Evaluation of Linkers’ Influence on Peptide-Based Piezoelectric Biosensors’ Sensitivity to Aldehydes in the Gas Phase
Abstract
1. Introduction
2. Results
2.1. In Silico Docking Simulations
2.2. Peptides Deposition
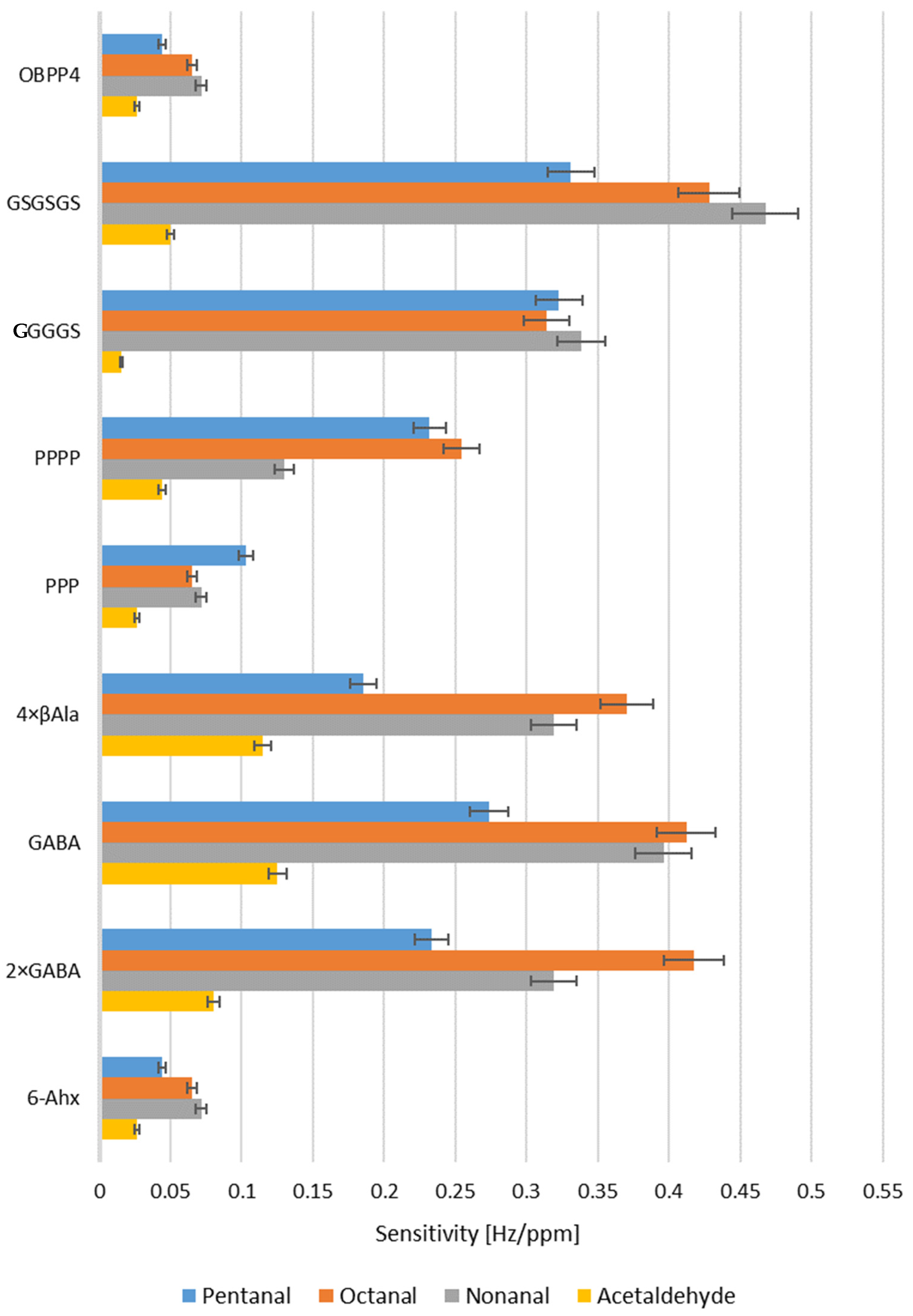
2.3. Biosensors’ Responses to Aldehydes in Gas Phase
3. Discussion
4. Materials and Methods
4.1. Peptides Design and Molecular Modelling
4.2. Peptide Synthesis and Deposition on QCM Transducers
4.3. Measurement Setup
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- World Health Organization. WHO The Top 10 Causes of Death. Available online: https://www.who.int/news-room/fact-sheets/detail/the-top-10-causes-of-death (accessed on 9 December 2020).
- Karimi-Maleh, H.; Liu, Y.; Li, Z.; Darabi, R.; Orooji, Y.; Karaman, C.; Karimi, F.; Baghayeri, M.; Rouhi, J.; Fu, L.; et al. Calf thymus ds-DNA intercalation with pendimethalin herbicide at the surface of ZIF-8/Co/rGO/C3N4/ds-DNA/SPCE; A bio-sensing approach for pendimethalin quantification confirmed by molecular docking study. Chemosphere 2023, 332, 138815. [Google Scholar] [CrossRef] [PubMed]
- Feng, Z.; Pepe, M.S. Adding rigor to biomarker evaluations-EDRN experience. Cancer Epidemiol. Biomark. Prev. 2020, 29, 2575–2582. [Google Scholar] [CrossRef] [PubMed]
- Wasilewski, T.; Neubauer, D.; Kamysz, W.; Gębicki, J. Recent progress in the development of peptide-based gas biosensors for environmental monitoring. Case Stud. Chem. Environ. Eng. 2022, 5, 100197. [Google Scholar] [CrossRef]
- Gaggiotti, S.; Palmieri, S.; Della Pelle, F.; Sergi, M.; Cichelli, A.; Mascini, M.; Compagnone, D. Piezoelectric peptide-hpDNA based electronic nose for the detection of terpenes; Evaluation of the aroma profile in different Cannabis sativa L. (hemp) samples. Sens. Actuators B Chem. 2020, 308, 127697. [Google Scholar] [CrossRef]
- Masoumi, S.; Hajghassem, H. Design of the trinitrotoluene biosensor using polydiacetylene conjugated with peptide receptors coated on GR-FETs with colorimetric response. Sens. Rev. 2019, 39, 819–827. [Google Scholar] [CrossRef]
- Pathak, A.K.; Swargiary, K.; Kongsawang, N.; Jitpratak, P. Recent Advances in Sensing Materials Targeting Clinical Volatile Organic Compound (VOC) Biomarkers: A Review. Biosensors 2023, 13, 114. [Google Scholar] [CrossRef]
- Wasilewski, T.; Gębicki, J.; Kamysz, W. Advances in olfaction-inspired biomaterials applied to bioelectronic noses. Sens. Actuators B Chem. 2018, 257, 511–537. [Google Scholar] [CrossRef]
- Tertis, M.; Hosu, O.; Feier, B.; Cernat, A.; Florea, A.; Cristea, C. Electrochemical Peptide-Based Sensors for Foodborne Pathogens Detection. Molecules 2021, 26, 3200. [Google Scholar] [CrossRef]
- Alves, L.M.; Barros, H.L.S.; Flauzino, J.M.R.; Guedes, P.H.G.; Pereira, J.M.; Fujiwara, R.T.; Mineo, T.W.P.; Mineo, J.R.; de Oliveira, R.J.; Madurro, J.M.; et al. A novel peptide-based sensor platform for detection of anti-Toxoplasma gondii immunoglobulins. J. Pharm. Biomed. Anal. 2019, 175, 112778. [Google Scholar] [CrossRef]
- Cernat, A.; Canciu, A.; Tertis, M.; Graur, F.; Cristea, C. Synergic action of thermosensitive hydrogel and Au/Ag nanoalloy for sensitive and selective detection of pyocyanin. Anal. Bioanal. Chem. 2019, 411, 3829–3838. [Google Scholar] [CrossRef]
- Migoń, D.; Wasilewski, T.; Suchy, D. Application of QCM in Peptide and Protein-Based Drug Product Development. Molecules 2020, 25, 3950. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Q.; Zheng, C.; Zhu, L.; Wang, J. A review on rapid detection of modified quartz crystal microbalance sensors for food: Contamination, flavour and adulteration. TrAC Trends Anal. Chem. 2022, 157, 116805. [Google Scholar] [CrossRef]
- Wasilewski, T.; Szulczyński, B.; Dobrzyniewski, D.; Jakubaszek, W.; Gębicki, J.; Kamysz, W. Development and Assessment of Regeneration Methods for Peptide-Based QCM Biosensors in VOCs Analysis Applications. Biosensors 2022, 12, 309. [Google Scholar] [CrossRef]
- Barbosa, A.J.M.; Oliveira, A.R.; Roque, A.C.A. Protein- and Peptide-Based Biosensors in Artificial Olfaction. Trends Biotechnol. 2018, 36, 1244–1258. [Google Scholar] [CrossRef]
- Kotlowski, C.; Larisika, M.; Guerin, P.M.; Kleber, C.; Kröber, T.; Mastrogiacomo, R.; Nowak, C.; Pelosi, P.; Schütz, S.; Schwaighofer, A.; et al. Chemical Fine discrimination of volatile compounds by graphene-immobilized odorant-binding proteins. Sens. Actuators B Chem. 2018, 256, 564–572. [Google Scholar] [CrossRef]
- Khadka, R.; Aydemir, N.; Carraher, C.; Hamiaux, C.; Colbert, D.; Cheema, J.; Malmström, J.; Kralicek, A.; Travas-Sejdic, J. An ultrasensitive electrochemical impedance-based biosensor using insect odorant receptors to detect odorants. Biosens. Bioelectron. 2019, 126, 207–213. [Google Scholar] [CrossRef] [PubMed]
- Pelosi, P.; Zhu, J.; Knoll, W. Odorant-binding proteins as sensing elements for odour monitoring. Sensors 2018, 18, 3248. [Google Scholar] [CrossRef]
- Wasilewski, T.; Szulczyński, B.; Wojciechowski, M.; Kamysz, W.; Gębicki, J. A Highly Selective Biosensor Based on Peptide Directly Derived from the HarmOBP7 Aldehyde Binding Site. Sensors 2019, 19, 4284. [Google Scholar] [CrossRef] [PubMed]
- Floss, M.A.; Fink, T.; Maurer, F.; Volk, T.; Kreuer, S.; Müller-Wirtz, L.M. Exhaled Aldehydes as Biomarkers for Lung Diseases: A Narrative Review. Molecules 2022, 27, 5258. [Google Scholar] [CrossRef] [PubMed]
- Nasim, F.; Sabath, B.F.; Eapen, G.A. Lung Cancer. Med. Clin. N. Am. 2019, 103, 463–473. [Google Scholar] [CrossRef]
- Fuchs, P.; Loeseken, C.; Schubert, J.K.; Miekisch, W. Breath gas aldehydes as biomarkers of lung cancer. Int. J. Cancer 2010, 126, 2663–2670. [Google Scholar] [CrossRef]
- Yang, G.; Xiao, Z.; Tang, C.; Deng, Y.; Huang, H.; He, Z. Recent advances in biosensor for detection of lung cancer biomarkers. Biosens. Bioelectron. 2019, 141, 111416. [Google Scholar] [CrossRef]
- Španěl, P.; Smith, D. Quantification of volatile metabolites in exhaled breath by selected ion flow tube mass spectrometry, SIFT-MS. Clin. Mass Spectrom. 2020, 16, 18–24. [Google Scholar] [CrossRef]
- Zhang, J.; Zhang, X.; Wei, X.; Xue, Y.; Wan, H.; Wang, P. Recent advances in acoustic wave biosensors for the detection of disease-related biomarkers: A review. Anal. Chim. Acta 2021, 1164, 338321. [Google Scholar] [CrossRef] [PubMed]
- Broza, Y.Y.; Zhou, X.; Yuan, M.; Qu, D.; Zheng, Y.; Vishinkin, R.; Khatib, M.; Wu, W.; Haick, H. Disease Detection with Molecular Biomarkers: From Chemistry of Body Fluids to Nature-Inspired Chemical Sensors. Chem. Rev. 2019, 119, 11761–11817. [Google Scholar] [CrossRef]
- Sánchez, C.; Santos, J.P.; Lozano, J. Use of electronic noses for diagnosis of digestive and respiratory diseases through the breath. Biosensors 2019, 9, 35. [Google Scholar] [CrossRef] [PubMed]
- Li, P.; Lee, G.H.; Kim, S.Y.; Kwon, S.Y.; Kim, H.R.; Park, S. From Diagnosis to Treatment: Recent Advances in Patient-Friendly Biosensors and Implantable Devices. ACS Nano 2021, 15, 1960–2004. [Google Scholar] [CrossRef] [PubMed]
- Thorn, R.M.S.; Greenman, J. Microbial volatile compounds in health and disease conditions. J. Breath Res. 2012, 6, 24001. [Google Scholar] [CrossRef] [PubMed]
- Wasilewski, T.; Brito, N.F.; Szulczyński, B.; Wojciechowski, M.; Buda, N.; Melo, A.C.A.; Kamysz, W.; Gębicki, J. Olfactory receptor-based biosensors as potential future tools in medical diagnosis. TrAC Trends Anal. Chem. 2022, 150, 116599. [Google Scholar] [CrossRef]
- Mcnerney, R.; Mallard, K.; Okolo, P.I.; Turner, C. Production of volatile organic compounds by mycobacteria. FEMS Microbiol. Lett. 2012, 328, 150–156. [Google Scholar] [CrossRef]
- Davis, V.W.; Bathe, O.F.; Schiller, D.E.; Slupsky, C.M.; Sawyer, M.B. Metabolomics and surgical oncology: Potential role for small molecule biomarkers. J. Surg. Oncol. 2011, 103, 451–459. [Google Scholar] [CrossRef] [PubMed]
- Noreldeen, H.A.A.; Liu, X.; Xu, G. Metabolomics of lung cancer: Analytical platforms and their applications. J. Sep. Sci. 2020, 43, 120–133. [Google Scholar] [CrossRef] [PubMed]
- Zhao, X.; Gao, W.; Yin, J.; Fan, W.; Wang, Z.; Hu, K.; Mai, Y.; Luan, A.; Xu, B.; Jin, Q. A high-precision thermometry microfluidic chip for real-time monitoring of the physiological process of live tumour cells. Talanta 2021, 226, 122101. [Google Scholar] [CrossRef]
- Xia, Z.; Li, D.; Deng, W. Identification and Detection of Volatile Aldehydes as Lung Cancer Biomarkers by Vapor Generation Combined with Paper-Based Thin-Film Microextraction. Anal. Chem. 2021, 93, 4924–4931. [Google Scholar] [CrossRef]
- Zhang, F.; Sadovski, O.; Xin, S.J.; Woolley, G.A. Stabilization of Folded Peptide and Protein Structures via Distance Matching with a Long, Rigid Cross-Linker. J. Am. Chem. Soc. 2007, 129, 14154–14155. [Google Scholar] [CrossRef]
- Wang, J.; Sakai, K.; Kiwa, T. Rational Design of Peptides Derived from Odorant-Binding Proteins for SARS-CoV-2-Related Volatile Organic Compounds Recognition. Molecules 2022, 27, 3917. [Google Scholar] [CrossRef] [PubMed]
- Rózycki, B.; Cazade, P.A.; O’Mahony, S.; Thompson, D.; Cieplak, M. The length but not the sequence of peptide linker modules exerts the primary influence on the conformations of protein domains in cellulosome multi-enzyme complexes. Phys. Chem. Chem. Phys. 2017, 19, 21414–21425. [Google Scholar] [CrossRef]
- Vazana, Y.; Barak, Y.; Unger, T.; Peleg, Y.; Shamshoum, M.; Ben-Yehezkel, T.; Mazor, Y.; Shapiro, E.; Lamed, R.; Bayer, E.A. A synthetic biology approach for evaluating the functional contribution of designer cellulosome components to deconstruction of cellulosic substrates. Biotechnol. Biofuels 2013, 6, 182. [Google Scholar] [CrossRef]
- Li, G.; Huang, Z.; Zhang, C.; Dong, B.J.; Guo, R.H.; Yue, H.W.; Yan, L.T.; Xing, X.H. Construction of a linker library with widely controllable flexibility for fusion protein design. Appl. Microbiol. Biotechnol. 2016, 100, 215–225. [Google Scholar] [CrossRef]
- Nowinski, A.K.; Sun, F.; White, A.D.; Keefe, A.J.; Jiang, S. Sequence, structure, and function of peptide self-assembled monolayers. J. Am. Chem. Soc. 2012, 134, 6000–6005. [Google Scholar] [CrossRef]
- Zhang, L.; Navaratna, T.; Liao, J.; Thurber, G.M. Dual-purpose linker for alpha helix stabilization and imaging agent conjugation to glucagon-like peptide-1 receptor ligands. Bioconjug. Chem. 2015, 26, 329–337. [Google Scholar] [CrossRef] [PubMed]
- Fujimoto, K.; Kajino, M.; Inouye, M. Development of a Series of Cross-Linking Agents that Effectively Stabilize α-Helical Structures in Various Short Peptides. Chem. Eur. J. 2008, 14, 857–863. [Google Scholar] [CrossRef] [PubMed]
- Henchey, L.K.; Jochim, A.L.; Arora, P.S. Contemporary strategies for the stabilization of peptides in the α-helical conformation. Curr. Opin. Chem. Biol. 2008, 12, 692–697. [Google Scholar] [CrossRef]
- Schmidt, S.; Debant, A. Aptamer-derived peptide inhibitors of rho guanine nucleotide exchange factors. Enzymes 2013, 33, 147–168. [Google Scholar]
- Leo, N.; Liu, J.; Archbold, I.; Tang, Y.; Zeng, X. Ionic Strength, Surface Charge, and Packing Density Effects on the Properties of Peptide Self-Assembled Monolayers. Langmuir 2017, 33, 2050–2058. [Google Scholar] [CrossRef] [PubMed]
- Singh, M.; Kaur, N.; Comini, E. The role of self-assembled monolayers in electronic devices. J. Mater. Chem. C 2020, 8, 3938–3955. [Google Scholar] [CrossRef]
- Groß, A.; Hashimoto, C.; Sticht, H.; Eichler, J. Synthetic Peptides as Protein Mimics. Front. Bioeng. Biotechnol. 2016, 3, 211. [Google Scholar] [CrossRef]
- González-Fernández, E.; Staderini, M.; Avlonitis, N.; Murray, A.F.; Mount, A.R.; Bradley, M. Effect of spacer length on the performance of peptide-based electrochemical biosensors for protease detection. Sens. Actuators B Chem. 2018, 255, 3040–3046. [Google Scholar] [CrossRef]
- Shen, Y.; Maupetit, J.; Derreumaux, P.; Tufféry, P. Improved PEP-FOLD Approach for Peptide and Miniprotein Structure Prediction. J. Chem. Theory Comput. 2014, 10, 4745–4758. [Google Scholar] [CrossRef]
- Thévenet, P.; Shen, Y.; Maupetit, J.; Guyon, F.; Derreumaux, P.; Tufféry, P. PEP-FOLD: An updated de novo structure prediction server for both linear and disulfide bonded cyclic peptides. Nucleic Acids Res. 2012, 40, 288–293. [Google Scholar] [CrossRef]
- Van Der Spoel, D.; Lindahl, E.; Hess, B.; Groenhof, G.; Mark, A.E.; Berendsen, H.J.C. GROMACS: Fast, flexible, and free. J. Comput. Chem. 2005, 26, 1701–1718. [Google Scholar] [CrossRef]
- Kutzner, C.; Páll, S.; Fechner, M.; Esztermann, A.; de Groot, B.L.; Grubmüller, H. More bang for your buck: Improved use of GPU nodes for GROMACS 2018. J. Comput. Chem. 2019, 40, 2418–2431. [Google Scholar] [CrossRef]
- Futera, Z.; Blumberger, J. Adsorption of Amino Acids on Gold: Assessing the Accuracy of the GolP-CHARMM Force Field and Parametrization of Au-S Bonds. J. Chem. Theory Comput. 2019, 15, 613–624. [Google Scholar] [CrossRef] [PubMed]
- Froimowitz, M. HyperChem(TM): A software package for computational chemistry and molecular modeling. Biotechniques 1993, 14, 1010–1013. [Google Scholar]
- Butt, S.S.; Badshah, Y.; Shabbir, M.; Rafiq, M. Molecular Docking Using Chimera and Autodock Vina Software for Nonbioinformaticians. JMIR Bioinf. Biotechnol. 2020, 1, e14232. [Google Scholar] [CrossRef]
- Morris, G.M.; Huey, R.; Lindstrom, W.; Sanner, M.F.; Belew, R.K.; Goodsell, D.S.; Olson, A.J. AutoDock4 and AutoDockTools4: Automated docking with selective receptor flexibility. J. Comput. Chem. 2009, 30, 2785–2791. [Google Scholar] [CrossRef]
- Wojciechowski, M. Simplified AutoDock force field for hydrated binding sites. J. Mol. Graph. Model. 2017, 78, 74–80. [Google Scholar] [CrossRef] [PubMed]
- Wasilewski, T.; Szulczyński, B.; Kamysz, W.; Gębicki, J.; Namieśnik, J. Evaluation of Three Peptide Immobilization Techniques on a QCM Surface Related to Acetaldehyde Responses in the Gas Phase. Sensors 2018, 18, 3942. [Google Scholar] [CrossRef]
- Wasilewski, T.; Szulczyński, B.; Wojciechowski, M.; Kamysz, W.; Gębicki, J. Determination of long-chain aldehydes using a novel quartz crystal microbalance sensor based on a biomimetic peptide. Microchem. J. 2020, 154, 104509. [Google Scholar] [CrossRef]
- Skountzos, E.N.; von Wrochem, F.; Mavrantzas, V.G. Structure and Conformation of a Crystalline P3HT Film Adsorbed on an Alkanethiol Self-Assembled Monolayer Deposited on Gold. Macromol. Theory Simul. 2020, 29, 202000010. [Google Scholar] [CrossRef]
- Van Rosmalen, M.; Krom, M.; Merkx, M. Tuning the Flexibility of Glycine-Serine Linkers to Allow Rational Design of Multidomain Proteins. Biochemistry 2017, 56, 6565–6574. [Google Scholar] [CrossRef] [PubMed]
- Leo, N.; Shang, Y.; Yu, J.; Zeng, X. Characterization of Self-Assembled Monolayers of Peptide Mimotopes of CD20 Antigen and Their Binding with Rituximab. Langmuir 2015, 31, 13764–13772. [Google Scholar] [CrossRef] [PubMed]
- Lim, J.M.; Ryu, M.Y.; Kim, J.H.; Cho, C.H.; Park, T.J.; Park, J.P. An electrochemical biosensor for detection of the sepsis-related biomarker procalcitonin. RSC Adv. 2017, 7, 36562–36565. [Google Scholar] [CrossRef]
- Markowska, A.; Markowski, A.R.; Jarocka-Karpowicz, I. The Importance of 6-Aminohexanoic Acid as a Hydrophobic, Flexible Structural Element. Int. J. Mol. Sci. 2021, 22, 12122. [Google Scholar] [CrossRef] [PubMed]
- Guo, H.; Hathaway, H.; Royce, M.E.; Prossnitz, E.R.; Miao, Y. Influences of hydrocarbon linkers on the receptor binding affinities of gonadotropin-releasing hormone peptides. Bioorganic Med. Chem. Lett. 2013, 23, 5484–5487. [Google Scholar] [CrossRef] [PubMed]
- Sancho, V.; Di Florio, A.; Moody, T.W.; Jensen, R.T. Bombesin Receptor-Mediated Imaging and Cytotoxicity: Review and Current Status. Curr. Drug Deliv. 2011, 8, 79–134. [Google Scholar] [CrossRef] [PubMed]
- Shahabi, M.; Hajihosseini, R.; Nau, W.M.; Noghabi, K.A.; Norouzy, A. Augmenting Peptide Flexibility by Inserting Gamma-Aminobutyric Acid (GABA) in Their Sequence. Int. J. Pept. Res. Ther. 2020, 26, 2633–2640. [Google Scholar] [CrossRef]
- Li, H.; Cao, Y.; Wu, X.; Ye, Z.; Li, G. Peptide-based electrochemical biosensor for amyloid β 1–42 soluble oligomer assay. Talanta 2012, 93, 358–363. [Google Scholar] [CrossRef]
- Trzeciakiewicz, H.; Esteves-Villanueva, J.; Soudy, R.; Kaur, K.; Martic-Milne, S. Electrochemical Characterization of Protein Adsorption onto YNGRT-Au and VLGXE-Au Surfaces. Sensors 2015, 15, 19429–19442. [Google Scholar] [CrossRef]
- Xu, H.; Lu, J.R.; Williams, D.E. Effect of surface packing density of interfacially adsorbed monoclonal antibody on the binding of hormonal antigen human chorionic gonadotrophin. J. Phys. Chem. B 2006, 110, 1907–1914. [Google Scholar] [CrossRef]
- Xu, H.; Williams, D.; Lu, J. The effect of antibody surface packing density on its antigen binding capacity. Prog. Nat. Sci. 2005, 15, 139–144. [Google Scholar] [CrossRef]
- Daneshkhah, A.; Vij, S.; Siegel, A.P.; Agarwal, M. Polyetherimide/carbon black composite sensors demonstrate selective detection of medium-chain aldehydes including nonanal. Chem. Eng. J. 2019, 383, 123104. [Google Scholar] [CrossRef]
- Masuda, Y.; Kato, K.; Kida, M.; Otsuka, J. Selective nonanal molecular recognition with SnO2 nanosheets for lung cancer sensor. Int. J. Appl. Ceram. Technol. 2019, 16, 1807–1811. [Google Scholar] [CrossRef]
- Masuda, Y.; Itoh, T.; Shin, W.; Kato, K. SnO2 nanosheet/nanoparticle detector for the sensing of 1-nonanal gas produced by lung cancer. Sci. Rep. 2015, 5, 10122. [Google Scholar] [CrossRef]
- Itoh, T.; Nakashima, T.; Izu, N.; Shin, W.; Setoguchi, Y.; Kato, K.; Toyota, M. Noble Metal Added Tin Oxide VOC Sensors as Nonanal Detection for Exhaled Breath Air Monitoring. In Proceedings of the IMCS 2012, Nuremberg, Germany, 20–23 May 2012; AMA Service GmbH: Wunstorf, Germany, 2012; pp. 547–550. [Google Scholar]
- Tsujiguchi, M.; Aitoku, T.; Takase, H.; Maruo, Y.Y. Nonanal Sensor Fabrication Using Aldol Condensation Reaction Inside Alkali-Resistant Porous Glass. IEEE Sens. J. 2021, 21, 8868–8877. [Google Scholar] [CrossRef]
- Jahangiri-Manesh, A.; Mousazadeh, M.; Nikkhah, M.; Abbasian, S.; Moshaii, A.; Masroor, M.J.; Norouzi, P. Molecularly imprinted polymer-based chemiresistive sensor for detection of nonanal as a cancer related biomarker. Microchem. J. 2022, 173, 106988. [Google Scholar] [CrossRef]
- Aasi, A.; Ebrahimi Bajgani, S.; Panchapakesan, B. A first-principles investigation on the adsorption of octanal and nonanal molecules with decorated monolayer WS2as promising gas sensing platform. AIP Adv. 2023, 13, 025157. [Google Scholar] [CrossRef]
- Liu, B.; Huang, Y.; Kam, K.W.; Cheung, W.-F.; Zhao, N.; Zheng, B. Functionalized graphene-based chemiresistive electronic nose for discrimination of disease-related volatile organic compounds. Biosens. Bioelectron. X 2019, 1, 100016. [Google Scholar] [CrossRef]
- Liu, C.; Wyszynski, B.; Yatabe, R.; Hayashi, K.; Toko, K. Molecularly imprinted sol-gel-based QCM sensor arrays for the detection and recognition of volatile aldehydes. Sensors 2017, 17, 382. [Google Scholar] [CrossRef]
- Neubauer, D.; Jaśkiewicz, M.; Bauer, M.; Gołacki, K.; Kamysz, W. Ultrashort cationic lipopeptides–effect of N-terminal amino acid and fatty acid type on antimicrobial activity and hemolysis. Molecules 2020, 25, 257. [Google Scholar] [CrossRef]
- Zhang, H. Surface characterization techniques for polyurethane biomaterials. In Advances in Polyurethane Biomaterials; Elsevier: Amsterdam, The Netherlands, 2016; pp. 23–73. ISBN 9780081006221. [Google Scholar]
- Latif, U.; Can, S.; Hayden, O.; Grillberger, P.; Dickert, F.L. Sauerbrey and anti-Sauerbrey behavioral studies in QCM sensors—Detection of bioanalytes. Sens. Actuators B Chem. 2013, 176, 825–830. [Google Scholar] [CrossRef]
- Brothers, M.C.; Moore, D.; St. Lawrence, M.; Harris, J.; Joseph, R.M.; Ratcliff, E.; Ruiz, O.N.; Glavin, N.; Kim, S.S. Impact of Self-Assembled Monolayer Design and Electrochemical Factors on Impedance-Based Biosensing. Sensors 2020, 20, 2246. [Google Scholar] [CrossRef] [PubMed]
- Walensky, L.D.; Bird, G.H. Hydrocarbon-Stapled Peptides: Principles, Practice, and Progress. J. Med. Chem. 2014, 57, 6275–6288. [Google Scholar] [CrossRef] [PubMed]







| Linker | Number of AA Residues | Number of Atoms Between Amino Group and Terminal Carbonyl Carbon Atom | Calculated Length [nm] |
|---|---|---|---|
| Group I—proteinogenic | |||
| GSGSGS | 6 | 16 | 0.94 |
| GGGGS | 5 | 13 | 1.40 |
| PPPP | 4 | 10 | 1.14 |
| PPP | 3 | 7 | 0.84 |
| Group II—non-proteinogenic | |||
| 4 × βAla | 4 | 14 | 0.95 |
| GABA | 1 | 3 | 0.56 |
| 2 × GABA | 2 | 8 | 0.90 |
| 6-Ahx | 1 | 5 | 0.66 |
| Peptide | Sensitivity [Hz/ppm] LOD [ppm] | |||
|---|---|---|---|---|
| Pentanal | Octanal | Nonanal | Acetaldehyde | |
| OBPP4 | 0.0442 | 0.0649 | 0.0715 | 0.0265 |
| 79 | 43 | 18 | 187 | |
| GSGSGS | 0.3312 | 0.4281 | 0.4676 | 0.0503 |
| 11 | 5 | 2 | 352 | |
| GGGGS | 0.3228 | 0.3141 | 0.3382 | 0.0155 |
| 13 | 10 | 11 | - | |
| PPPP | 0.2319 | 0.2544 | 0.1298 | 0.0442 |
| 35 | 32 | 65 | 320 | |
| PPP | 0.1028 | 0.0649 | 0.0715 | 0.0265 |
| 58 | 289 | 255 | - | |
| 4 × βAla | 0.1853 | 0.3702 | 0.3188 | 0.1149 |
| 39 | 15 | 13 | 59 | |
| GABA | 0.2738 | 0.4123 | 0.3961 | 0.1250 |
| 29 | 7 | 9 | 50 | |
| 2 × GABA | 0.2331 | 0.4176 | 0.3188 | 0.0802 |
| 22 | 5 | 11 | 322 | |
| 6-Ahx | 0.0442 | 0.0649 | 0.0715 | 0.0265 |
| 211 | 259 | 287 | - | |
| Recognition | LOD [ppm] | Ref. |
|---|---|---|
| Chemoresistive, SnO2 nanosheets; SnO2 nanosheets and nanoparticles | 0.1; 0.05 | [74,75] |
| Chemoresistive, Pt-, Pd-, and Au-loaded SnO2 thick films | 0.05 | [76] |
| Chemoresistive, functionalized rGO | 25 | [80] |
| Chemoresistive, Polyetherimide/carbon black | 1 | [73] |
| Chemoresistive, MIP-AuNPs | 4.5 | [78] |
| Optical, vanillin | 0.125 | [77] |
| Piezoelectric, molecularly imprinted sol-gel | Several ppm | [81] |
| Piezoelectric, peptide | 14 | [60] |
| Piezoelectric, peptide | 2 | This study |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wasilewski, T.; Neubauer, D.; Wojciechowski, M.; Szulczyński, B.; Gębicki, J.; Kamysz, W. Evaluation of Linkers’ Influence on Peptide-Based Piezoelectric Biosensors’ Sensitivity to Aldehydes in the Gas Phase. Int. J. Mol. Sci. 2023, 24, 10610. https://doi.org/10.3390/ijms241310610
Wasilewski T, Neubauer D, Wojciechowski M, Szulczyński B, Gębicki J, Kamysz W. Evaluation of Linkers’ Influence on Peptide-Based Piezoelectric Biosensors’ Sensitivity to Aldehydes in the Gas Phase. International Journal of Molecular Sciences. 2023; 24(13):10610. https://doi.org/10.3390/ijms241310610
Chicago/Turabian StyleWasilewski, Tomasz, Damian Neubauer, Marek Wojciechowski, Bartosz Szulczyński, Jacek Gębicki, and Wojciech Kamysz. 2023. "Evaluation of Linkers’ Influence on Peptide-Based Piezoelectric Biosensors’ Sensitivity to Aldehydes in the Gas Phase" International Journal of Molecular Sciences 24, no. 13: 10610. https://doi.org/10.3390/ijms241310610
APA StyleWasilewski, T., Neubauer, D., Wojciechowski, M., Szulczyński, B., Gębicki, J., & Kamysz, W. (2023). Evaluation of Linkers’ Influence on Peptide-Based Piezoelectric Biosensors’ Sensitivity to Aldehydes in the Gas Phase. International Journal of Molecular Sciences, 24(13), 10610. https://doi.org/10.3390/ijms241310610










