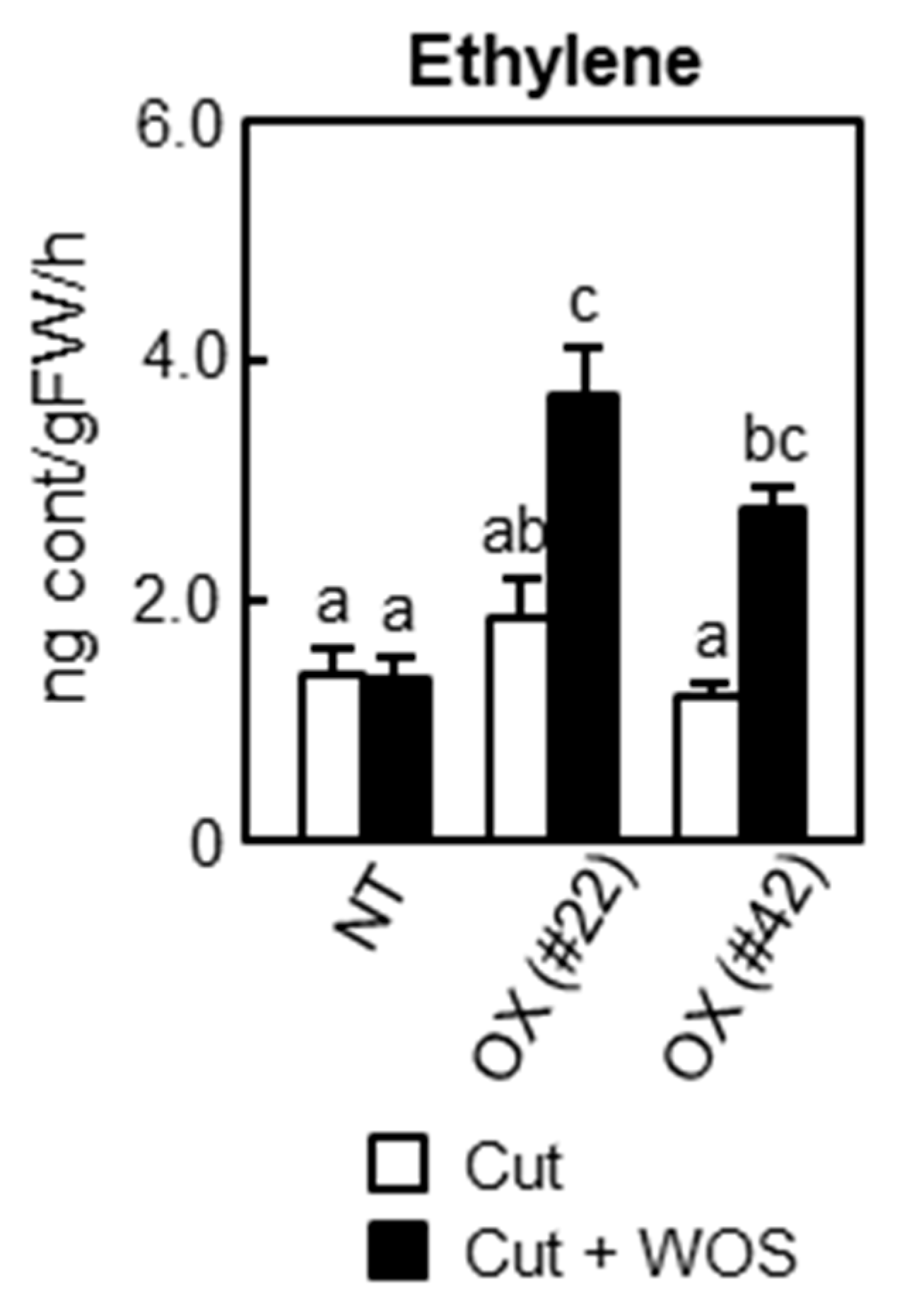
2.5. Defense-Related Gene Expression and Mechanical Defenses
2.6. Phytohormone Biosynthesis in BSR1-OX
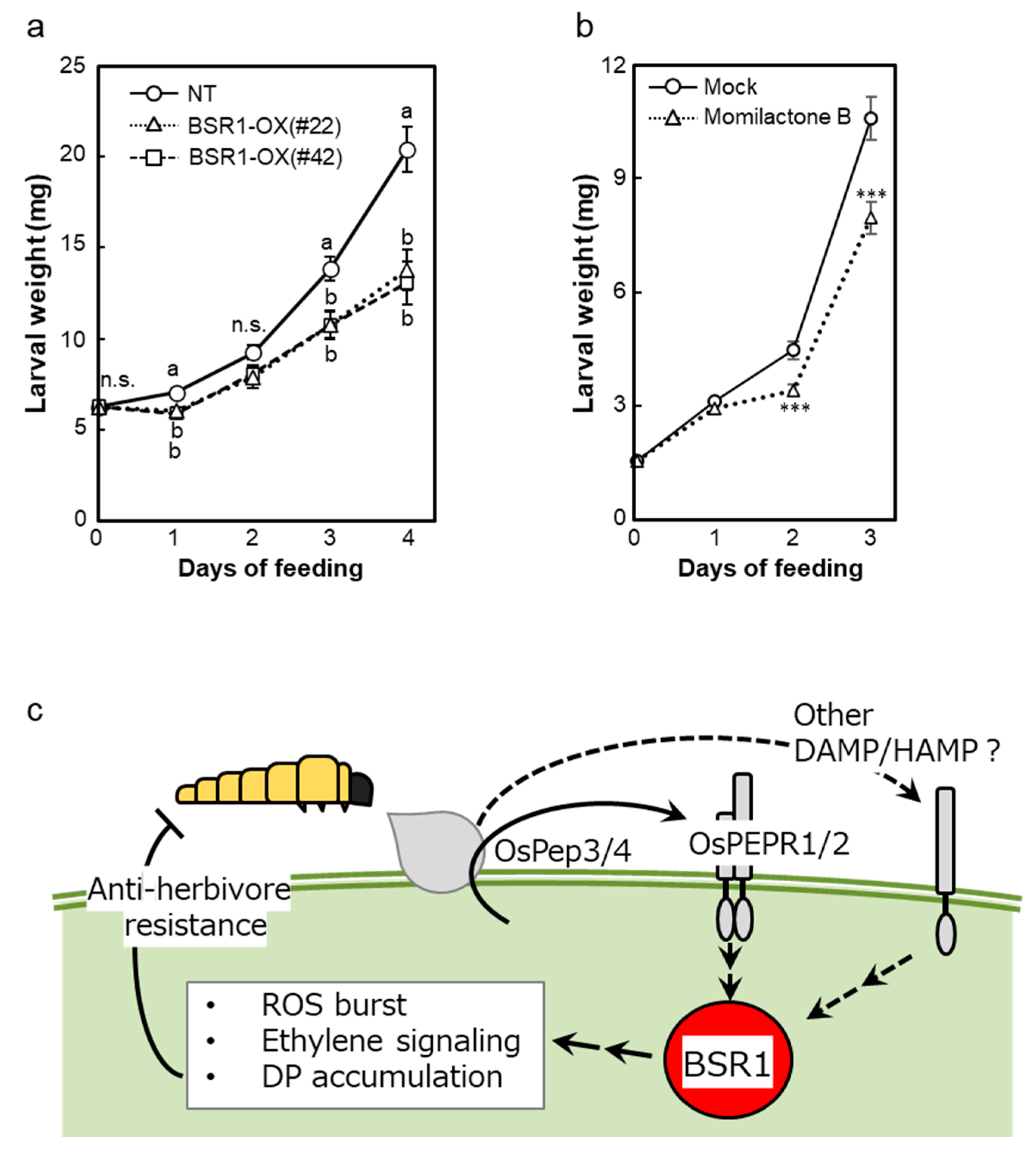
2.7. BSR1 Overexpression and Momilactone B Suppress the Larval Performance of M. loreyi
3. Discussion
4. Materials and Methods
4.1. Plant and Lepidopteran Materials
4.2. Collection of Oral Secretions
4.3. Preparation of Suspension-Cultured Cells and Leaf Strips
4.4. Measurement of H2O2
4.5. Reverse Transcription-Quantitative PCR
4.6. Treaements with Wounding and Oral Secretions as Simulated Herbivory
4.7. Herbivore Feeding Assay
4.8. Recovery and Analysis of Secondary Metabolites
4.9. Phytohormone Measurement
4.10. Scanning Electron Microscopy and Histochemical Staining
4.11. Statistical Data Analyses
4.12. Accession Numbers
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Oerke, E. Crop losses to pests. J. Agric. Sci. 2006, 144, 31–43. [Google Scholar] [CrossRef]
- Stokstad, E. New crop pest takes Africa at lightning speed. Science 2017, 356, 473–474. [Google Scholar] [CrossRef] [PubMed]
- Ogunwolu, E.; Reagan, T.; Flynn, J.; Hensley, S. Effects of Diatraea saccharalis (F.)(Lepidoptera: Pyralidae) damage and stalk rot fungi on sugarcane yield in Louisiana. Crop Protect. 1991, 10, 57–61. [Google Scholar] [CrossRef]
- Franco, F.P.; Tuler, A.C.; Gallan, D.Z.; Goncalves, F.G.; Favaris, A.P.; Penaflor, M.; Leal, W.S.; Moura, D.S.; Bento, J.M.S.; Silva-Filho, M.C. Fungal phytopathogen modulates plant and insect responses to promote its dissemination. ISME J 2021, 15, 3522–3533. [Google Scholar] [CrossRef] [PubMed]
- Wielkopolan, B.; Jakubowska, M.; Obrepalska-Steplowska, A. Beetles as Plant Pathogen Vectors. Front. Plant Sci. 2021, 12, 748093. [Google Scholar] [CrossRef]
- Boller, T.; Felix, G. A Renaissance of Elicitors: Perception of Microbe-Associated Molecular Patterns and Danger Signals by Pattern-Recognition Receptors. Annu. Rev. Plant Biol. 2009, 60, 379–406. [Google Scholar] [CrossRef]
- Saijo, Y.; Loo, E.; Yasuda, S. Pattern recognition receptors and signaling in plant-microbe interactions. Plant J. 2018, 93, 592–613. [Google Scholar] [CrossRef]
- Huffaker, A.; Pearce, G.; Ryan, C. An endogenous peptide signal in Arabidopsis activates components of the innate immune response. Proc. Natl. Acad. Sci. USA 2006, 103, 10098–10103. [Google Scholar] [CrossRef]
- Yamaguchi, Y.; Pearce, G.; Ryan, C.A. The cell surface leucine-rich repeat receptor for At Pep1, an endogenous peptide elicitor in Arabidopsis, is functional in transgenic tobacco cells. Proc. Natl. Acad. Sci. USA 2006, 103, 10104–10109. [Google Scholar] [CrossRef]
- Bartels, S.; Boller, T. Quo vadis, Pep? Plant elicitor peptides at the crossroads of immunity, stress, and development. J. Exp. Bot. 2015, 66, 5183–5193. [Google Scholar] [CrossRef]
- Huffaker, A.; Pearce, G.; Veyrat, N.; Erb, M.; Turlings, T.; Sartor, R.; Shen, Z.; Briggs, S.; Vaughan, M.; Alborn, H.; et al. Plant elicitor peptides are conserved signals regulating direct and indirect antiherbivore defense. Proc. Natl. Acad. Sci. USA 2013, 110, 5707–5712. [Google Scholar] [CrossRef] [PubMed]
- Shinya, T.; Yasuda, S.; Hyodo, K.; Tani, R.; Hojo, Y.; Fujiwara, Y.; Hiruma, K.; Ishizaki, T.; Fujita, Y.; Saijo, Y.; et al. Integration of danger peptide signals with herbivore-associated molecular pattern signaling amplifies anti-herbivore defense responses in rice. Plant J. 2018, 94, 626–637. [Google Scholar] [CrossRef] [PubMed]
- Shen, W.; Zhang, X.; Liu, J.; Tao, K.; Li, C.; Xiao, S.; Zhang, W.; Li, J.F. Plant elicitor peptide signalling confers rice resistance to piercing-sucking insect herbivores and pathogens. Plant Biotechnol. J. 2022, 20, 991–1005. [Google Scholar] [CrossRef] [PubMed]
- Macho, A.; Zipfel, C. Plant PRRs and the Activation of Innate Immune Signaling. Mol. Cell 2014, 54, 263–272. [Google Scholar] [CrossRef]
- Schmelz, E.; Huffaker, A.; Sims, J.; Christensen, S.; Lu, X.; Okada, K.; Peters, R. Biosynthesis, elicitation and roles of monocot terpenoid phytoalexins. Plant J. 2014, 79, 659–678. [Google Scholar] [CrossRef]
- Stahl, E.; Hilfiker, O.; Reymond, P. Plant-arthropod interactions: Who is the winner? Plant J. 2018, 93, 703–728. [Google Scholar] [CrossRef]
- Erb, M.; Reymond, P. Molecular Interactions Between Plants and Insect Herbivores. Annu. Rev. Plant Biol. 2019, 70, 527–557. [Google Scholar] [CrossRef]
- Reymond, P. Receptor kinases in plant responses to herbivory. Curr. Opin. Biotechnol. 2021, 70, 143–150. [Google Scholar] [CrossRef]
- Snoeck, S.; Guayazan-Palacios, N.; Steinbrenner, A. Molecular tug-of-war: Plant immune recognition of herbivory. Plant Cell 2022, 34, 1497–1513. [Google Scholar] [CrossRef]
- Steinbrenner, A.; Munoz-Amatriain, M.; Chaparro, A.; Aguilar-Venegas, J.; Lo, S.; Okuda, S.; Glauser, G.; Dongiovanni, J.; Shi, D.; Hall, M.; et al. A receptor-like protein mediates plant immune responses to herbivore-associated molecular patterns. Proc. Natl. Acad. Sci. USA 2020, 117, 31510–31518. [Google Scholar] [CrossRef]
- Hu, L.; Ye, M.; Kuai, P.; Ye, M.; Erb, M.; Lou, Y. OsLRR-RLK1, an early responsive leucine-rich repeat receptor-like kinase, initiates rice defense responses against a chewing herbivore. New Phytol. 2018, 219, 1097–1111. [Google Scholar] [CrossRef] [PubMed]
- Uemura, T.; Hachisu, M.; Desaki, Y.; Ito, A.; Hoshino, R.; Sano, Y.; Nozawa, A.; Mujiono, K.; Galis, I.; Yoshida, A.; et al. Soy and Arabidopsis receptor-like kinases respond to polysaccharide signals from Spodoptera species and mediate herbivore resistance. Commun. Biol. 2020, 3, 224. [Google Scholar] [CrossRef] [PubMed]
- Shiu, S.; Karlowski, W.; Pan, R.; Tzeng, Y.; Mayer, K.; Li, W. Comparative analysis of the receptor-like kinase family in Arabidopsis and rice. Plant Cell 2004, 16, 1220–1234. [Google Scholar] [CrossRef] [PubMed]
- Vij, S.; Giri, J.; Dansana, P.; Kapoor, S.; Tyagi, A. The receptor-like cytoplasmic kinase (OsRLCK) gene family in rice: Organization, phylogenetic relationship, and expression during development and stress. Mol. Plant 2008, 1, 732–750. [Google Scholar] [CrossRef]
- Liang, X.; Zhou, J.M. Receptor-Like Cytoplasmic Kinases: Central Players in Plant Receptor Kinase-Mediated Signaling. Annu. Rev. Plant Biol. 2018, 69, 267–299. [Google Scholar] [CrossRef]
- Yamada, K.; Yamaguchi, K.; Shirakawa, T.; Nakagami, H.; Mine, A.; Ishikawa, K.; Fujiwara, M.; Narusaka, M.; Narusaka, Y.; Ichimura, K.; et al. The Arabidopsis CERK1-associated kinase PBL27 connects chitin perception to MAPK activation. EMBO J. 2016, 35, 2468–2483. [Google Scholar] [CrossRef]
- Wang, C.; Wang, G.; Zhang, C.; Zhu, P.; Dai, H.; Yu, N.; He, Z.; Xu, L.; Wang, E. OsCERK1-Mediated Chitin Perception and Immune Signaling Requires Receptor-like Cytoplasmic Kinase 185 to Activate an MAPK Cascade in Rice. Mol. Plant 2017, 10, 619–633. [Google Scholar] [CrossRef]
- Yamada, K.; Yamaguchi, K.; Yoshimura, S.; Terauchi, A.; Kawasaki, T. Conservation of Chitin-Induced MAPK Signaling Pathways in Rice and Arabidopsis. Plant Cell Physiol. 2017, 58, 993–1002. [Google Scholar] [CrossRef]
- Kadota, Y.; Sklenar, J.; Derbyshire, P.; Stransfeld, L.; Asai, S.; Ntoukakis, V.; Jones, J.; Shirasu, K.; Menke, F.; Jones, A.; et al. Direct Regulation of the NADPH Oxidase RBOHD by the PRR-Associated Kinase BIK1 during Plant Immunity. Mol. Cell 2014, 54, 43–55. [Google Scholar] [CrossRef]
- Li, L.; Li, M.; Yu, L.; Zhou, Z.; Liang, X.; Liu, Z.; Cai, G.; Gao, L.; Zhang, X.; Wang, Y.; et al. The FLS2-Associated Kinase BIK1 Directly Phosphorylates the NADPH Oxidase RbohD to Control Plant Immunity. Cell Host Microbe 2014, 15, 329–338. [Google Scholar] [CrossRef]
- Fan, J.; Bai, P.; Ning, Y.; Wang, J.; Shi, X.; Xiong, Y.; Zhang, K.; He, F.; Zhang, C.; Wang, R.; et al. The Monocot-Specific Receptor-like Kinase SDS2 Controls Cell Death and Immunity in Rice. Cell Host Microbe 2018, 23, 498–510. [Google Scholar] [CrossRef] [PubMed]
- Dubouzet, J.G.; Maeda, S.; Sugano, S.; Ohtake, M.; Hayashi, N.; Ichikawa, T.; Kondou, Y.; Kuroda, H.; Horii, Y.; Matsui, M.; et al. Screening for resistance against Pseudomonas syringae in rice-FOX Arabidopsis lines identified a putative receptor-like cytoplasmic kinase gene that confers resistance to major bacterial and fungal pathogens in Arabidopsis and rice. Plant Biotechnol. J. 2011, 9, 466–485. [Google Scholar] [CrossRef] [PubMed]
- Sugano, S.; Maeda, S.; Hayashi, N.; Kajiwara, H.; Inoue, H.; Jiang, C.; Takatsuji, H.; Mori, M. Tyrosine phosphorylation of a receptor-like cytoplasmic kinase, BSR1, plays a crucial role in resistance to multiple pathogens in rice. Plant J. 2018, 96, 1137–1147. [Google Scholar] [CrossRef]
- Kanda, Y.; Yokotani, N.; Maeda, S.; Nishizawa, Y.; Kamakura, T.; Mori, M. The receptor-like cytoplasmic kinase BSR1 mediates chitin-induced defense signaling in rice cells. Biosci. Biotechnol. Biochem. 2017, 81, 1497–1502. [Google Scholar] [CrossRef] [PubMed]
- Kanda, Y.; Nakagawa, H.; Nishizawa, Y.; Kamakura, T.; Mori, M. Broad-Spectrum Disease Resistance Conferred by the Overexpression of Rice RLCK BSR1 Results from an Enhanced Immune Response to Multiple MAMPs. Int. J. Mol. Sci. 2019, 20, 5523. [Google Scholar] [CrossRef] [PubMed]
- Maeda, S.; Hayashi, N.; Sasaya, T.; Mori, M. Overexpression of BSR1 confers broad-spectrum resistance against two bacterial diseases and two major fungal diseases in rice. Breed. Sci. 2016, 66, 396–406. [Google Scholar] [CrossRef]
- Maeda, S.; Ackley, W.; Yokotani, N.; Sasaki, K.; Ohtsubo, N.; Oda, K.; Mori, M. Enhanced Resistance to Fungal and Bacterial Diseases Due to Overexpression of BSR1, a Rice RLCK, in Sugarcane, Tomato, and Torenia. Int. J. Mol. Sci. 2023, 24, 3644. [Google Scholar] [CrossRef]
- Arimura, G.I. Making Sense of the Way Plants Sense Herbivores. Trends Plant Sci. 2021, 26, 288–298. [Google Scholar] [CrossRef]
- Shinya, T.; Hojo, Y.; Desaki, Y.; Christeller, J.; Okada, K.; Shibuya, N.; Galis, I. Modulation of plant defense responses to herbivores by simultaneous recognition of different herbivore-associated elicitors in rice. Sci. Rep. 2016, 6, 32537. [Google Scholar] [CrossRef]
- Valea, I.; Motegi, A.; Kawamura, N.; Kawamoto, K.; Miyao, A.; Ozawa, R.; Takabayashi, J.; Gomi, K.; Nemoto, K.; Nozawa, A.; et al. The rice wound-inducible transcription factor RERJ1 sharing same signal transduction pathway with OsMYC2 is necessary for defense response to herbivory and bacterial blight. Plant Mol. Biol. 2022, 109, 651–666. [Google Scholar] [CrossRef]
- Andama, J.; Mujiono, K.; Hojo, Y.; Shinya, T.; Galis, I. Nonglandular silicified trichomes are essential for rice defense against chewing herbivores. Plant Cell Environ. 2020, 43, 2019–2032. [Google Scholar] [CrossRef] [PubMed]
- Mujiono, K.; Tohi, T.; Sobhy, I.; Hojo, Y.; Ho, N.; Shinya, T.; Galis, I. Ethylene functions as a suppressor of volatile production in rice. J. Exp. Bot. 2020, 71, 6491–6511. [Google Scholar] [CrossRef] [PubMed]
- Alamgir, K.; Hojo, Y.; Christeller, J.; Fukumoto, K.; Isshiki, R.; Shinya, T.; Baldwin, I.; Galis, I. Systematic analysis of rice (Oryza sativa) metabolic responses to herbivory. Plant Cell Environ. 2016, 39, 453–466. [Google Scholar] [CrossRef] [PubMed]
- Kanno, H.; Hasegawa, M.; Kodama, O. Accumulation of salicylic acid, jasmonic acid and phytoalexins in rice, Oryza sativa, infested by the white-backed planthopper, Sogatella furcifera (Hemiptera: Delphacidae). Appl. Entomol. Zool. 2012, 47, 27–34. [Google Scholar] [CrossRef]
- Zhang, J.; Li, W.; Xiang, T.; Liu, Z.; Laluk, K.; Ding, X.; Zou, Y.; Gao, M.; Zhang, X.; Chen, S.; et al. Receptor-like Cytoplasmic Kinases Integrate Signaling from Multiple Plant Immune Receptors and Are Targeted by a Pseudomonas syringae Effector. Cell Host Microbe 2010, 7, 290–301. [Google Scholar] [CrossRef] [PubMed]
- Liu, Z.; Wu, Y.; Yang, F.; Zhang, Y.; Chen, S.; Xie, Q.; Tian, X.; Zhou, J. BIK1 interacts with PEPRs to mediate ethylene-induced immunity. Proc. Natl. Acad. Sci. USA 2013, 110, 6205–6210. [Google Scholar] [CrossRef]
- Klauser, D.; Desurmont, G.; Glauser, G.; Vallat, A.; Flury, P.; Boller, T.; Turlings, T.; Bartels, S. The Arabidopsis Pep-PEPR system is induced by herbivore feeding and contributes to JA-mediated plant defence against herbivory. J. Exp. Bot. 2015, 66, 5327–5336. [Google Scholar] [CrossRef]
- Mujiono, K.; Tohi, T.; Sobhy, I.; Hojo, Y.; Shinya, T.; Galis, I. Herbivore-induced and constitutive volatiles are controlled by different oxylipin-dependent mechanisms in rice. Plant Cell Environ. 2021, 44, 2687–2699. [Google Scholar] [CrossRef]
- von Dahl, C.; Winz, R.; Halitschke, R.; Kuhnemann, F.; Gase, K.; Baldwin, I. Tuning the herbivore-induced ethylene burst: The role of transcript accumulation and ethylene perception in Nicotiana attenuata. Plant J. 2007, 51, 293–307. [Google Scholar] [CrossRef]
- Schmelz, E.; Engelberth, J.; Alborn, H.; Tumlinson, J.; Teal, P. Phytohormone-based activity mapping of insect herbivore-produced elicitors. Proc. Natl. Acad. Sci. USA 2009, 106, 653–657. [Google Scholar] [CrossRef]
- Diezel, C.; Allmann, S.; Baldwin, I.T. Mechanisms of optimal defense patterns in Nicotiana attenuata: Flowering attenuates herbivory-elicited ethylene and jasmonate signaling. J. Integr. Plant Biol. 2011, 53, 971–983. [Google Scholar] [CrossRef] [PubMed]
- Mewis, I.; Appel, H.; Hom, A.; Raina, R.; Schultz, J. Major signaling pathways modulate Arabidopsis glucosinolate accumulation and response to both phloem-feeding and chewing insects. Plant Physiol. 2005, 138, 1149–1162. [Google Scholar] [CrossRef] [PubMed]
- Bodenhausen, N.; Reymond, P. Signaling pathways controlling induced resistance to insect herbivores in Arabidopsis. Mol. Plant-Microbe Interact. 2007, 20, 1406–1420. [Google Scholar] [CrossRef]
- Harfouche, A.; Shivaji, R.; Stocker, R.; Williams, P.; Luthe, D. Ethylene signaling mediates a maize defense response to insect herbivory. Mol. Plant-Microbe Interact. 2006, 19, 189–199. [Google Scholar] [CrossRef]
- Lu, J.; Li, J.; Ju, H.; Liu, X.; Erb, M.; Wang, X.; Lou, Y. Contrasting Effects of Ethylene Biosynthesis on Induced Plant Resistance against a Chewing and a Piercing-Sucking Herbivore in Rice. Mol. Plant 2014, 7, 1670–1682. [Google Scholar] [CrossRef] [PubMed]
- Tanabe, K.; Hojo, Y.; Shinya, T.; Galis, I. Molecular evidence for biochemical diversification of phenolamide biosynthesis in rice plants. J. Integr. Plant Biol. 2016, 58, 903–913. [Google Scholar] [CrossRef] [PubMed]
- Lu, X.; Zhang, J.; Brown, B.; Li, R.; Rodriguez-Romero, J.; Berasategui, A.; Liu, B.; Xu, M.; Luo, D.; Pan, Z.; et al. Inferring Roles in Defense from Metabolic Allocation of Rice Diterpenoids([OPEN]). Plant Cell 2018, 30, 1119–1131. [Google Scholar] [CrossRef]
- Zhang, J.; Li, R.; Xu, M.; Hoffmann, R.; Zhang, Y.; Liu, B.; Zhang, M.; Yang, B.; Li, Z.; Peters, R. A (conditional) role for labdane-related diterpenoid natural products in rice stomatal closure. New Phytol. 2021, 230, 698–709. [Google Scholar] [CrossRef]
- Desmedt, W.; Kudjordjie, E.N.; Chavan, S.N.; Zhang, J.; Li, R.; Yang, B.; Nicolaisen, M.; Mori, M.; Peters, R.J.; Vanholme, B.; et al. Rice diterpenoid phytoalexins are involved in defence against parasitic nematodes and shape rhizosphere nematode communities. New Phytol. 2022, 235, 1231–1245. [Google Scholar] [CrossRef]
- Yamamura, C.; Mizutani, E.; Okada, K.; Nakagawa, H.; Fukushima, S.; Tanaka, A.; Maeda, S.; Kamakura, T.; Yamane, H.; Takatsuji, H.; et al. Diterpenoid phytoalexin factor, a bHLH transcription factor, plays a central role in the biosynthesis of diterpenoid phytoalexins in rice. Plant J. 2015, 84, 1100–1113. [Google Scholar] [CrossRef]
- Kim, S.; Park, H.; Park, E.; Lee, S. Cytotoxic and antitumor activity of momilactone B from rice hulls. J. Agric. Food Chem. 2007, 55, 1702–1706. [Google Scholar] [CrossRef]
- Park, C.; Jeong, N.Y.; Kim, G.-Y.; Han, M.H.; Chung, I.-M.; Kim, W.-J.; Yoo, Y.H.; Choi, Y.H. Momilactone B induces apoptosis and G1 arrest of the cell cycle in human monocytic leukemia U937 cells through downregulation of pRB phosphorylation and induction of the cyclin-dependent kinase inhibitor p21Waf1/Cip1. Oncol. Rep. 2014, 31, 1653–1660. [Google Scholar] [CrossRef] [PubMed]
- Schmelz, E.A.; Huffaker, A.; Carroll, M.J.; Alborn, H.T.; Ali, J.G.; Teal, P.E. An amino acid substitution inhibits specialist herbivore production of an antagonist effector and recovers insect-induced plant defenses. Plant Physiol. 2012, 160, 1468–1478. [Google Scholar] [CrossRef] [PubMed]
- Ye, W.; Yu, H.; Jian, Y.; Zeng, J.; Ji, R.; Chen, H.; Lou, Y. A salivary EF-hand calcium-binding protein of the brown planthopper Nilaparvata lugens functions as an effector for defense responses in rice. Sci. Rep. 2017, 7, 40498. [Google Scholar] [CrossRef] [PubMed]
- Qi, Y.; Katagiri, F. Purification of low-abundance Arabidopsis plasma-membrane protein complexes and identification of candidate components. Plant J. 2009, 57, 932–944. [Google Scholar] [CrossRef]
- Goto, S.; Sasakura-Shimoda, F.; Suetsugu, M.; Selvaraj, M.G.; Hayashi, N.; Yamazaki, M.; Ishitani, M.; Shimono, M.; Sugano, S.; Matsushita, A.; et al. Development of disease-resistant rice by optimized expression of WRKY45. Plant Biotechnol. J. 2015, 13, 753–765. [Google Scholar] [CrossRef]
- Goto, S.; Sasakura-Shimoda, F.; Yamazaki, M.; Hayashi, N.; Suetsugu, M.; Ochiai, H.; Takatsuji, H. Development of disease-resistant rice by pathogen-responsive expression of WRKY45. Plant Biotechnol. J. 2016, 14, 1127–1138. [Google Scholar] [CrossRef]
- Toki, S.; Hara, N.; Ono, K.; Onodera, H.; Tagiri, A.; Oka, S.; Tanaka, H. Early infection of scutellum tissue with Agrobacterium allows high-speed transformation of rice. Plant J. 2006, 47, 969–976. [Google Scholar] [CrossRef]
- Kouzai, Y.; Nakajima, K.; Hayafune, M.; Ozawa, K.; Kaku, H.; Shibuya, N.; Minami, E.; Nishizawa, Y. CEBiP is the major chitin oligomer-binding protein in rice and plays a main role in the perception of chitin oligomers. Plant Mol. Biol. 2014, 84, 519–528. [Google Scholar] [CrossRef]
- Desaki, Y.; Shimada, H.; Takahashi, S.; Sakurayama, C.; Kawai, M.; Kaku, H.; Shibuya, N. Handmade leaf cutter for efficient and reliable ROS assay. Plant Biotechnol. 2019, 36, 275–278. [Google Scholar] [CrossRef]
- Kanda, Y.; Nishizawa, Y.; Kamakura, T.; Mori, M. Overexpressed BSR1-Mediated Enhancement of Disease Resistance Depends on the MAMP-Recognition System. Int. J. Mol. Sci. 2020, 21, 5397. [Google Scholar] [CrossRef] [PubMed]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]
- Jiang, C.; Shimono, M.; Sugano, S.; Kojima, M.; Yazawa, K.; Yoshida, R.; Inoue, H.; Hayashi, N.; Sakakibara, H.; Takatsuji, H. Abscisic Acid Interacts Antagonistically with Salicylic Acid Signaling Pathway in Rice-Magnaporthe grisea Interaction. Mol. Plant-Microbe Interact. 2010, 23, 791–798. [Google Scholar] [CrossRef] [PubMed]
- Fukumoto, K.; Alamgir, K.; Yamashita, Y.; Mori, I.; Matsuura, H.; Galis, I. Response of Rice to Insect Elicitors and the Role of OsJAR1 in Wound and Herbivory-Induced JA-Ile Accumulation. J. Integr. Plant Biol. 2013, 55, 775–784. [Google Scholar] [CrossRef] [PubMed]
- Gulzar, A.; Wright, D.J. Sub-lethal effects of Vip3A toxin on survival, development and fecundity of Heliothis virescens and Plutella xylostella. Ecotoxicology 2015, 24, 1815–1822. [Google Scholar] [CrossRef]
- Tomita, K.; Yashiroda, Y.; Matsuo, Y.; Piotrowski, J.; Li, S.; Okamoto, R.; Yoshimura, M.; Kimura, H.; Kawamura, Y.; Kawamukai, M.; et al. Genome-wide screening of genes associated with momilactone B sensitivity in the fission yeast Schizosaccharomyces pombe. G3-Genes Genomes Genet. 2021, 11, jkab156. [Google Scholar] [CrossRef]
- Wari, D.; Kabir, M.A.; Mujiono, K.; Hojo, Y.; Shinya, T.; Tani, A.; Nakatani, H.; Galis, I. Honeydew-associated microbes elicit defense responses against brown planthopper in rice. J. Exp. Bot. 2019, 70, 1683–1696. [Google Scholar] [CrossRef]
- Sobhy, I.; Miyake, A.; Shinya, T.; Galis, I. Oral Secretions Affect HIPVs Induced by Generalist (Mythimna loreyi) and Specialist (Parnara guttata) Herbivores in Rice. J. Chem. Ecol. 2017, 43, 929–943. [Google Scholar] [CrossRef]
- Ambavaram, M.; Krishnan, A.; Trijatmiko, K.; Pereira, A. Coordinated Activation of Cellulose and Repression of Lignin Biosynthesis Pathways in Rice. Plant Physiol. 2011, 155, 916–931. [Google Scholar] [CrossRef]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kanda, Y.; Shinya, T.; Maeda, S.; Mujiono, K.; Hojo, Y.; Tomita, K.; Okada, K.; Kamakura, T.; Galis, I.; Mori, M. BSR1, a Rice Receptor-like Cytoplasmic Kinase, Positively Regulates Defense Responses to Herbivory. Int. J. Mol. Sci. 2023, 24, 10395. https://doi.org/10.3390/ijms241210395
Kanda Y, Shinya T, Maeda S, Mujiono K, Hojo Y, Tomita K, Okada K, Kamakura T, Galis I, Mori M. BSR1, a Rice Receptor-like Cytoplasmic Kinase, Positively Regulates Defense Responses to Herbivory. International Journal of Molecular Sciences. 2023; 24(12):10395. https://doi.org/10.3390/ijms241210395
Chicago/Turabian StyleKanda, Yasukazu, Tomonori Shinya, Satoru Maeda, Kadis Mujiono, Yuko Hojo, Keisuke Tomita, Kazunori Okada, Takashi Kamakura, Ivan Galis, and Masaki Mori. 2023. "BSR1, a Rice Receptor-like Cytoplasmic Kinase, Positively Regulates Defense Responses to Herbivory" International Journal of Molecular Sciences 24, no. 12: 10395. https://doi.org/10.3390/ijms241210395
APA StyleKanda, Y., Shinya, T., Maeda, S., Mujiono, K., Hojo, Y., Tomita, K., Okada, K., Kamakura, T., Galis, I., & Mori, M. (2023). BSR1, a Rice Receptor-like Cytoplasmic Kinase, Positively Regulates Defense Responses to Herbivory. International Journal of Molecular Sciences, 24(12), 10395. https://doi.org/10.3390/ijms241210395






