Heterologous DNA Prime- Subunit Protein Boost with Chikungunya Virus E2 Induces Neutralizing Antibodies and Cellular-Mediated Immunity
Abstract
1. Introduction
2. Results
2.1. Characterization of E2-Based Vaccines
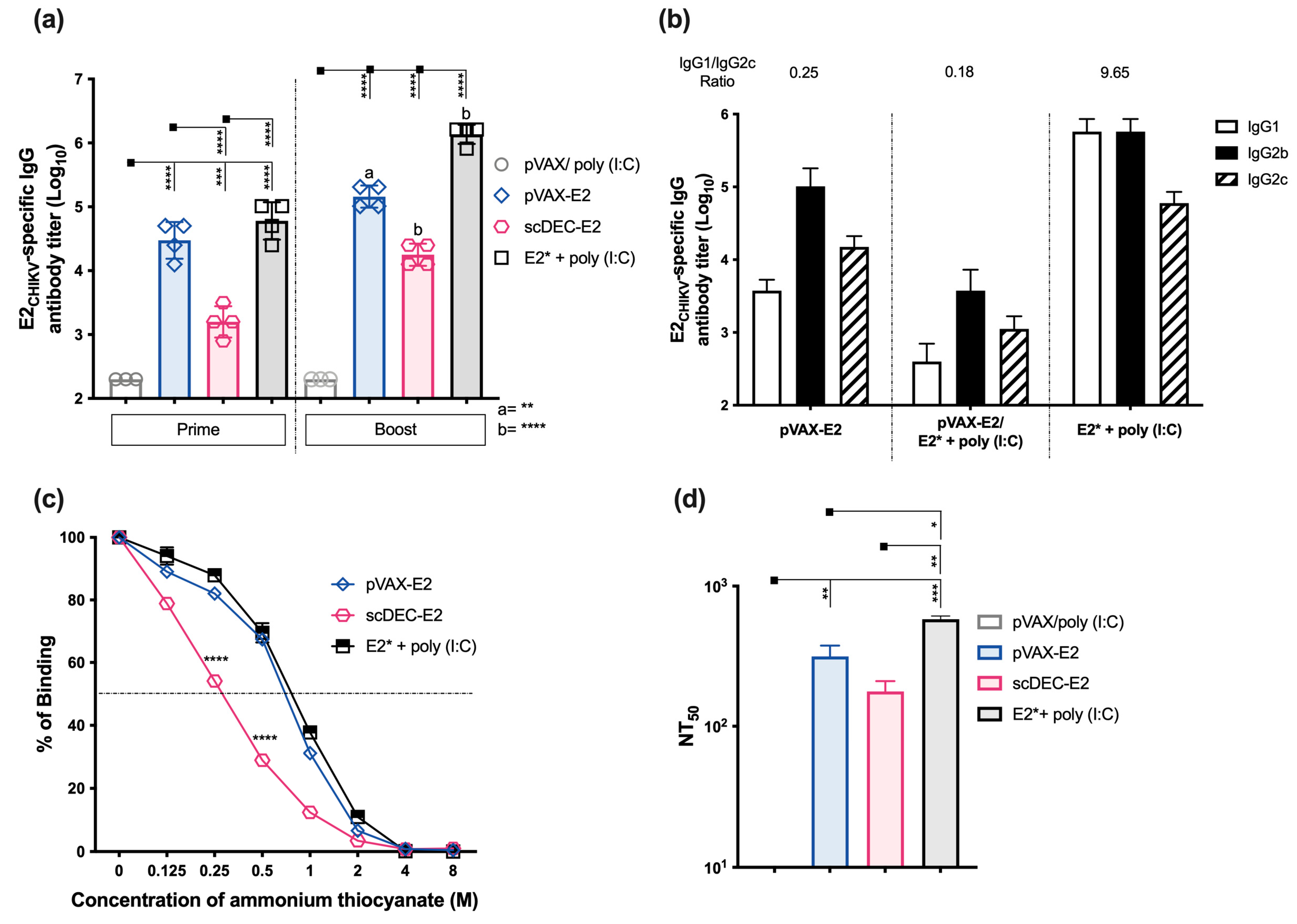
2.2. Immunization with E2CHIKV-Based Vaccines Induces Robust Specific Humoral Responses
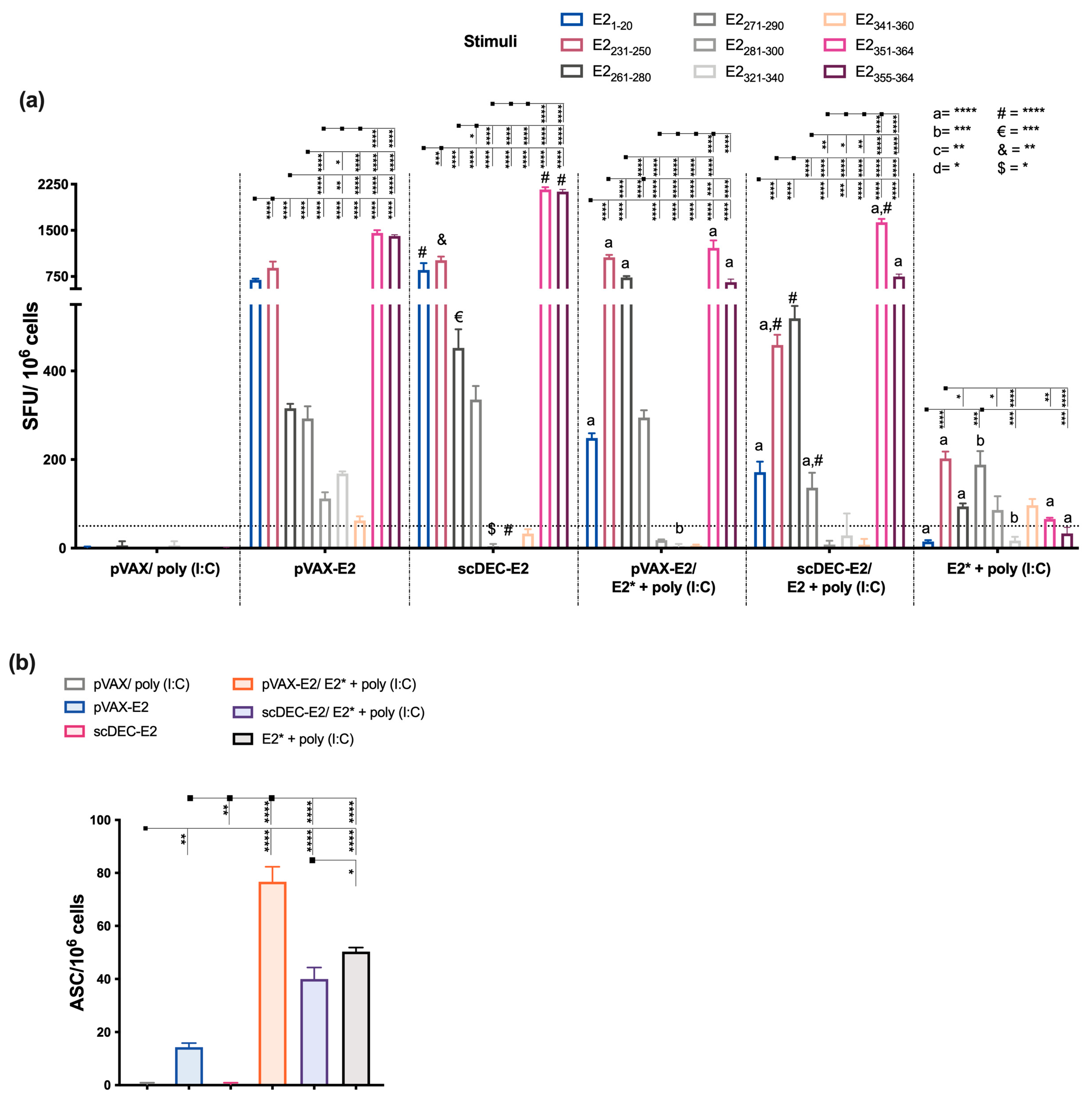
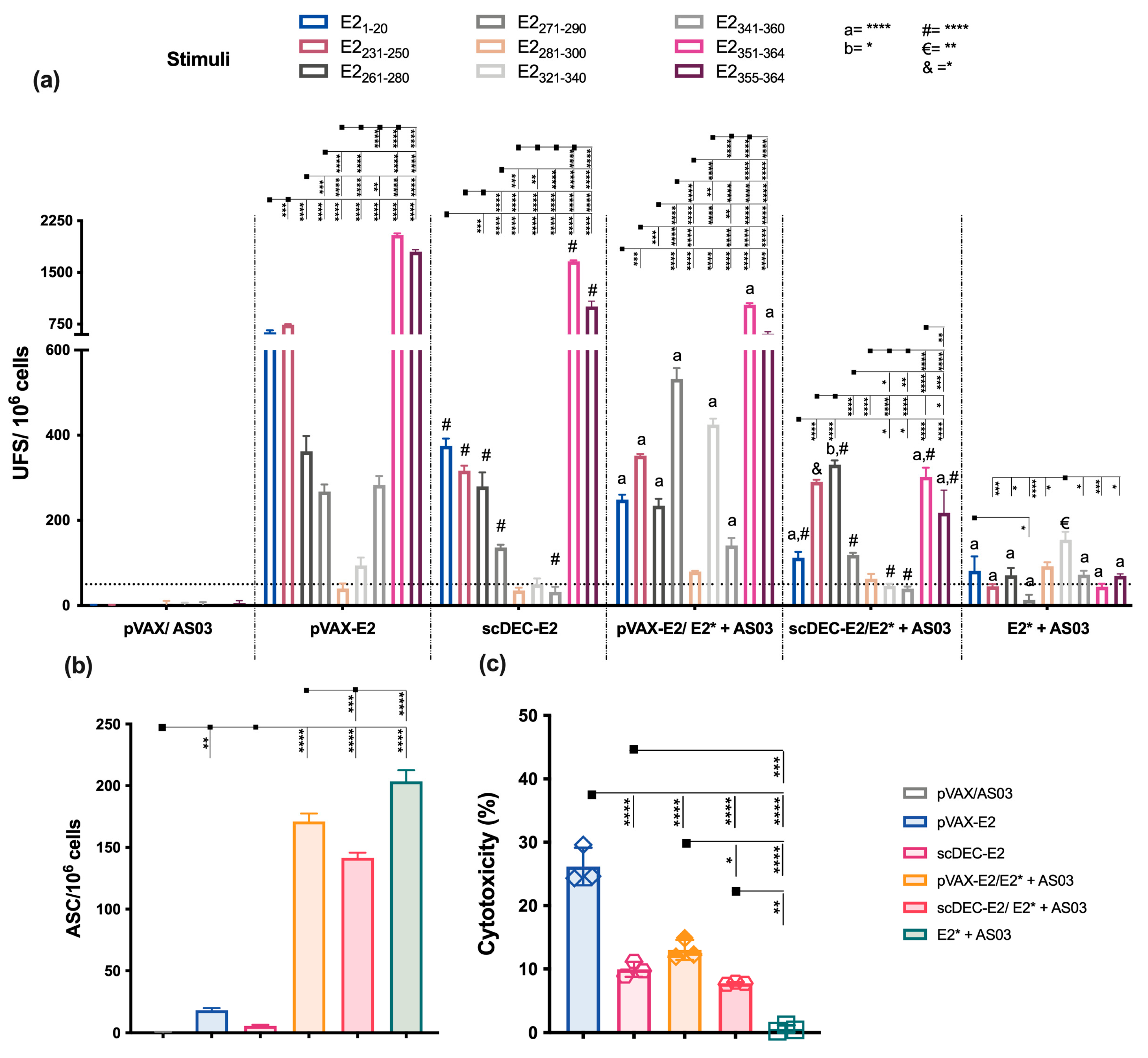
2.3. E2CHIKV T Cell Epitope Mapping
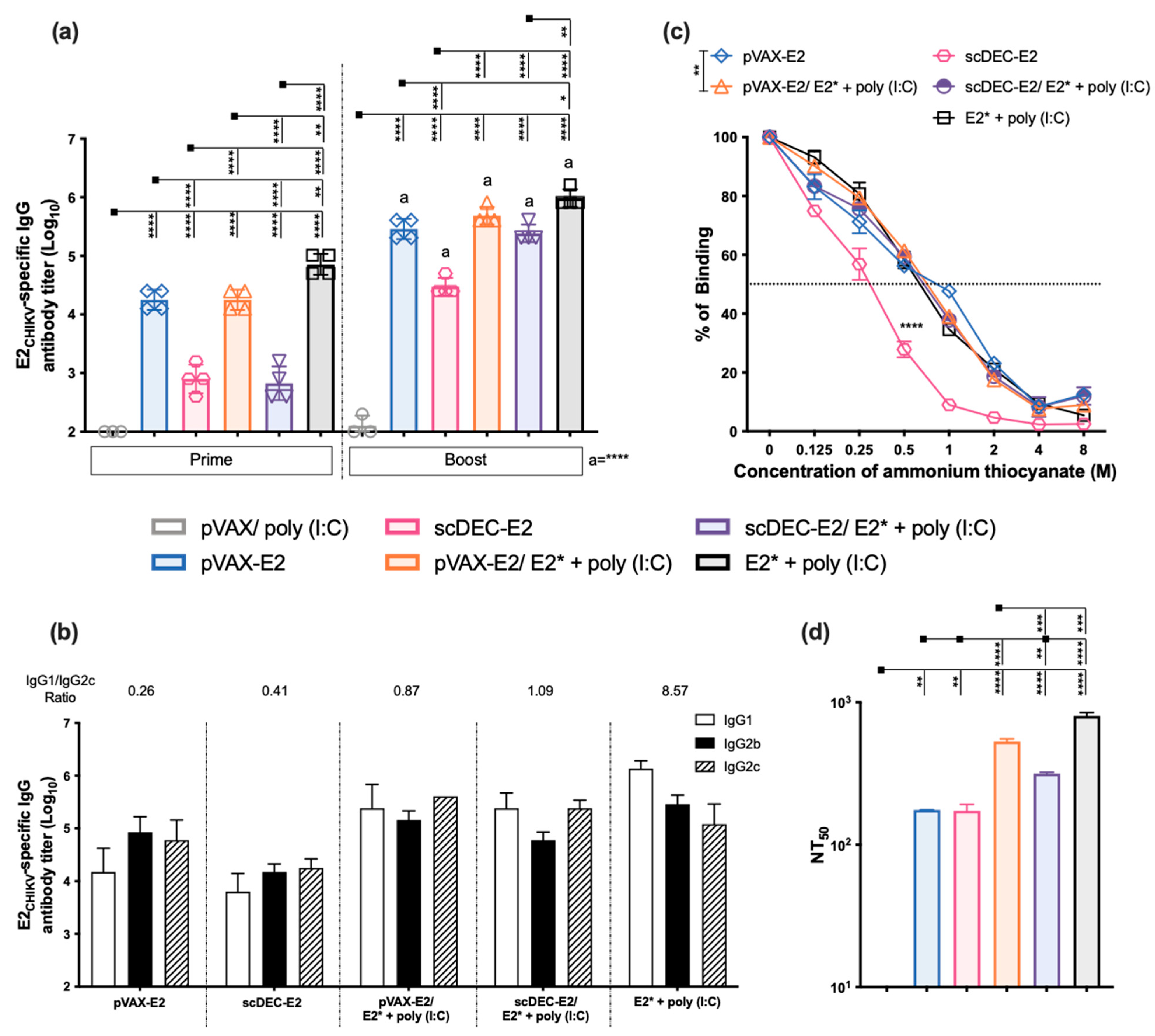
2.4. The Heterologous DNA Prime-E2*CHIKV Protein Boost Strategy Induces Both Humoral and Cellular Immune Responses
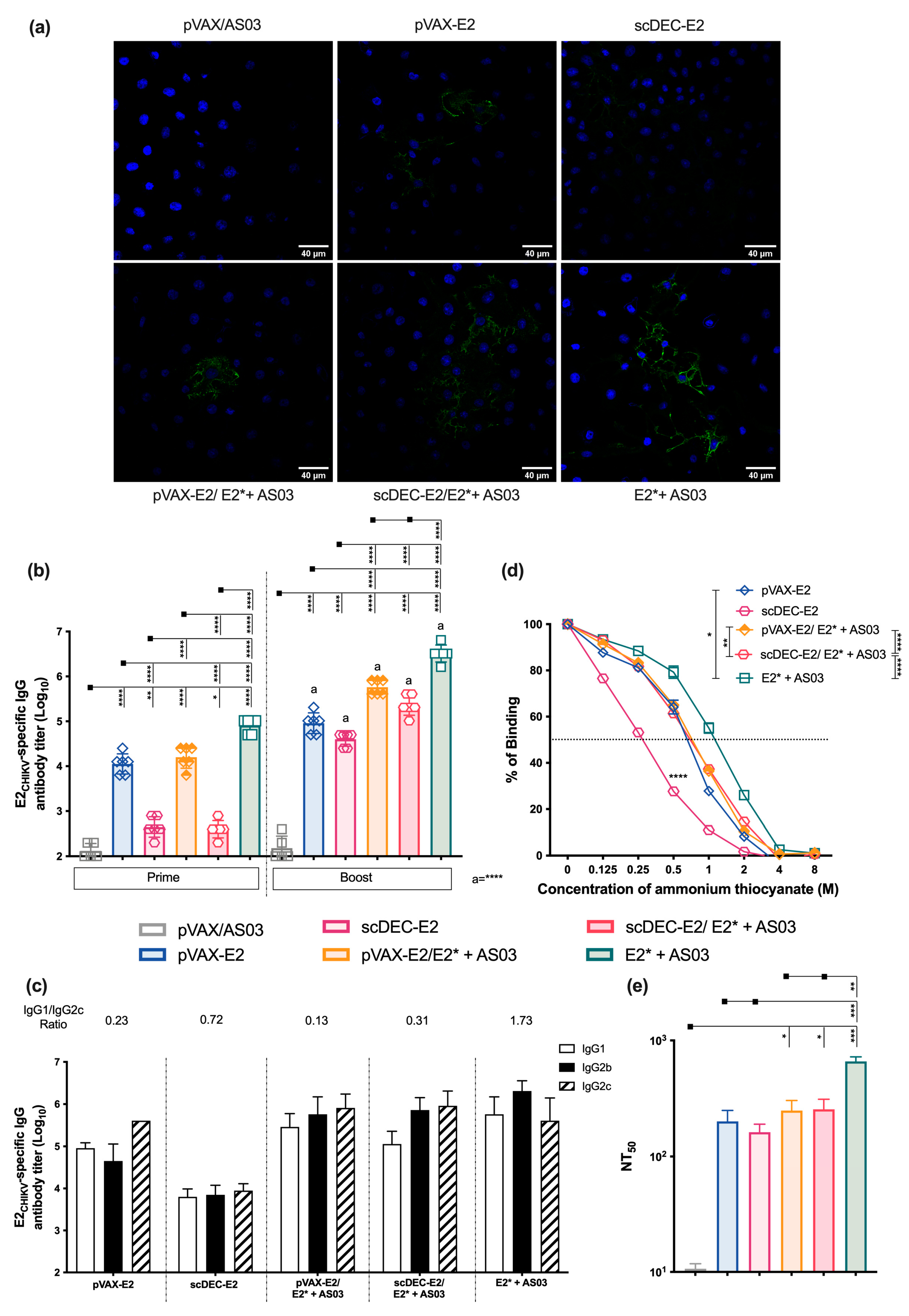
2.5. Immunization with Recombinant E2* Protein in the Presence of Different Adjuvant Formulations Elicits Neutralizing Antibodies
2.6. AS03 Induces Robust Responses in Heterologous DNA prime-E2* Protein Boost
3. Discussion
4. Materials and Methods
4.1. Design and Construction of Vaccines
4.2. E2* Protein Expression and Purification in Bacteria
4.3. DNA Vaccines In Vitro Expression
4.4. Mice and Immunization
4.5. Immunoblot
4.6. ELISA
4.7. Plaque Reduction Neutralization Assay (PRNT)
4.8. Immunofluorescence
4.9. Spleen and Lymph Node Cell Suspension
4.10. Peptide Library
4.11. T Cell ELISpot Assay
4.12. B Cell ELISpot Assay
4.13. Polyfuncional E2CHIKV-Specific T Cell Response by Flow Cytometry
4.14. In Vivo Cytotoxicity Assay
4.15. Data Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Robinson, M.C. An epidemic of virus disease in Southern Province, Tanganyika Territory, in 1952-53. I. Clinical features. Trans. R. Soc. Trop. Med. Hyg. 1955, 49, 28–32. [Google Scholar] [CrossRef]
- Schuffenecker, I.; Iteman, I.; Michault, A.; Murri, S.; Frangeul, L.; Vaney, M.C.; Lavenir, R.; Pardigon, N.; Reynes, J.M.; Pettinelli, F.; et al. Genome microevolution of chikungunya viruses causing the Indian Ocean outbreak. PLoS Med. 2006, 3, e263. [Google Scholar] [CrossRef]
- Silva, L.A.; Dermody, T.S. Chikungunya virus: Epidemiology, replication, disease mechanisms, and prospective intervention strategies. J. Clin. Investig. 2017, 127, 737–749. [Google Scholar] [CrossRef]
- Leparc-Goffart, I.; Nougairede, A.; Cassadou, S.; Prat, C.; de Lamballerie, X. Chikungunya in the Americas. Lancet 2014, 383, 514. [Google Scholar] [CrossRef]
- Rezza, G.; Weaver, S.C. Chikungunya as a paradigm for emerging viral diseases: Evaluating disease impact and hurdles to vaccine development. PLoS Negl. Trop. Dis. 2019, 13, e0006919. [Google Scholar] [CrossRef]
- Nunes, M.R.; Faria, N.R.; de Vasconcelos, J.M.; Golding, N.; Kraemer, M.U.; de Oliveira, L.F.; Azevedo Rdo, S.; da Silva, D.E.; da Silva, E.V.; da Silva, S.P.; et al. Emergence and potential for spread of Chikungunya virus in Brazil. BMC Med. 2015, 13, 102. [Google Scholar] [CrossRef]
- Rodrigues, A.M.; Souza, R.R.M.; Fonseca, L.; Rolo, C.A.; Carvalho, R.H.; Sardi, S.I.; Campos, G.S. Genomic surveillance of the Chikungunya Virus (CHIKV) in Northeast Brazil after the first outbreak in 2014. Rev. Soc. Bras. Med. Trop. 2020, 53, e20190583. [Google Scholar] [CrossRef]
- Xavier, J.; Fonseca, V.; Bezerra, J.F.; do Monte Alves, M.; Mares-Guia, M.A.; Claro, I.M.; de Jesus, R.; Adelino, T.; Araujo, E.; Cavalcante, K.; et al. Chikungunya virus ECSA lineage reintroduction in the northeasternmost region of Brazil. Int. J. Infect. Dis. 2021, 105, 120–123. [Google Scholar] [CrossRef]
- Burt, F.J.; Chen, W.; Miner, J.J.; Lenschow, D.J.; Merits, A.; Schnettler, E.; Kohl, A.; Rudd, P.A.; Taylor, A.; Herrero, L.J.; et al. Chikungunya virus: An update on the biology and pathogenesis of this emerging pathogen. Lancet Infect. Dis. 2017, 17, e107–e117. [Google Scholar] [CrossRef]
- Weaver, S.C.; Lecuit, M. Chikungunya Virus Infections. N. Engl. J. Med. 2015, 373, 94–95. [Google Scholar] [CrossRef]
- Bouquillard, E.; Fianu, A.; Bangil, M.; Charlette, N.; Ribera, A.; Michault, A.; Favier, F.; Simon, F.; Flipo, R.M. Rheumatic manifestations associated with Chikungunya virus infection: A study of 307 patients with 32-month follow-up (RHUMATOCHIK study). Jt. Bone Spine 2018, 85, 207–210. [Google Scholar] [CrossRef]
- Dupuis-Maguiraga, L.; Noret, M.; Brun, S.; Le Grand, R.; Gras, G.; Roques, P. Chikungunya disease: Infection-associated markers from the acute to the chronic phase of arbovirus-induced arthralgia. PLoS Negl. Trop. Dis. 2012, 6, e1446. [Google Scholar] [CrossRef]
- Feldstein, L.R.; Ellis, E.M.; Rowhani-Rahbar, A.; Hennessey, M.J.; Staples, J.E.; Halloran, M.E.; Weaver, M.R. Estimating the cost of illness and burden of disease associated with the 2014-2015 chikungunya outbreak in the U.S. Virgin Islands. PLoS Negl. Trop. Dis. 2019, 13, e0007563. [Google Scholar] [CrossRef]
- Solignat, M.; Gay, B.; Higgs, S.; Briant, L.; Devaux, C. Replication cycle of chikungunya: A re-emerging arbovirus. Virology 2009, 393, 183–197. [Google Scholar] [CrossRef]
- Weaver, S.C.; Lecuit, M. Chikungunya virus and the global spread of a mosquito-borne disease. N. Engl. J. Med. 2015, 372, 1231–1239. [Google Scholar] [CrossRef]
- Sun, S.; Xiang, Y.; Akahata, W.; Holdaway, H.; Pal, P.; Zhang, X.; Diamond, M.S.; Nabel, G.J.; Rossmann, M.G. Structural analyses at pseudo atomic resolution of Chikungunya virus and antibodies show mechanisms of neutralization. Elife 2013, 2, e00435. [Google Scholar] [CrossRef]
- Voss, J.E.; Vaney, M.C.; Duquerroy, S.; Vonrhein, C.; Girard-Blanc, C.; Crublet, E.; Thompson, A.; Bricogne, G.; Rey, F.A. Glycoprotein organization of Chikungunya virus particles revealed by X-ray crystallography. Nature 2010, 468, 709–712. [Google Scholar] [CrossRef]
- Poo, Y.S.; Rudd, P.A.; Gardner, J.; Wilson, J.A.; Larcher, T.; Colle, M.A.; Le, T.T.; Nakaya, H.I.; Warrilow, D.; Allcock, R.; et al. Multiple immune factors are involved in controlling acute and chronic chikungunya virus infection. PLoS Negl. Trop. Dis. 2014, 8, e3354. [Google Scholar] [CrossRef]
- Hawman, D.W.; Fox, J.M.; Ashbrook, A.W.; May, N.A.; Schroeder, K.M.S.; Torres, R.M.; Crowe, J.E., Jr.; Dermody, T.S.; Diamond, M.S.; Morrison, T.E. Pathogenic Chikungunya Virus Evades B Cell Responses to Establish Persistence. Cell Rep. 2016, 16, 1326–1338. [Google Scholar] [CrossRef]
- Lum, F.M.; Teo, T.H.; Lee, W.W.; Kam, Y.W.; Renia, L.; Ng, L.F. An essential role of antibodies in the control of Chikungunya virus infection. J. Immunol. 2013, 190, 6295–6302. [Google Scholar] [CrossRef]
- Rudd, P.A.; Wilson, J.; Gardner, J.; Larcher, T.; Babarit, C.; Le, T.T.; Anraku, I.; Kumagai, Y.; Loo, Y.M.; Gale, M., Jr.; et al. Interferon response factors 3 and 7 protect against Chikungunya virus hemorrhagic fever and shock. J. Virol. 2012, 86, 9888–9898. [Google Scholar] [CrossRef]
- Schilte, C.; Couderc, T.; Chretien, F.; Sourisseau, M.; Gangneux, N.; Guivel-Benhassine, F.; Kraxner, A.; Tschopp, J.; Higgs, S.; Michault, A.; et al. Type I IFN controls chikungunya virus via its action on nonhematopoietic cells. J. Exp. Med. 2010, 207, 429–442. [Google Scholar] [CrossRef]
- Fox, J.M.; Long, F.; Edeling, M.A.; Lin, H.; van Duijl-Richter, M.K.S.; Fong, R.H.; Kahle, K.M.; Smit, J.M.; Jin, J.; Simmons, G.; et al. Broadly Neutralizing Alphavirus Antibodies Bind an Epitope on E2 and Inhibit Entry and Egress. Cell 2015, 163, 1095–1107. [Google Scholar] [CrossRef]
- Lam, S.; Nyo, M.; Phuektes, P.; Yew, C.W.; Tan, Y.J.; Chu, J.J. A potent neutralizing IgM mAb targeting the N218 epitope on E2 protein protects against Chikungunya virus pathogenesis. MAbs 2015, 7, 1178–1194. [Google Scholar] [CrossRef]
- Smith, S.A.; Silva, L.A.; Fox, J.M.; Flyak, A.I.; Kose, N.; Sapparapu, G.; Khomandiak, S.; Ashbrook, A.W.; Kahle, K.M.; Fong, R.H.; et al. Isolation and Characterization of Broad and Ultrapotent Human Monoclonal Antibodies with Therapeutic Activity against Chikungunya Virus. Cell Host Microbe 2015, 18, 86–95. [Google Scholar] [CrossRef]
- Clayton, A.M. Monoclonal Antibodies as Prophylactic and Therapeutic Agents Against Chikungunya Virus. J. Infect. Dis. 2016, 214, S506–S509. [Google Scholar] [CrossRef]
- Schmidt, C.; Schnierle, B.S. Chikungunya Vaccine Candidates: Current Landscape and Future Prospects. Drug Des. Dev. Ther. 2022, 16, 3663–3673. [Google Scholar] [CrossRef]
- Montalvo Zurbia-Flores, G.; Reyes-Sandoval, A.; Kim, Y.C. Chikungunya Virus: Priority Pathogen or Passing Trend? Vaccines 2023, 11, 568. [Google Scholar] [CrossRef] [PubMed]
- Harrison, V.R.; Eckels, K.H.; Bartelloni, P.J.; Hampton, C. Production and evaluation of a formalin-killed Chikungunya vaccine. J. Immunol. 1971, 107, 643–647. [Google Scholar] [CrossRef]
- Metz, S.W.; Geertsema, C.; Martina, B.E.; Andrade, P.; Heldens, J.G.; van Oers, M.M.; Goldbach, R.W.; Vlak, J.M.; Pijlman, G.P. Functional processing and secretion of Chikungunya virus E1 and E2 glycoproteins in insect cells. Virol. J. 2011, 8, 353. [Google Scholar] [CrossRef]
- Kumar, M.; Sudeep, A.B.; Arankalle, V.A. Evaluation of recombinant E2 protein-based and whole-virus inactivated candidate vaccines against chikungunya virus. Vaccine 2012, 30, 6142–6149. [Google Scholar] [CrossRef]
- Metz, S.W.; Martina, B.E.; van den Doel, P.; Geertsema, C.; Osterhaus, A.D.; Vlak, J.M.; Pijlman, G.P. Chikungunya virus-like particles are more immunogenic in a lethal AG129 mouse model compared to glycoprotein E1 or E2 subunits. Vaccine 2013, 31, 6092–6096. [Google Scholar] [CrossRef]
- Weber, C.; Buchner, S.M.; Schnierle, B.S. A small antigenic determinant of the Chikungunya virus E2 protein is sufficient to induce neutralizing antibodies which are partially protective in mice. PLoS Negl. Trop. Dis. 2015, 9, e0003684. [Google Scholar] [CrossRef]
- Rossi, S.L.; Comer, J.E.; Wang, E.; Azar, S.R.; Lawrence, W.S.; Plante, J.A.; Ramsauer, K.; Schrauf, S.; Weaver, S.C. Immunogenicity and Efficacy of a Measles Virus-Vectored Chikungunya Vaccine in Nonhuman Primates. J. Infect. Dis. 2019, 220, 735–742. [Google Scholar] [CrossRef]
- Lopez-Camacho, C.; Kim, Y.C.; Blight, J.; Lazaro Moreli, M.; Montoya-Diaz, E.; Huiskonen, J.T.; Kummerer, B.M.; Reyes-Sandoval, A. Assessment of Immunogenicity and Neutralisation Efficacy of Viral-Vectored Vaccines Against Chikungunya Virus. Viruses 2019, 11, 322. [Google Scholar] [CrossRef] [PubMed]
- Muthumani, K.; Lankaraman, K.M.; Laddy, D.J.; Sundaram, S.G.; Chung, C.W.; Sako, E.; Wu, L.; Khan, A.; Sardesai, N.; Kim, J.J.; et al. Immunogenicity of novel consensus-based DNA vaccines against Chikungunya virus. Vaccine 2008, 26, 5128–5134. [Google Scholar] [CrossRef]
- Mallilankaraman, K.; Shedlock, D.J.; Bao, H.; Kawalekar, O.U.; Fagone, P.; Ramanathan, A.A.; Ferraro, B.; Stabenow, J.; Vijayachari, P.; Sundaram, S.G.; et al. A DNA vaccine against chikungunya virus is protective in mice and induces neutralizing antibodies in mice and nonhuman primates. PLoS Negl. Trop. Dis. 2011, 5, e928. [Google Scholar] [CrossRef] [PubMed]
- Muthumani, K.; Block, P.; Flingai, S.; Muruganantham, N.; Chaaithanya, I.K.; Tingey, C.; Wise, M.; Reuschel, E.L.; Chung, C.; Muthumani, A.; et al. Rapid and Long-Term Immunity Elicited by DNA-Encoded Antibody Prophylaxis and DNA Vaccination Against Chikungunya Virus. J. Infect. Dis. 2016, 214, 369–378. [Google Scholar] [CrossRef]
- Du, L.; He, Y.; Jiang, S.; Zheng, B.J. Development of subunit vaccines against severe acute respiratory syndrome. Drugs Today 2008, 44, 63–73. [Google Scholar] [CrossRef] [PubMed]
- Zhang, N.; Tang, J.; Lu, L.; Jiang, S.; Du, L. Receptor-binding domain-based subunit vaccines against MERS-CoV. Virus Res. 2015, 202, 151–159. [Google Scholar] [CrossRef]
- Vijayachari, P.; Vedhagiri, K.; Mallilankaraman, K.; Mathur, P.P.; Sardesai, N.Y.; Weiner, D.B.; Ugen, K.E.; Muthumani, K. Immunogenicity of a novel enhanced consensus DNA vaccine encoding the leptospiral protein LipL45. Hum. Vaccines Immunother. 2015, 11, 1945–1953. [Google Scholar] [CrossRef] [PubMed]
- Bonifaz, L.; Bonnyay, D.; Mahnke, K.; Rivera, M.; Nussenzweig, M.C.; Steinman, R.M. Efficient targeting of protein antigen to the dendritic cell receptor DEC-205 in the steady state leads to antigen presentation on major histocompatibility complex class I products and peripheral CD8+ T cell tolerance. J. Exp. Med. 2002, 196, 1627–1638. [Google Scholar] [CrossRef]
- Drabick, J.J.; Glasspool-Malone, J.; King, A.; Malone, R.W. Cutaneous transfection and immune responses to intradermal nucleic acid vaccination are significantly enhanced by in vivo electropermeabilization. Mol. Ther. 2001, 3, 249–255. [Google Scholar] [CrossRef]
- Simon, A.J.; Casimiro, D.R.; Finnefrock, A.C.; Davies, M.E.; Tang, A.; Chen, M.; Chastain, M.; Kath, G.S.; Chen, L.; Shiver, J.W. Enhanced in vivo transgene expression and immunogenicity from plasmid vectors following electrostimulation in rodents and primates. Vaccine 2008, 26, 5202–5209. [Google Scholar] [CrossRef]
- Kardani, K.; Bolhassani, A.; Shahbazi, S. Prime-boost vaccine strategy against viral infections: Mechanisms and benefits. Vaccine 2016, 34, 413–423. [Google Scholar] [CrossRef]
- Trumpfheller, C.; Finke, J.S.; Lopez, C.B.; Moran, T.M.; Moltedo, B.; Soares, H.; Huang, Y.; Schlesinger, S.J.; Park, C.G.; Nussenzweig, M.C.; et al. Intensified and protective CD4+ T cell immunity in mice with anti-dendritic cell HIV gag fusion antibody vaccine. J. Exp. Med. 2006, 203, 607–617. [Google Scholar] [CrossRef] [PubMed]
- Tewari, K.; Flynn, B.J.; Boscardin, S.B.; Kastenmueller, K.; Salazar, A.M.; Anderson, C.A.; Soundarapandian, V.; Ahumada, A.; Keler, T.; Hoffman, S.L.; et al. Poly(I:C) is an effective adjuvant for antibody and multi-functional CD4+ T cell responses to Plasmodium falciparum circumsporozoite protein (CSP) and alphaDEC-CSP in non human primates. Vaccine 2010, 28, 7256–7266. [Google Scholar] [CrossRef]
- Cruz, L.J.; Rosalia, R.A.; Kleinovink, J.W.; Rueda, F.; Lowik, C.W.; Ossendorp, F. Targeting nanoparticles to CD40, DEC-205 or CD11c molecules on dendritic cells for efficient CD8(+) T cell response: A comparative study. J. Control Release 2014, 192, 209–218. [Google Scholar] [CrossRef] [PubMed]
- Demangel, C.; Zhou, J.; Choo, A.B.; Shoebridge, G.; Halliday, G.M.; Britton, W.J. Single chain antibody fragments for the selective targeting of antigens to dendritic cells. Mol. Immunol. 2005, 42, 979–985. [Google Scholar] [CrossRef]
- Nchinda, G.; Kuroiwa, J.; Oks, M.; Trumpfheller, C.; Park, C.G.; Huang, Y.; Hannaman, D.; Schlesinger, S.J.; Mizenina, O.; Nussenzweig, M.C.; et al. The efficacy of DNA vaccination is enhanced in mice by targeting the encoded protein to dendritic cells. J. Clin. Investig. 2008, 118, 1427–1436. [Google Scholar] [CrossRef]
- Ngu, L.N.; Nji, N.N.; Ambada, G.E.; Sagnia, B.; Sake, C.N.; Tchadji, J.C.; Njambe Priso, G.D.; Lissom, A.; Tchouangueu, T.F.; Manga Tebit, D.; et al. In vivo targeting of protein antigens to dendritic cells using anti-DEC-205 single chain antibody improves HIV Gag specific CD4(+) T cell responses protecting from airway challenge with recombinant vaccinia-gag virus. Immun. Inflamm. Dis. 2019, 7, 55–67. [Google Scholar] [CrossRef] [PubMed]
- Tenbusch, M.; Ignatius, R.; Nchinda, G.; Trumpfheller, C.; Salazar, A.M.; Topfer, K.; Sauermann, U.; Wagner, R.; Hannaman, D.; Tenner-Racz, K.; et al. Immunogenicity of DNA vaccines encoding simian immunodeficiency virus antigen targeted to dendritic cells in rhesus macaques. PLoS ONE 2012, 7, e39038. [Google Scholar] [CrossRef]
- Agarwal, A.; Sharma, A.K.; Sukumaran, D.; Parida, M.; Dash, P.K. Two novel epistatic mutations (E1:K211E and E2:V264A) in structural proteins of Chikungunya virus enhance fitness in Aedes aegypti. Virology 2016, 497, 59–68. [Google Scholar] [CrossRef] [PubMed]
- Verma, A.; Chandele, A.; Nayak, K.; Kaja, M.K.; Arulandu, A.; Lodha, R.; Ray, P. High yield expression and purification of Chikungunya virus E2 recombinant protein and its evaluation for serodiagnosis. J. Virol. Methods 2016, 235, 73–79. [Google Scholar] [CrossRef]
- Zaneti, A.B.; Yamamoto, M.M.; Sulczewski, F.B.; Almeida, B.D.S.; Souza, H.F.S.; Ferreira, N.S.; Maeda, D.; Sales, N.S.; Rosa, D.S.; Ferreira, L.C.S.; et al. Dendritic Cell Targeting Using a DNA Vaccine Induces Specific Antibodies and CD4(+) T Cells to the Dengue Virus Envelope Protein Domain III. Front. Immunol. 2019, 10, 59. [Google Scholar] [CrossRef]
- Valvena. Valneva Announces Positive Phase 3 Pivotal Results for its Single-Shot Chikungunya Vaccine Candidate. 2021. Available online: https://valneva.com/press-release/valneva-announces-positive-phase-3-pivotal-results-for-its-single-shot-chikungunya-vaccine-candidate/ (accessed on 6 June 2023).
- Niezold, T.; Storcksdieck Genannt Bonsmann, M.; Maaske, A.; Temchura, V.; Heinecke, V.; Hannaman, D.; Buer, J.; Ehrhardt, C.; Hansen, W.; Uberla, K.; et al. DNA vaccines encoding DEC205-targeted antigens: Immunity or tolerance? Immunology 2015, 145, 519–533. [Google Scholar] [CrossRef]
- Do, Y.; Park, C.G.; Kang, Y.S.; Park, S.H.; Lynch, R.M.; Lee, H.; Powell, B.S.; Steinman, R.M. Broad T cell immunity to the LcrV virulence protein is induced by targeted delivery to DEC-205/CD205-positive mouse dendritic cells. Eur. J. Immunol. 2008, 38, 20–29. [Google Scholar] [CrossRef]
- Boscardin, S.B.; Hafalla, J.C.; Masilamani, R.F.; Kamphorst, A.O.; Zebroski, H.A.; Rai, U.; Morrot, A.; Zavala, F.; Steinman, R.M.; Nussenzweig, R.S.; et al. Antigen targeting to dendritic cells elicits long-lived T cell help for antibody responses. J. Exp. Med. 2006, 203, 599–606. [Google Scholar] [CrossRef]
- Rampazo, E.V.; Amorim, K.N.; Yamamoto, M.M.; Panatieri, R.H.; Rodrigues, M.M.; Boscardin, S.B. Antigen targeting to dendritic cells allows the identification of a CD4 T-cell epitope within an immunodominant Trypanosoma cruzi antigen. PLoS ONE 2015, 10, e0117778. [Google Scholar] [CrossRef]
- Silva-Sanchez, A.; Meza-Perez, S.; Flores-Langarica, A.; Donis-Maturano, L.; Estrada-Garcia, I.; Calderon-Amador, J.; Hernandez-Pando, R.; Idoyaga, J.; Steinman, R.M.; Flores-Romo, L. ESAT-6 Targeting to DEC205+ Antigen Presenting Cells Induces Specific-T Cell Responses against ESAT-6 and Reduces Pulmonary Infection with Virulent Mycobacterium tuberculosis. PLoS ONE 2015, 10, e0124828. [Google Scholar] [CrossRef] [PubMed]
- Lakhrif, Z.; Moreau, A.; Herault, B.; Di-Tommaso, A.; Juste, M.; Moire, N.; Dimier-Poisson, I.; Mevelec, M.N.; Aubrey, N. Targeted Delivery of Toxoplasma gondii Antigens to Dendritic Cells Promote Immunogenicity and Protective Efficiency against Toxoplasmosis. Front. Immunol. 2018, 9, 317. [Google Scholar] [CrossRef]
- Zuhaida, A.A.; Ali, A.M.; Tamilselvan, S.; Alitheen, N.B.; Hamid, M.; Noor, A.M.; Yeap, S.K. Construction of single-chain variable fragment antibodies against MCF-7 breast cancer cells. Genet. Mol. Res. 2013, 12, 5547–5559. [Google Scholar] [CrossRef] [PubMed]
- Lambricht, L.; Lopes, A.; Kos, S.; Sersa, G.; Preat, V.; Vandermeulen, G. Clinical potential of electroporation for gene therapy and DNA vaccine delivery. Expert Opin. Drug Deliv. 2016, 13, 295–310. [Google Scholar] [CrossRef]
- Vasan, S.; Hurley, A.; Schlesinger, S.J.; Hannaman, D.; Gardiner, D.F.; Dugin, D.P.; Boente-Carrera, M.; Vittorino, R.; Caskey, M.; Andersen, J.; et al. In vivo electroporation enhances the immunogenicity of an HIV-1 DNA vaccine candidate in healthy volunteers. PLoS ONE 2011, 6, e19252. [Google Scholar] [CrossRef]
- Xu, Z.; Patel, A.; Tursi, N.J.; Zhu, X.; Muthumani, K.; Kulp, D.W.; Weiner, D.B. Harnessing Recent Advances in Synthetic DNA and Electroporation Technologies for Rapid Vaccine Development Against COVID-19 and Other Emerging Infectious Diseases. Front. Med. Technol. 2020, 2, 571030. [Google Scholar] [CrossRef] [PubMed]
- Kam, Y.W.; Lum, F.M.; Teo, T.H.; Lee, W.W.; Simarmata, D.; Harjanto, S.; Chua, C.L.; Chan, Y.F.; Wee, J.K.; Chow, A.; et al. Early neutralizing IgG response to Chikungunya virus in infected patients targets a dominant linear epitope on the E2 glycoprotein. EMBO Mol. Med. 2012, 4, 330–343. [Google Scholar] [CrossRef]
- Kam, Y.W.; Pok, K.Y.; Eng, K.E.; Tan, L.K.; Kaur, S.; Lee, W.W.; Leo, Y.S.; Ng, L.C.; Ng, L.F. Sero-prevalence and cross-reactivity of chikungunya virus specific anti-E2EP3 antibodies in arbovirus-infected patients. PLoS Negl. Trop. Dis. 2015, 9, e3445. [Google Scholar] [CrossRef] [PubMed]
- Kam, Y.W.; Simarmata, D.; Chow, A.; Her, Z.; Teng, T.S.; Ong, E.K.; Renia, L.; Leo, Y.S.; Ng, L.F. Early appearance of neutralizing immunoglobulin G3 antibodies is associated with chikungunya virus clearance and long-term clinical protection. J. Infect. Dis. 2012, 205, 1147–1154. [Google Scholar] [CrossRef]
- Yoon, I.K.; Alera, M.T.; Lago, C.B.; Tac-An, I.A.; Villa, D.; Fernandez, S.; Thaisomboonsuk, B.; Klungthong, C.; Levy, J.W.; Velasco, J.M.; et al. High rate of subclinical chikungunya virus infection and association of neutralizing antibody with protection in a prospective cohort in the Philippines. PLoS Negl. Trop. Dis. 2015, 9, e0003764. [Google Scholar] [CrossRef]
- Selvarajah, S.; Sexton, N.R.; Kahle, K.M.; Fong, R.H.; Mattia, K.A.; Gardner, J.; Lu, K.; Liss, N.M.; Salvador, B.; Tucker, D.F.; et al. A neutralizing monoclonal antibody targeting the acid-sensitive region in chikungunya virus E2 protects from disease. PLoS Negl. Trop. Dis. 2013, 7, e2423. [Google Scholar] [CrossRef]
- Khan, M.; Dhanwani, R.; Rao, P.V.; Parida, M. Subunit vaccine formulations based on recombinant envelope proteins of Chikungunya virus elicit balanced Th1/Th2 response and virus-neutralizing antibodies in mice. Virus Res. 2012, 167, 236–246. [Google Scholar] [CrossRef] [PubMed]
- Snell, L.M.; Osokine, I.; Yamada, D.H.; De la Fuente, J.R.; Elsaesser, H.J.; Brooks, D.G. Overcoming CD4 Th1 Cell Fate Restrictions to Sustain Antiviral CD8 T Cells and Control Persistent Virus Infection. Cell Rep. 2016, 16, 3286–3296. [Google Scholar] [CrossRef]
- Amaral, M.C.; Coirada, F.C.; Apostolico, J.S.; Tomita, N.; Fernandes, E.R.; Souza, H.F.S.; Chura-Chambi, R.M.; Morganti, L.; Boscardin, S.B.; Rosa, D.S. Prime-boost with Chikungunya virus E2 envelope protein combined with Poly (I:C) induces specific humoral and cellular immune responses. Curr. Res. Immunol. 2021, 2, 23–31. [Google Scholar] [CrossRef]
- Nagar, P.K.; Pradhan, S.; Verma, P.; Joshi, G.; Singh, A.; Rao, D.N. Mapping and Immunological Response of Immunodominant B and T cell Epitopes of E2 Glycoprotein of Chikungunya Virus. MOJ Immunol. 2016, 4, 00117. [Google Scholar] [CrossRef]
- Teo, T.H.; Chan, Y.H.; Lee, W.W.; Lum, F.M.; Amrun, S.N.; Her, Z.; Rajarethinam, R.; Merits, A.; Rotzschke, O.; Renia, L.; et al. Fingolimod treatment abrogates chikungunya virus-induced arthralgia. Sci. Transl. Med. 2017, 9, eaal1333. [Google Scholar] [CrossRef] [PubMed]
- Vidhya, R.V.; Nair, A.S.; Dhar, P.K.; Nayarisseri, A. Epitope characterization and docking studies on Chikungunya viral Envelope 2 protein. Int. J. Sci. Res. Publ. 2015, 5. [Google Scholar]
- Brown, S.A.; Surman, S.L.; Sealy, R.; Jones, B.G.; Slobod, K.S.; Branum, K.; Lockey, T.D.; Howlett, N.; Freiden, P.; Flynn, P.; et al. Heterologous Prime-Boost HIV-1 Vaccination Regimens in Pre-Clinical and Clinical Trials. Viruses 2010, 2, 435–467. [Google Scholar] [CrossRef]
- Levine, M.Z.; Holiday, C.; Jefferson, S.; Gross, F.L.; Liu, F.; Li, S.; Friel, D.; Boutet, P.; Innis, B.L.; Mallett, C.P.; et al. Heterologous prime-boost with A(H5N1) pandemic influenza vaccines induces broader cross-clade antibody responses than homologous prime-boost. NPJ Vaccines 2019, 4, 22. [Google Scholar] [CrossRef]
- Ledford, H. Could mixing COVID vaccines boost immune response? Nature 2021, 590, 375–376. [Google Scholar] [CrossRef]
- Roques, P.; Ljungberg, K.; Kummerer, B.M.; Gosse, L.; Dereuddre-Bosquet, N.; Tchitchek, N.; Hallengard, D.; Garcia-Arriaza, J.; Meinke, A.; Esteban, M.; et al. Attenuated and vectored vaccines protect nonhuman primates against Chikungunya virus. JCI Insight 2017, 2, e83527. [Google Scholar] [CrossRef]
- Hallengard, D.; Lum, F.M.; Kummerer, B.M.; Lulla, A.; Lulla, V.; Garcia-Arriaza, J.; Fazakerley, J.K.; Roques, P.; Le Grand, R.; Merits, A.; et al. Prime-boost immunization strategies against Chikungunya virus. J. Virol. 2014, 88, 13333–13343. [Google Scholar] [CrossRef] [PubMed]
- Knudsen, M.L.; Ljungberg, K.; Kakoulidou, M.; Kostic, L.; Hallengard, D.; Garcia-Arriaza, J.; Merits, A.; Esteban, M.; Liljestrom, P. Kinetic and phenotypic analysis of CD8+ T cell responses after priming with alphavirus replicons and homologous or heterologous booster immunizations. J. Virol. 2014, 88, 12438–12451. [Google Scholar] [CrossRef]
- Del Giudice, G.; Rappuoli, R.; Didierlaurent, A.M. Correlates of adjuvanticity: A review on adjuvants in licensed vaccines. Semin. Immunol. 2018, 39, 14–21. [Google Scholar] [CrossRef] [PubMed]
- Khurana, S.; Verma, N.; Yewdell, J.W.; Hilbert, A.K.; Castellino, F.; Lattanzi, M.; Del Giudice, G.; Rappuoli, R.; Golding, H. MF59 adjuvant enhances diversity and affinity of antibody-mediated immune response to pandemic influenza vaccines. Sci. Transl. Med. 2011, 3, 85ra48. [Google Scholar] [CrossRef]
- Lindblad, E.B. Aluminium compounds for use in vaccines. Immunol. Cell Biol. 2004, 82, 497–505. [Google Scholar] [CrossRef]
- Martins, K.A.; Bavari, S.; Salazar, A.M. Vaccine adjuvant uses of poly-IC and derivatives. Expert Rev. Vaccines 2015, 14, 447–459. [Google Scholar] [CrossRef]
- Wilkins, A.L.; Kazmin, D.; Napolitani, G.; Clutterbuck, E.A.; Pulendran, B.; Siegrist, C.A.; Pollard, A.J. AS03- and MF59-Adjuvanted Influenza Vaccines in Children. Front. Immunol. 2017, 8, 1760. [Google Scholar] [CrossRef]
- Jang, J.; Chu, H.; Kim, S. Chikungunya Virus Methods and Protocols–Methods in Molecular Biology; Springer: Berlin/Heidelberg, Germany, 2016. [Google Scholar] [CrossRef]
- de Mello, L.R.; Porosk, L.; Lourenco, T.C.; Garcia, B.B.M.; Costa, C.A.R.; Han, S.W.; de Souza, J.S.; Langel, U.; da Silva, E.R. Amyloid-like Self-Assembly of a Hydrophobic Cell-Penetrating Peptide and Its Use as a Carrier for Nucleic Acids. ACS Appl. Bio Mater. 2021, 4, 6404–6416. [Google Scholar] [CrossRef]
- Precopio, M.L.; Butterfield, T.R.; Casazza, J.P.; Little, S.J.; Richman, D.D.; Koup, R.A.; Roederer, M. Optimizing peptide matrices for identifying T-cell antigens. Cytom. Part A 2008, 73, 1071–1078. [Google Scholar] [CrossRef]
- Clemente, T.; Dominguez, M.R.; Vieira, N.J.; Rodrigues, M.M.; Amarante-Mendes, G.P. In vivo assessment of specific cytotoxic T lymphocyte killing. Methods 2013, 61, 105–109. [Google Scholar] [CrossRef] [PubMed]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Coirada, F.C.; Fernandes, E.R.; Mello, L.R.d.; Schuch, V.; Soares Campos, G.; Braconi, C.T.; Boscardin, S.B.; Santoro Rosa, D. Heterologous DNA Prime- Subunit Protein Boost with Chikungunya Virus E2 Induces Neutralizing Antibodies and Cellular-Mediated Immunity. Int. J. Mol. Sci. 2023, 24, 10517. https://doi.org/10.3390/ijms241310517
Coirada FC, Fernandes ER, Mello LRd, Schuch V, Soares Campos G, Braconi CT, Boscardin SB, Santoro Rosa D. Heterologous DNA Prime- Subunit Protein Boost with Chikungunya Virus E2 Induces Neutralizing Antibodies and Cellular-Mediated Immunity. International Journal of Molecular Sciences. 2023; 24(13):10517. https://doi.org/10.3390/ijms241310517
Chicago/Turabian StyleCoirada, Fernanda Caroline, Edgar Ruz Fernandes, Lucas Rodrigues de Mello, Viviane Schuch, Gúbio Soares Campos, Carla Torres Braconi, Silvia Beatriz Boscardin, and Daniela Santoro Rosa. 2023. "Heterologous DNA Prime- Subunit Protein Boost with Chikungunya Virus E2 Induces Neutralizing Antibodies and Cellular-Mediated Immunity" International Journal of Molecular Sciences 24, no. 13: 10517. https://doi.org/10.3390/ijms241310517
APA StyleCoirada, F. C., Fernandes, E. R., Mello, L. R. d., Schuch, V., Soares Campos, G., Braconi, C. T., Boscardin, S. B., & Santoro Rosa, D. (2023). Heterologous DNA Prime- Subunit Protein Boost with Chikungunya Virus E2 Induces Neutralizing Antibodies and Cellular-Mediated Immunity. International Journal of Molecular Sciences, 24(13), 10517. https://doi.org/10.3390/ijms241310517




