KDM6B Negatively Regulates the Neurogenesis Potential of Apical Papilla Stem Cells via HES1
Abstract
:1. Introduction
2. Results
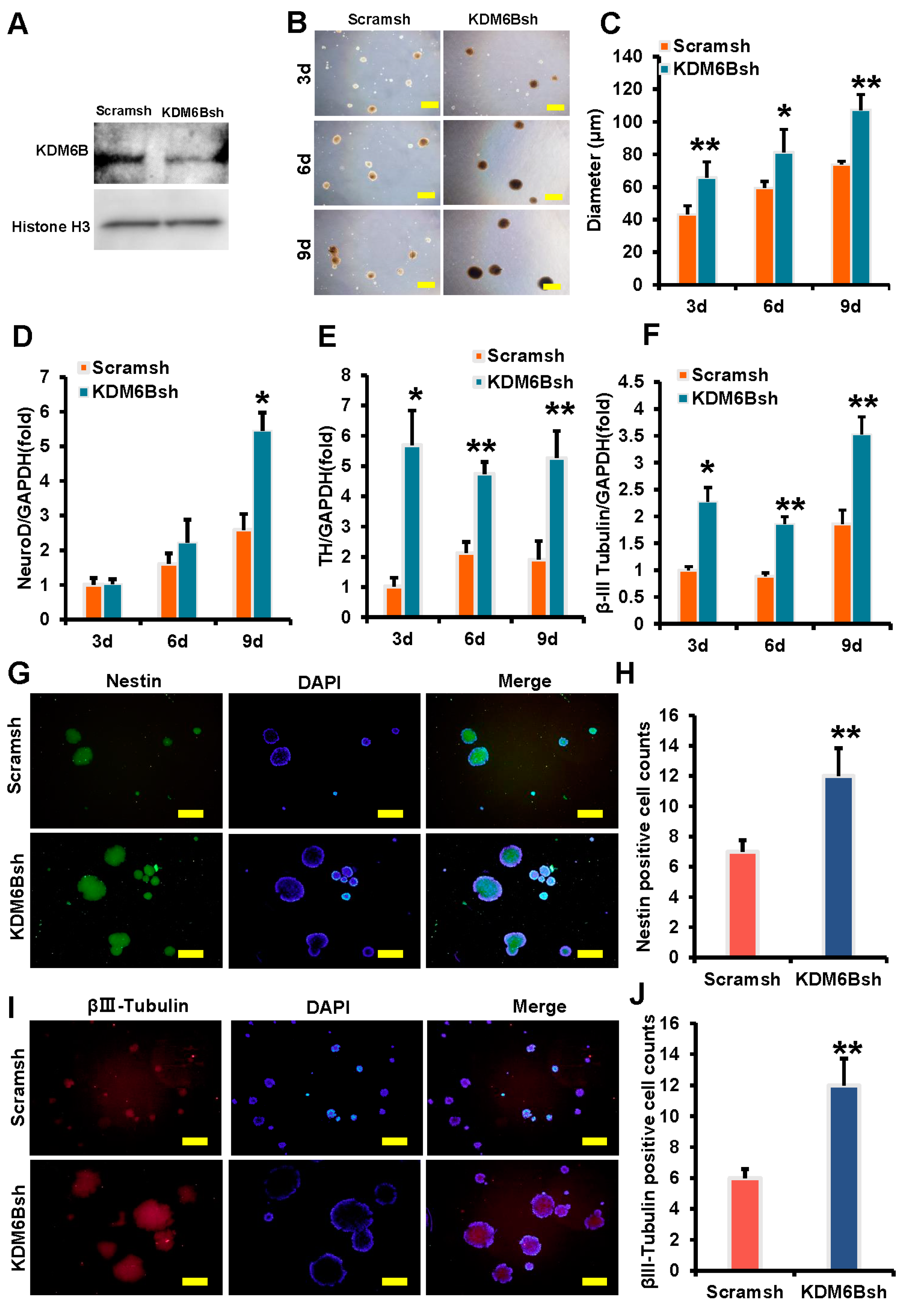
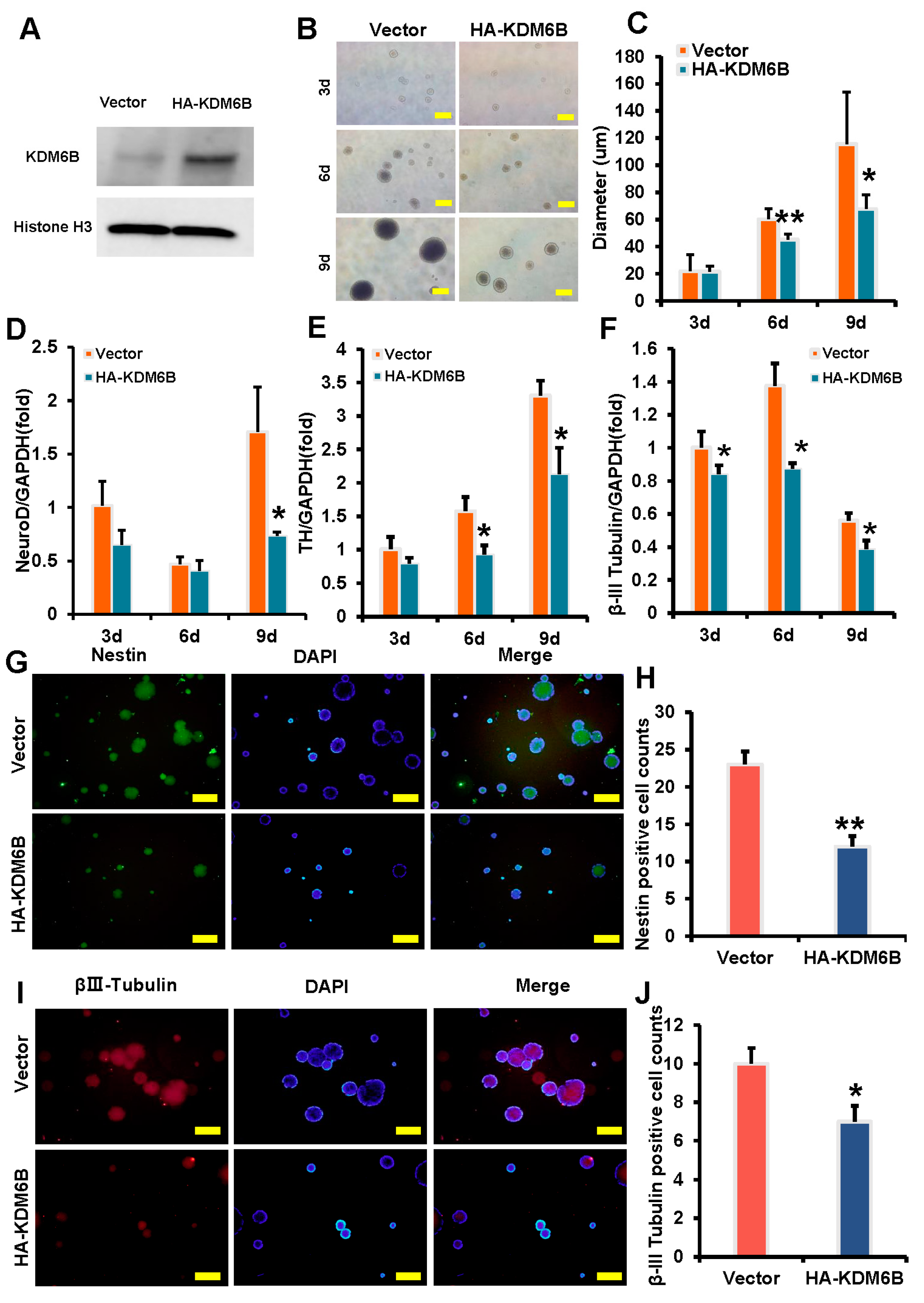
2.1. KDM6B Inhibited the Expression of Neural Markers in SCAPs
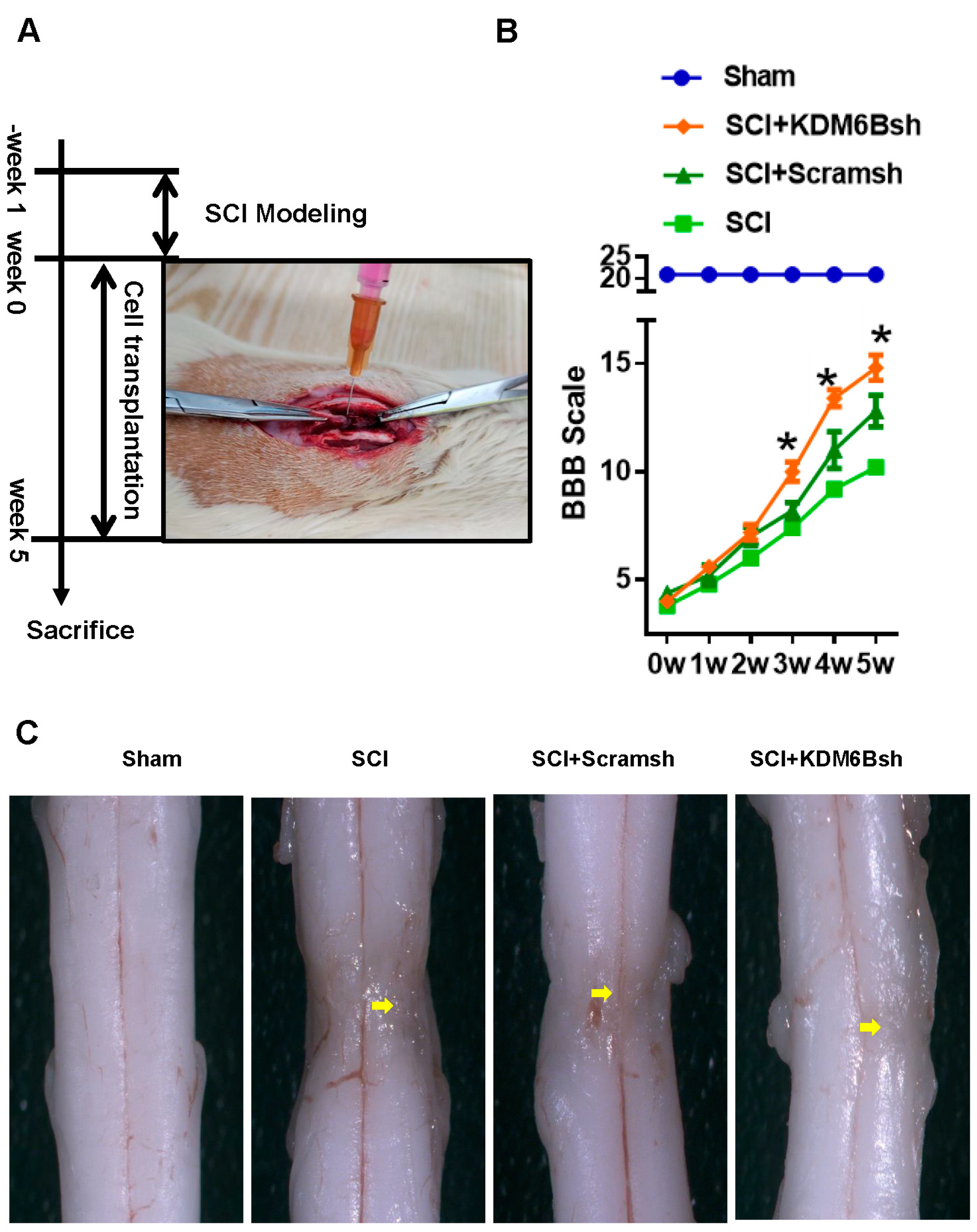
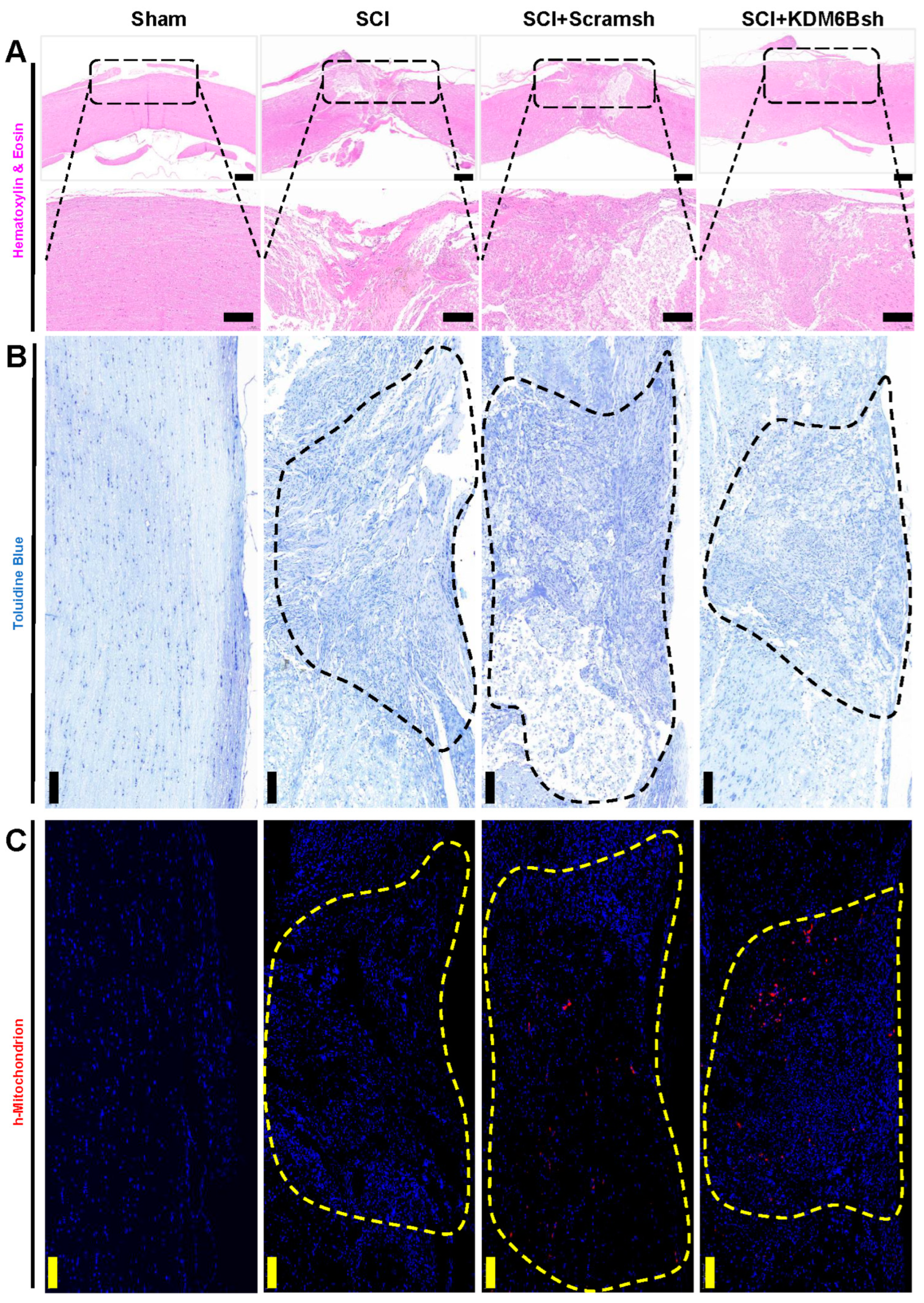
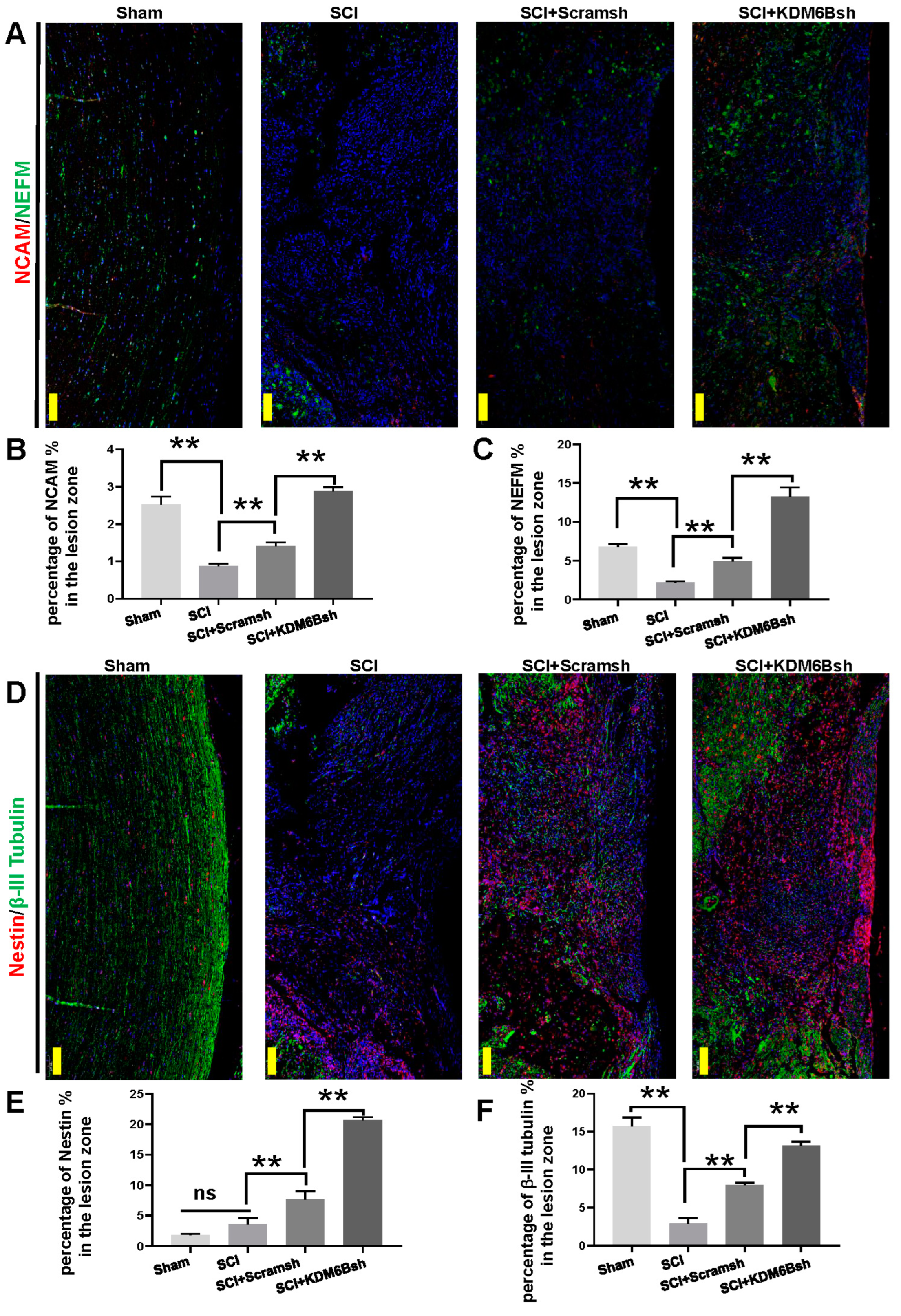
2.2. KDM6B Knockdown Significantly Promoted SCAP-Mediated Locomotor Recovery after Complete Transection of the Rat Spinal Cord
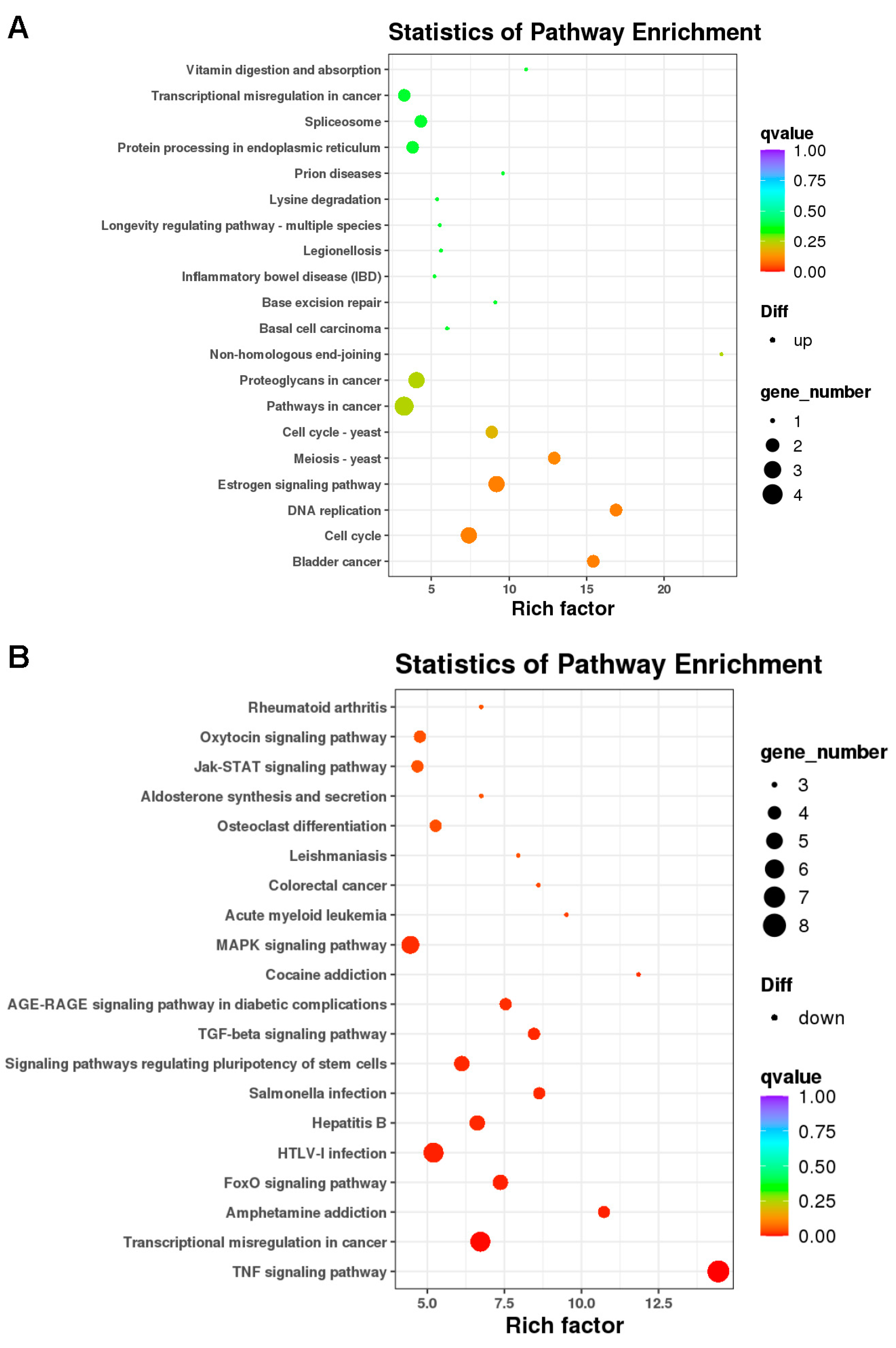
2.3. Differentially Expressed Genes and Bioinformatic Analysis in KDM6B Overexpressed SCAPs
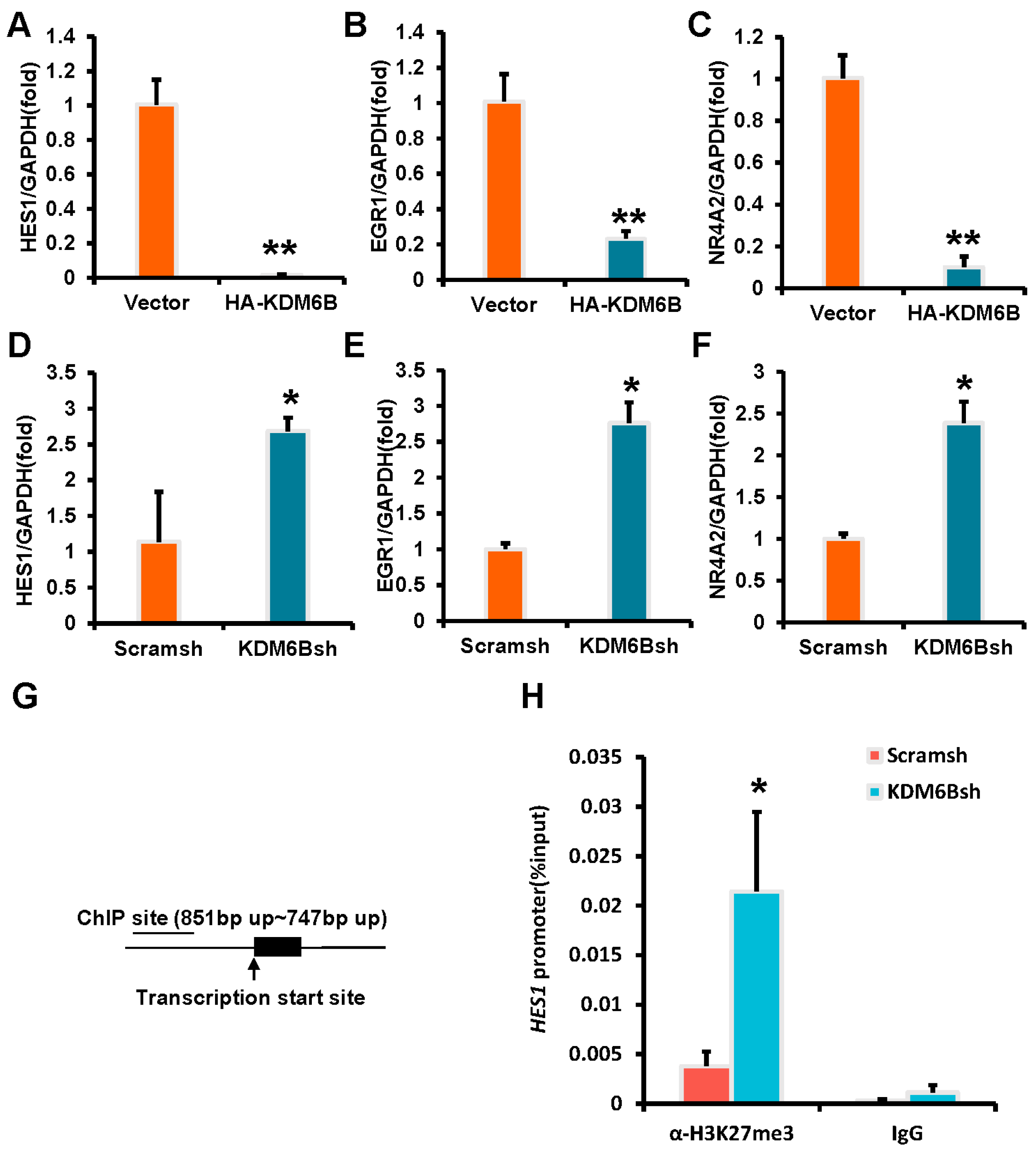
2.4. KDM6B Negatively Regulated the HES1 by Binding to the HES1 Promoter in SCAPs
3. Discussion
4. Materials and Methods
4.1. Cell Cultures
4.2. Plasmid Construction and Viral Infection
4.3. Neurogenic Differentiation and Immunofluorescence Staining
4.4. Western Blot Analysis
4.5. Real-Time Reverse Transcriptase-Polymerase Chain Reaction (Real-Time RT-PCR)
4.6. Spinal Cord Injury Model
4.7. Behavioral Testing
4.8. Histological Analyses and Toluidine Blue Staining
4.9. Microarray Analysis
4.10. Chromatin Immunoprecipitation (ChIP) Assays
4.11. Statistical Analyses
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- National Spinal Cord Injury Statistical Center. Spinal cord injury facts and figures at a glance. J. Spinal Cord Med. 2013, 36, 568–569. [Google Scholar] [CrossRef] [PubMed]
- Merritt, C.H.; Taylor, M.A.; Yelton, C.J.; Ray, S.K. Economic impact of traumatic spinal cord injuries in the United States. Neuroimmunol. Neuroinflamm. 2019, 6, 9. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mahabaleshwarkar, R.; Khanna, R. National hospitalization burden associated with spinal cord injuries in the United States. Spinal Cord 2014, 52, 139–144. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fehlings, M.G.; Tetreault, L.A.; Wilson, J.R.; Kwon, B.K.; Burns, A.S.; Martin, A.R.; Hawryluk, G.; Harrop, J.S. A Clinical Practice Guideline for the Management of Acute Spinal Cord Injury: Introduction, Rationale, and Scope. Glob. Spine J. 2017, 7 (Suppl. 3), 84s–94s. [Google Scholar] [CrossRef]
- Kadoya, K.; Lu, P.; Nguyen, K.; Lee-Kubli, C.; Kumamaru, H.; Yao, L.; Knackert, J.; Poplawski, G.; Dulin, J.N.; Strobl, H.; et al. Spinal cord reconstitution with homologous neural grafts enables robust corticospinal regeneration. Nat. Med. 2016, 22, 479–487. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, L.; Xiao, B.; Mu, J.; Zhang, Y.; Zhang, C.; Cao, H.; Chen, R. A MnO(2) Nanoparticle-Dotted Hydrogel Promotes Spinal Cord Repair via Regulating Reactive Oxygen Species Microenvironment and Synergizing with Mesenchymal Stem Cells. ACS Nano 2019, 13, 14283–14293. [Google Scholar] [CrossRef]
- An, H.; Li, Q.; Wen, J. Bone marrow mesenchymal stem cells encapsulated thermal-responsive hydrogel network bridges combined photo-plasmonic nanoparticulate system for the treatment of urinary bladder dysfunction after spinal cord injury. J. Photochem. Photobiol. B Biol. 2020, 203, 111741. [Google Scholar] [CrossRef]
- Guo, S.; Perets, N.; Betzer, O. Intranasal Delivery of Mesenchymal Stem Cell Derived Exosomes Loaded with Phosphatase and Tensin Homolog siRNA Repairs Complete Spinal Cord Injury. ACS Nano 2019, 13, 10015–10028. [Google Scholar] [CrossRef]
- Park, J.; Lee, K.; Kim, K. The role of histone modifications: From neurodevelopment to neurodiseases. Signal Transduct. Target Ther. 2022, 7, 217. [Google Scholar] [CrossRef]
- Sessa, A.; Fagnocchi, L.; Mastrototaro, G.; Massimino, L.; Zaghi, M.; Indrigo, M.; Cattaneo, S.; Martini, D.; Gabellini, C.; Pucci, C.; et al. SETD5 Regulates Chromatin Methylation State and Preserves Global Transcriptional Fidelity during Brain Development and Neuronal Wiring. Neuron 2019, 104, 271–289.e13. [Google Scholar] [CrossRef]
- Franz, H.; Villarreal, A.; Heidrich, S.; Videm, P.; Kilpert, F.; Mestres, I.; Calegari, F.; Backofen, R.; Manke, T.; Vogel, T. DOT1L promotes progenitor proliferation and primes neuronal layer identity in the developing cerebral cortex. Nucleic Acids Res. 2019, 47, 168–183. [Google Scholar] [CrossRef] [PubMed]
- Hirano, K.; Namihira, M. LSD1 Mediates Neuronal Differentiation of Human Fetal Neural Stem Cells by Controlling the Expression of a Novel Target Gene, HEYL. Stem Cells (Dayt. Ohio) 2016, 34, 1872–1882. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pappa, S.; Padilla, N.; Iacobucci, S. PHF2 histone demethylase prevents DNA damage and genome instability by controlling cell cycle progression of neural progenitors. Proc. Natl. Acad. Sci. USA 2019, 116, 19464–19473. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Park, D.H.; Hong, S.J.; Salinas, R.D.; Liu, S.J.; Sun, S.W.; Sgualdino, J.; Testa, G.; Matzuk, M.M.; Iwamori, N.; Lim, D.A. Activation of neuronal gene expression by the JMJD3 demethylase is required for postnatal and adult brain neurogenesis. Cell Rep. 2014, 8, 1290–1299. [Google Scholar] [CrossRef] [Green Version]
- Jepsen, K.; Solum, D.; Zhou, T.; McEvilly, R.J.; Kim, H.J.; Glass, C.K.; Hermanson, O.; Rosenfeld, M.G. SMRT-mediated repression of an H3K27 demethylase in progression from neural stem cell to neuron. Nature 2007, 450, 415–419. [Google Scholar] [CrossRef]
- Burgold, T.; Spreafico, F.; De Santa, F.; Totaro, M.G.; Prosperini, E.; Natoli, G.; Testa, G. The histone H3 lysine 27-specific demethylase Jmjd3 is required for neural commitment. PLoS ONE 2008, 3, e3034. [Google Scholar] [CrossRef]
- Hoffman, R.M. Tissue Culture☆. In Reference Module in Life Sciences; Elsevier: Amsterdam, The Netherlands, 2017. [Google Scholar]
- Wang, W.; Cho, H.; Lee, J.W. The histone demethylase Kdm6b regulates subtype diversification of mouse spinal motor neurons during development. Nat. Commun. 2022, 13, 958. [Google Scholar] [CrossRef]
- Meng, Y.; Zhang, T.; Zheng, R.; Ding, S.; Yang, J.; Liu, R.; Jiang, Y.; Jiang, W. Depletion of Demethylase KDM6 Enhances Early Neuroectoderm Commitment of Human PSCs. Front. Cell Dev. Biol. 2021, 9, 702462. [Google Scholar] [CrossRef]
- de Almeida, J.F.A.; Chen, P.; Henry, M.A.; Diogenes, A. Stem Cells of the Apical Papilla Regulate Trigeminal Neurite Outgrowth and Targeting through a BDNF-Dependent Mechanism. Tissue Eng. Part A 2014, 20, 3089–3100. [Google Scholar] [CrossRef] [Green Version]
- Kolar, M.K.; Itte, V.N.; Kingham, P.J.; Novikov, L.N.; Wiberg, M.; Kelk, P. The neurotrophic effects of different human dental mesenchymal stem cells. Sci. Rep. 2017, 7, 12605. [Google Scholar] [CrossRef] [Green Version]
- De Berdt, P.; Bottemanne, P.; Bianco, J.; Alhouayek, M.; Diogenes, A.; Lloyd, A.; Llyod, A.; Gerardo-Nava, J.; Brook, G.A.; Miron, V.; et al. Stem cells from human apical papilla decrease neuro-inflammation and stimulate oligodendrocyte progenitor differentiation via activin-A secretion. Cell. Mol. Life Sci. CMLS 2018, 75, 2843–2856. [Google Scholar] [CrossRef] [Green Version]
- Simonovic, J.; Toljic, B.; Nikolic, N.; Peric, M.; Vujin, J.; Panajotovic, R.; Gajic, R.; Bekyarova, E.; Cataldi, A.; Parpura, V.; et al. Differentiation of stem cells from apical papilla into neural lineage using graphene dispersion and single walled carbon nanotubes. J. Biomed. Mater. Res. Part A 2018, 106, 2653–2661. [Google Scholar] [CrossRef] [PubMed]
- Jendelova, P. Therapeutic Strategies for Spinal Cord Injury. Int. J. Mol. Sci. 2018, 19, 3200. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Suzuki, H.; Imajo, Y.; Funaba, M.; Ikeda, H.; Nishida, N.; Sakai, T. Current Concepts of Biomaterial Scaffolds and Regenerative Therapy for Spinal Cord Injury. Int. J. Mol. Sci. 2023, 24, 2528. [Google Scholar] [CrossRef]
- Wang, X.; Pei, Z.; Hossain, A.; Bai, Y.; Chen, G. Transcription factor-based gene therapy to treat glioblastoma through direct neuronal conversion. Cancer Biol. Med. 2021, 18, 860–874. [Google Scholar] [CrossRef] [PubMed]
- Shu, M.; Xue, X.; Nie, H.; Wu, X.; Sun, M.; Qiao, L.; Li, X.; Xu, B.; Xiao, Z.; Zhao, Y.; et al. Single-cell RNA sequencing reveals Nestin(+) active neural stem cells outside the central canal after spinal cord injury. Sci. China Life Sci. 2022, 65, 295–308. [Google Scholar] [CrossRef]
- Sanchez-Petidier, M.; Guerri, C.; Moreno-Manzano, V. Toll-like receptors 2 and 4 differentially regulate the self-renewal and differentiation of spinal cord neural precursor cells. Stem Cell Res. Ther. 2022, 13, 117. [Google Scholar] [CrossRef]
- Tutukova, S.; Tarabykin, V.; Hernandez-Miranda, L.R. The Role of Neurod Genes in Brain Development, Function, and Disease. Front. Mol. Neurosci. 2021, 14, 662774. [Google Scholar] [CrossRef]
- Desouza, L.A.; Ladiwala, U.; Daniel, S.M.; Agashe, S.; Vaidya, R.A.; Vaidya, V.A. Thyroid hormone regulates hippocampal neurogenesis in the adult rat brain. Mol. Cell. Neurosci. 2005, 29, 414–426. [Google Scholar] [CrossRef]
- Korzhevskii, D.E.; Karpenko, M.N.; Kirik, O.V. Microtubule-Associated Proteins as Indicators of Differentiation and the Functional State of Nerve Cells. Neurosci. Behav. Physiol. 2012, 42, 215–222. [Google Scholar] [CrossRef]
- Mattucci, S.; Speidel, J.; Liu, J.; Tetzlaff, W.; Oxland, T.R. Temporal Progression of Acute Spinal Cord Injury Mechanisms in a Rat Model: Contusion, Dislocation, and Distraction. J. Neurotrauma 2021, 38, 2103–2121. [Google Scholar] [CrossRef]
- Tan, Z.-J.; Peng, Y.; Song, H.-L.; Zheng, J.-J.; Yu, X. N-cadherin-dependent neuron–neuron interaction is required for the maintenance of activity-induced dendrite growth. Proc. Natl. Acad. Sci. USA 2010, 107, 9873–9878. [Google Scholar] [CrossRef] [Green Version]
- Kim, D.-H.; Cho, H.-J.; Park, C.-Y.; Cho, M.S.; Kim, D.-W. Transplantation of PSA-NCAM-Positive Neural Precursors from Human Embryonic Stem Cells Promotes Functional Recovery in an Animal Model of Spinal Cord Injury. Tissue Eng. Regen. Med. 2022, 19, 1349–1358. [Google Scholar] [CrossRef] [PubMed]
- Khalil, M.; Teunissen, C.E.; Otto, M.; Piehl, F.; Sormani, M.P.; Gattringer, T.; Barro, C.; Kappos, L.; Comabella, M.; Fazekas, F.; et al. Neurofilaments as biomarkers in neurological disorders. Nat. Rev. Neurol. 2018, 14, 577–589. [Google Scholar] [CrossRef] [PubMed]
- Liu, M.; Xu, P.; Guan, Z.; Qian, X.; Dockery, P.; Fitzgerald, U.; O’Brien, T.; Shen, S. Ulk4 deficiency leads to hypomyelination in mice. Glia 2018, 66, 175–190. [Google Scholar] [CrossRef]
- Myers, M.W.; Lazzarini, R.A.; Lee, V.M.; Schlaepfer, W.W.; Nelson, D.L. The human mid-size neurofilament subunit: A repeated protein sequence and the relationship of its gene to the intermediate filament gene family. EMBO J. 1987, 6, 1617–1626. [Google Scholar] [CrossRef] [PubMed]
- Liu, C.; Zhong, Y.; Apostolou, A.; Fang, S. Neural differentiation of human embryonic stem cells as an in vitro tool for the study of the expression patterns of the neuronal cytoskeleton during neurogenesis. Biochem. Biophys. Res. Commun. 2013, 439, 154–159. [Google Scholar] [CrossRef]
- Niu, Y.; Xia, X.; Song, P.; Fang, H.; Dong, F.; Tao, H.; Yang, C.; Shen, C. Bone mesenchymal stem cell-conditioned medium attenuates the effect of oxidative stress injury on NSCs by inhibiting the Notch1 signaling pathway. Cell Biol. Int. 2019, 43, 1267–1275. [Google Scholar] [CrossRef]
- Guo, H.; Xie, T.; Liu, H. 6.18—Notch in Human Cancers—A Complex Tale. In Comprehensive Pharmacology; Kenakin, T., Ed.; Elsevier: Oxford, UK, 2022; pp. 329–350. [Google Scholar]
- Chen, Y.; Lian, X.H.; Liao, L.Y.; Liu, Y.T.; Liu, S.L.; Gao, Q. Transplantation of bone marrow mesenchymal stem cells alleviates spinal cord injury via inhibiting Notch signaling. Eur. Rev. Med. Pharmacol. Sci. 2019, 23 (Suppl. 3), 31–38. [Google Scholar]
- Duclot, F.; Kabbaj, M. The Role of Early Growth Response 1 (EGR1) in Brain Plasticity and Neuropsychiatric Disorders. Front. Behav. Neurosci. 2017, 11, 35. [Google Scholar] [CrossRef] [Green Version]
- Luo, G.R.; Chen, Y.; Li, X.P.; Liu, T.X.; Le, W.D. Nr4a2 is essential for the differentiation of dopaminergic neurons during zebrafish embryogenesis. Mol. Cell. Neurosci. 2008, 39, 202–210. [Google Scholar] [CrossRef] [PubMed]
- Gao, R.; Dong, R.; Du, J.; Ma, P.; Wang, S.; Fan, Z. Depletion of histone demethylase KDM2A inhibited cell proliferation of stem cells from apical papilla by de-repression of p15INK4B and p27Kip1. Mol. Cell. Biochem. 2013, 379, 115–122. [Google Scholar] [CrossRef] [PubMed]
- Lin, X.; Dong, R.; Diao, S.; Yu, G.; Wang, L.; Li, J.; Fan, Z. SFRP2 enhanced the adipogenic and neuronal differentiation potentials of stem cells from apical papilla. Cell Biol. Int. 2017, 41, 534–543. [Google Scholar] [CrossRef] [PubMed]
- Campolo, M.; Filippone, A.; Biondo, C.; Mancuso, G.; Casili, G.; Lanza, M.; Cuzzocrea, S.; Esposito, E.; Paterniti, I. TLR7/8 in the Pathogenesis of Parkinson’s Disease. Int. J. Mol. Sci. 2020, 21, 9384. [Google Scholar] [CrossRef]
- Yang, Y.; Fan, Y.; Zhang, H.; Zhang, Q.; Zhao, Y.; Xiao, Z.; Liu, W.; Chen, B.; Gao, L.; Sun, Z.; et al. Small molecules combined with collagen hydrogel direct neurogenesis and migration of neural stem cells after spinal cord injury. Biomaterials 2021, 269, 120479. [Google Scholar] [CrossRef] [PubMed]
- Yan, W.; Li, L.; Ge, L.; Zhang, F.; Fan, Z.; Hu, L. The cannabinoid receptor I (CB1) enhanced the osteogenic differentiation of BMSCs by rescue impaired mitochondrial metabolism function under inflammatory condition. Stem Cell Res. Ther. 2022, 13, 22. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Zhang, C.; Yang, H.; Han, X.; Fan, Z.; Hou, B. Depletion of PRDM9 enhances proliferation, migration and chemotaxis potentials in human periodontal ligament stem cells. Connect Tissue Res. 2020, 61, 498–508. [Google Scholar] [CrossRef]
- Basso, D.M.; Beattie, M.S.; Bresnahan, J.C. A sensitive and reliable locomotor rating scale for open field testing in rats. J. Neurotrauma 1995, 12, 1–21. [Google Scholar] [CrossRef]
- Di Paola, R.; Fusco, R.; Gugliandolo, E.; Crupi, R.; Evangelista, M.; Granese, R.; Cuzzocrea, S. Co-micronized Palmitoylethanolamide/Polydatin Treatment Causes Endometriotic Lesion Regression in a Rodent Model of Surgically Induced Endometriosis. Front. Pharmacol. 2016, 7, 382. [Google Scholar] [CrossRef] [Green Version]
- Zhang, C.; Han, X.; Liang, Y.; Liu, H.; Fan, Z.; Zhang, J. The Histone Demethylase KDM3B Promotes Osteo-/Odontogenic Differentiation, Cell Proliferation, and Migration Potential of Stem Cells from the Apical Papilla. Stem Cells Int. 2020, 2020, 8881021. [Google Scholar] [CrossRef]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, C.; Ye, W.; Zhao, M.; Long, L.; Xia, D.; Fan, Z. KDM6B Negatively Regulates the Neurogenesis Potential of Apical Papilla Stem Cells via HES1. Int. J. Mol. Sci. 2023, 24, 10608. https://doi.org/10.3390/ijms241310608
Zhang C, Ye W, Zhao M, Long L, Xia D, Fan Z. KDM6B Negatively Regulates the Neurogenesis Potential of Apical Papilla Stem Cells via HES1. International Journal of Molecular Sciences. 2023; 24(13):10608. https://doi.org/10.3390/ijms241310608
Chicago/Turabian StyleZhang, Chen, Weilong Ye, Mengyao Zhao, Lujue Long, Dengsheng Xia, and Zhipeng Fan. 2023. "KDM6B Negatively Regulates the Neurogenesis Potential of Apical Papilla Stem Cells via HES1" International Journal of Molecular Sciences 24, no. 13: 10608. https://doi.org/10.3390/ijms241310608




