Non-Vesicular Lipid Transport Machinery in Leishmania donovani: Functional Implications in Host-Parasite Interaction
Abstract
1. Introduction
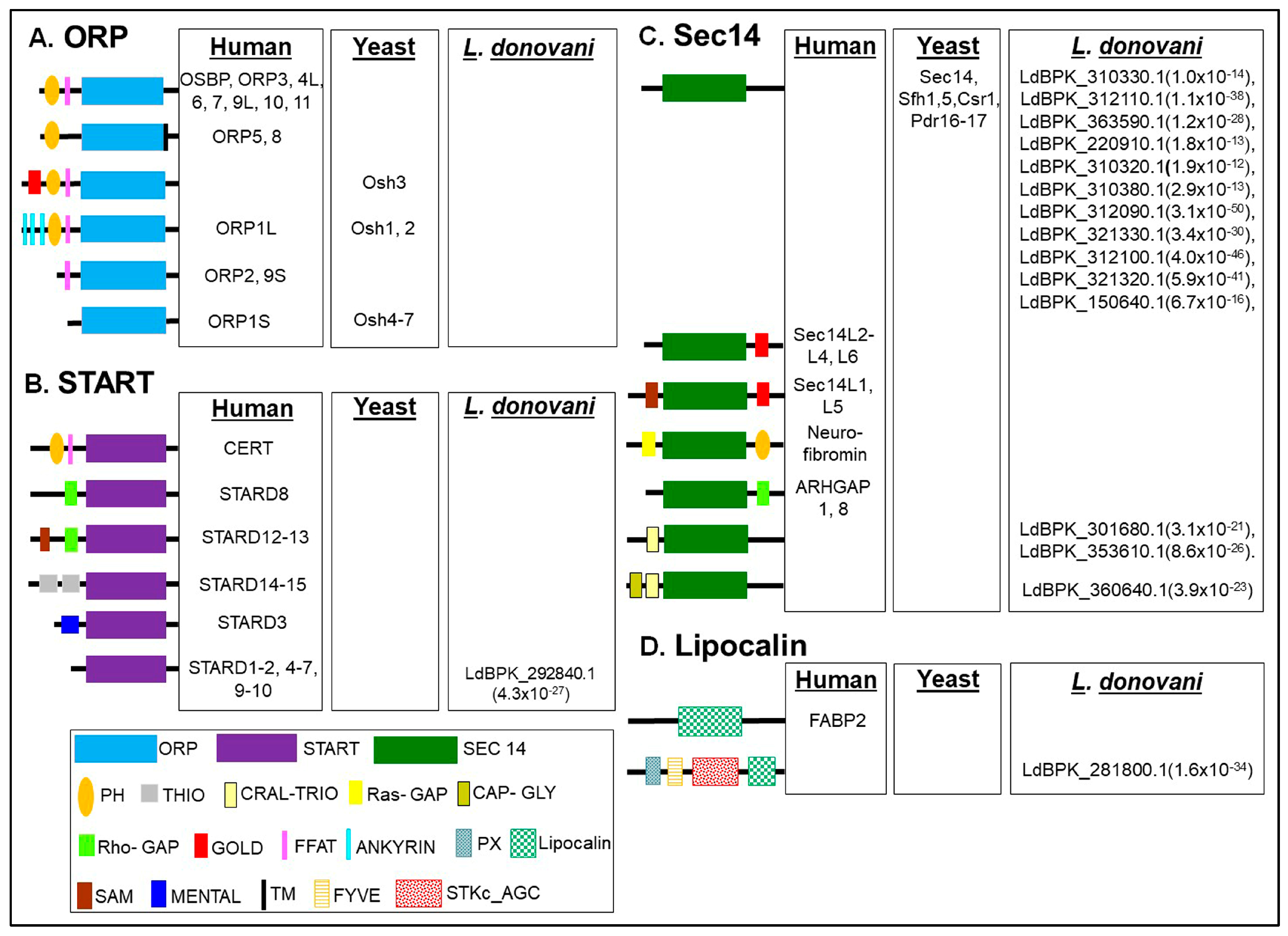
2. Overview on Domain Architecture of Lipid Transfer Proteins (LTPs)
3. Comparative Analysis of LTP Subtypes between L. donovani and Other Organisms
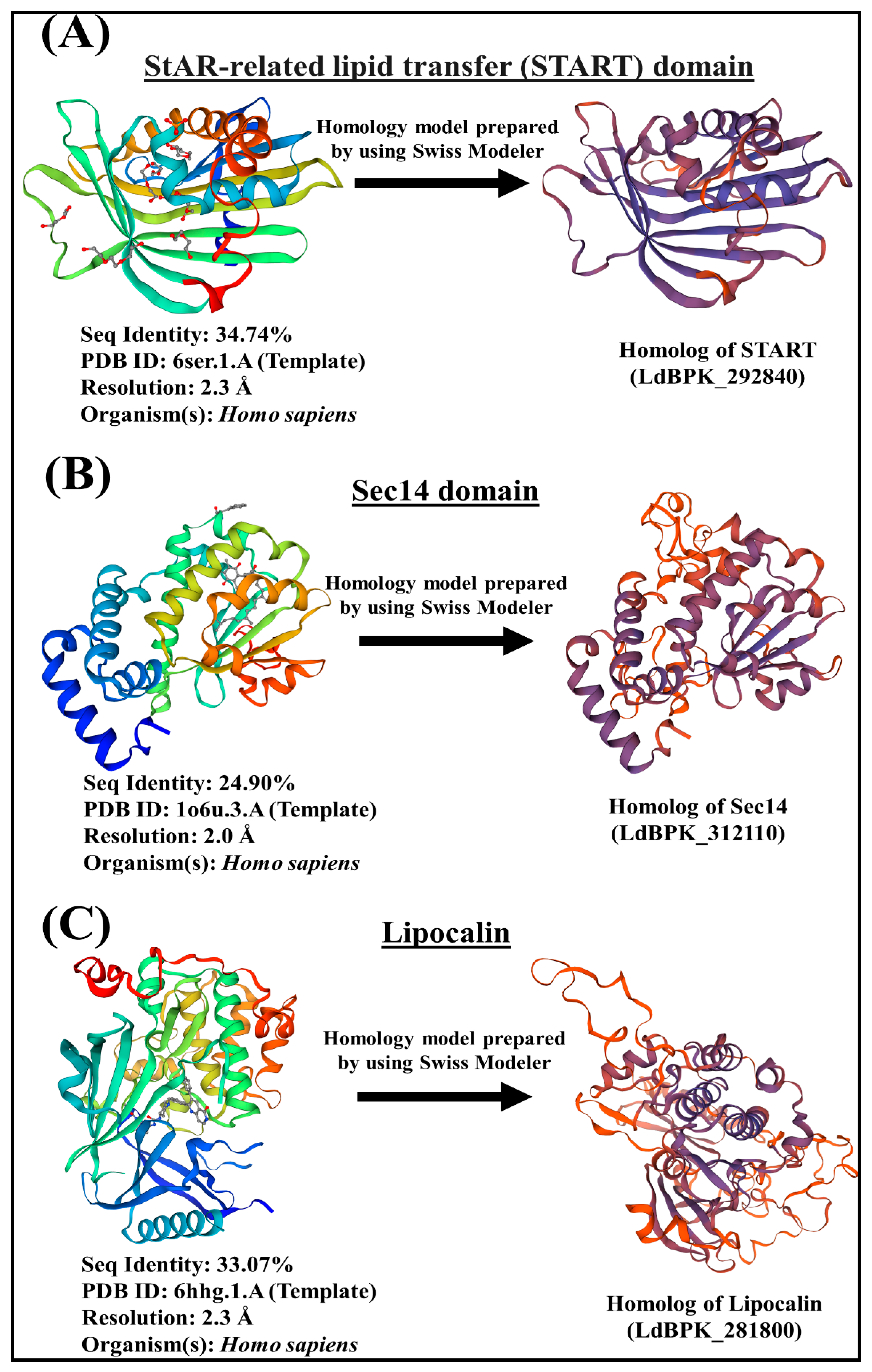
3.1. Identification, Domain Organization of LTP Homologs in L. donovani BPK282A1
3.1.1. START Domain Containing Proteins
3.1.2. Sec14s
3.1.3. Lipocalin
3.1.4. Other Proteins Known to Be Involved in Lipid Transfer in Other Organisms, but Missing in L. donovani
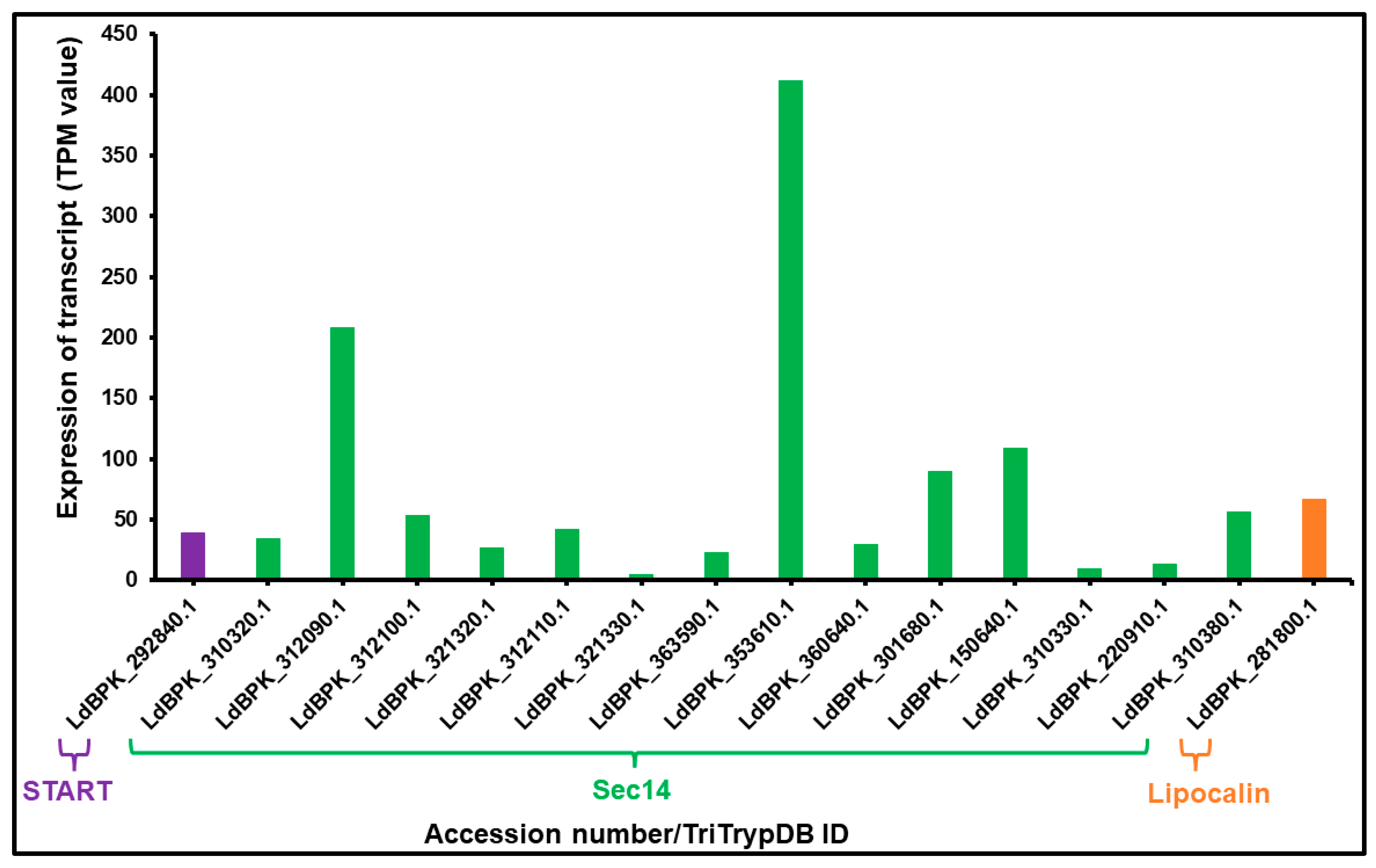
3.2. mRNA Expression of LTP Homologs L. donovani BPK282A1
4. LTPs and Their Biological Implications
4.1. Previous Reports on Biological Importance of LTPs in Other Eukaryotic Systems
4.2. Biological Significance of LTPs in L. donovani
4.2.1. Previous Reports on Lipids Homeostasis and Their Implications in Leishmania sp.
4.2.2. Proposed Implications of LTPs in the Biology of L. donovani
Role of LTPs in the Regulation of Vesicular Trafficking and Secretion of Virulence Factors
Implications of LTPs in Receptor-Ligand Interactions (Nutrient Sensing), Cytoskeleton Reorganization and Motility of the Parasite
Implications of LTPs in Intracellular Survival of L. donovani within Macrophages
5. Concluding Remarks
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| acyl-PG | acyl-phosphorylglycerol |
| BA | β-amylase |
| CL | cardiolipin, 1,3-bis(sn-3′-phosphatidyl)-sn-glycerol |
| DAG | diacyl glycerol |
| ER | endoplasmic reticulum |
| ERMES | ER–mitochondria encounter structure |
| HNF4 | hepatocyte nuclear factor 4 |
| IMM | inner mitochondrial membrane |
| LTD | lipid transfer domain |
| LTP | lipid transport protein |
| MCS | membrane contact sites |
| OMM | outer mitochondrial membrane |
| ORP | oxysterol-binding-protein-related proteins |
| PA | Phosphatidic acid |
| PC | phosphatidylcholine |
| PE | phosphatidylethanolamine |
| PH | pleckstrin homology |
| PI | phosphoinositide |
| PITP | phosphatidylinositol transfer protein |
| PKC | protein kinase C |
| PLA2 | phospholipase A2 |
| PM | plasma membrane |
| PRELI | protein of relevant evolutionary and lymphoid interest |
| PtdIn | phosphatidyl inositol |
| SHP | small heterodimer partner |
| SM | sphingomyelin |
| START | steroidogenic acute regulatory protein-related lipid transfer |
| TriTrypDB | database for kinetoplastid parasites |
References
- Holthuis, J.C.; van Meer, G.; Huitema, K. Lipid microdomains, lipid translocation and the organization of intracellular membrane transport. Mol. Membr. Biol. 2003, 20, 231–241. [Google Scholar] [CrossRef] [PubMed]
- Lev, S. Non-vesicular lipid transport by lipid-transfer proteins and beyond. Nat. Rev. Mol. Cell Biol. 2010, 11, 739–750. [Google Scholar] [CrossRef] [PubMed]
- Voelker, D.R. Organelle biogenesis and intracellular lipid transport in eukaryotes. Microbiol. Rev. 1991, 55, 543–560. [Google Scholar] [CrossRef] [PubMed]
- Sprong, H.; van der Sluijs, P.; van Meer, G. How proteins move lipids and lipids move proteins. Nat. Rev. Mol. Cell Biol. 2001, 2, 504–513. [Google Scholar] [CrossRef]
- Blom, T.; Somerharju, P.; Ikonen, E. Synthesis and Biosynthetic Trafficking of Membrane Lipids. Cold Spring Harb. Perspect. Biol. 2011, 3, a004713. [Google Scholar] [CrossRef]
- Kaplan, M.R.; Simoni, R.D. Intracellular transport of phosphatidylcholine to the plasma membrane. J. Cell Biol. 1985, 101, 441–445. [Google Scholar] [CrossRef]
- Vance, J.; Aasman, E.; Szarka, R. Brefeldin A does not inhibit the movement of phosphatidylethanolamine from its sites for synthesis to the cell surface. J. Biol. Chem. 1991, 266, 8241–8247. [Google Scholar] [CrossRef]
- Li, J.; Guizhen, G.; Kun, X.; Biyun, C.; Guixin, Y.; Feng, L.; Jiangwei, Q.; Tianyao, Z.; Xiaoming, W. Genome-wide survey and expression analysis of the putative nonspecific lipid transfer proteins in Brassica rapal. PLoS ONE 2014, 9, e84556. [Google Scholar]
- Levine, T. Short-range intracellular trafficking of small molecules across endoplasmic reticulum junctions. Trends Cell Biol. 2004, 14, 483–490. [Google Scholar] [CrossRef]
- Holthuis, J.; Levine, T.P. Lipid traffic: Floppy drives and a superhighway. Nat. Rev. Mol. Cell Biol. 2005, 6, 209–220. [Google Scholar] [CrossRef]
- Sleight, R.G. Intracellular lipid transport in eukaryotes. Annu. Rev. Physiol. 1987, 49, 193–208. [Google Scholar] [CrossRef] [PubMed]
- Van Meer, G. Lipid traffic in animal cells. Annu. Rev. Cell Biol. 1989, 5, 247–275. [Google Scholar] [CrossRef]
- Lev, S. Lipid homoeostasis and Golgi secretory function. Biochem. Soc. Trans. 2006, 34, 363–366. [Google Scholar] [CrossRef]
- Jones, J.D.; Thompson, T.E. Spontaneous phosphatidylcholine transfer by collision between vesicles at high lipid concentration. Biochemistry 1989, 28, 129–134. [Google Scholar] [CrossRef] [PubMed]
- Mesmin, B.; Maxfield, F.R. Intracellular sterol dynamics. Biochim. Biophys. Acta (BBA)-Mol. Cell Biol. Lipids 2009, 1791, 636–645. [Google Scholar] [CrossRef]
- Lev, S. Nonvesicular Lipid Transfer from the Endoplasmic Reticulum. Cold Spring Harb. Perspect. Biol. 2012, 4, a013300. [Google Scholar] [CrossRef] [PubMed]
- Helle, S.C.; Kanfer, G.; Kolar, K.; Lang, A.; Michel, A.H.; Kornmann, B. Organization and function of membrane contact sites. Biochim. Biophys. Acta (BBA) Mol. Cell Res. 2013, 1833, 2526–2541. [Google Scholar] [CrossRef]
- Ohashi, M.; De Vries, K.J.; Frank, R.; Snoek, G.; Bankaitis, V.; Wirtz, K.; Huttner, W.B. A role for phosphatidylinositol transfer protein in secretory vesicle formation. Nature 1995, 377, 544–547. [Google Scholar] [CrossRef]
- Kim, Y.J.; Guzman-Hernandez, M.-L.; Wisniewski, E.; Balla, T. Phosphatidylinositol-Phosphatidic Acid Exchange by Nir2 at ER-PM Contact Sites Maintains Phosphoinositide Signaling Competence. Dev. Cell 2015, 33, 549–561. [Google Scholar] [CrossRef]
- Casares, D.; Escribá, P.V.; Rosselló, C.A. Membrane Lipid Composition: Effect on Membrane and Organelle Structure, Function and Compartmentalization and Therapeutic Avenues. Int. J. Mol. Sci. 2019, 20, 2167. [Google Scholar] [CrossRef]
- Chiapparino, A.; Maeda, K.; Turei, D.; Saez-Rodriguez, J.; Gavin, A.-C. The orchestra of lipid-transfer proteins at the crossroads between metabolism and signaling. Prog. Lipid Res. 2016, 61, 30–39. [Google Scholar] [CrossRef] [PubMed]
- Wong, L.H.; Gatta, A.T.; Levine, T.P. Lipid transfer proteins: The lipid commute via shuttles, bridges and tubes. Nat. Rev. Mol. Cell Biol. 2018, 20, 85–101. [Google Scholar] [CrossRef]
- Neuman, S.D.; Levine, T.P.; Bashirullah, A. A novel superfamily of bridge-like lipid transfer proteins. Trends Cell Biol. 2022, 32, 962–974. [Google Scholar] [CrossRef] [PubMed]
- Levine, T.P. Sequence Analysis and Structural Predictions of Lipid Transfer Bridges in the Repeating Beta Groove (RBG) Superfamily Reveal Past and Present Domain Variations Affecting Form, Function and Interactions of VPS13, ATG2, SHIP164, Hobbit and Tweek. Contact 2022, 5, 251525642211343. [Google Scholar] [CrossRef] [PubMed]
- Alvar, J.; Vélez, I.D.; Bern, C.; Herrero, M.; Desjeux, P.; Cano, J.; Jannin, J.; den Boer, M.; the WHO Leishmaniasis Control Team. Leishmaniasis Worldwide and Global Estimates of Its Incidence. PLoS ONE 2012, 7, e35671. [Google Scholar] [CrossRef]
- Karunaweera, N.D. Leishmaniasis: Path toward elimination from the Indian subcontinent. Trop. Parasitol. 2016, 6, 2–4. [Google Scholar] [CrossRef]
- Thakur, L.; Singh, K.K.; Shanker, V.; Negi, A.; Jain, A.; Matlashewski, G.; Jain, M. Atypical leishmaniasis: A global perspective with emphasis on the Indian subcontinent. PLoS Negl. Trop. Dis. 2018, 12, e0006659. [Google Scholar] [CrossRef]
- Sundar, S.; Chakravarty, J. Liposomal amphotericin B and leishmaniasis: Dose and response. J. Glob. Infect. Dis. 2010, 2, 159–166. [Google Scholar] [CrossRef]
- Moradin, N.; Descoteaux, A. Leishmania promastigotes: Building a safe niche within macrophages. Front. Cell. Infect. Microbiol. 2012, 2, 121. [Google Scholar] [CrossRef]
- Colineau, L.; Lambertz, U.; Fornes, O.; Wasserman, W.W.; Reiner, N.E. c-Myc is a novel Leishmania virulence factor by proxy that targets the host miRNA system and is essential for survival in human macrophages. J. Biol. Chem. 2018, 293, 12805–12819. [Google Scholar] [CrossRef]
- Shah, Z.H.; Jones, D.R.; Sommer, L.; Foulger, R.; Bultsma, Y.; D’Santos, C.; Divecha, N. Nuclear phosphoinositides and their impact on nuclear functions. FEBS J. 2013, 280, 6295–6310. [Google Scholar] [CrossRef] [PubMed]
- Albi, E. Role of intranuclear lipids in health and disease. Clin. Lipidol. 2011, 6, 59–69. [Google Scholar] [CrossRef]
- Vaid, A.; Ranjan, R.; Smythe, W.A.; Hoppe, H.C.; Sharma, P. PfPI3K, a phosphatidylinositol-3 kinase from Plasmodium falciparum, is exported to the host erythrocyte and is involved in hemoglobin trafficking. Blood 2010, 115, 2500–2507. [Google Scholar] [CrossRef] [PubMed]
- Kumar, R.; Engwerda, C.R. Vaccines to prevent leishmaniasis. Clin. Transl. Immunol. 2014, 3, e13. [Google Scholar] [CrossRef]
- Zhang, K. Balancing de novo synthesis and salvage of lipids by Leishmania amastigotes. Curr. Opin. Microbiol. 2021, 63, 98–103. [Google Scholar] [CrossRef]
- Bouabid, C.; Yamaryo-Botté, Y.; Rabhi, S.; Bichiou, H.; Hkimi, C.; Bouglita, W.; Chaouach, M.; Eddaikra, N.; Ghedira, K.; Guizani-Tabbane, L.; et al. Fatty Acid Profiles of Leishmania major Derived from Human and Rodent Hosts in Endemic Cutaneous Leishmaniasis Areas of Tunisia and Algeria. Pathogens 2022, 11, 92. [Google Scholar] [CrossRef]
- Shakarian, A.M.; McGugan, G.C.; Joshi, M.B.; Stromberg, M.; Bowers, L.; Ganim, C.; Barowski, J.; Dwyer, D.M. Identification, characterization, and expression of a unique secretory lipase from the human pathogen Leishmania donovani. Mol. Cell. Biochem. 2010, 341, 17–31. [Google Scholar] [CrossRef]
- Deep, D.K.; Singh, R.; Kulshrestha, A.; Wajid, S.; Salotra, P. Lipase Precursor-Like Protein Promotes Miltefosine Tolerance in Leishmania donovani by Enhancing Parasite Infectivity and Eliciting Anti-inflammatory Responses in Host Macrophages. Antimicrob. Agents Chemother. 2018, 62, e00666-18. [Google Scholar] [CrossRef]
- Cestari, I.; Stuart, K. The phosphoinositide regulatory network in Trypanosoma brucei: Implications for cell-wide regulation in eukaryotes. PLoS Negl. Trop. Dis. 2020, 14, e0008689. [Google Scholar] [CrossRef]
- Watanabe, N.; Nakada-Tsukui, K.; Nozaki, T. Two isotypes of phosphatidylinositol 3-phosphate-binding sorting nexins play distinct roles in trogocytosis in Entamoeba histolytica. Cell. Microbiol. 2020, 22, e13144. [Google Scholar] [CrossRef]
- Nakada-Tsukui, K.; Watanabe, N.; Maehama, T.; Nozaki, T. Phosphatidylinositol Kinases and Phosphatases in Entamoeba histolytica. Front. Cell. Infect. Microbiol. 2019, 9, 150. [Google Scholar] [CrossRef] [PubMed]
- Cernikova, L.; Faso, C.; Hehl, A.B. Phosphoinositide-binding proteins mark, shape and functionally modulate highly-diverged endocytic compartments in the parasitic protist Giardia lamblia. PLoS Pathog. 2020, 16, e1008317. [Google Scholar] [CrossRef] [PubMed]
- Jana, A.; Sinha, A.; Sarkar, S. Phosphoinositide binding profiles of the PX domains of Giardia lamblia. Parasitol. Int. 2017, 66, 606–614. [Google Scholar] [CrossRef] [PubMed]
- Van Ooij, C.; Withers-Martinez, C.; Ringel, A.S.; Haldar, K.; Blackman, M.J. Identification of a Plasmodium falciparum phospholipid transfer protein. J. Biol. Chem. 2013, 288, 31971–31983. [Google Scholar] [CrossRef]
- Hill, R.J.; Ringel, A.; Knuepfer, E.; Moon, R.W.; Blackman, M.J.; van Ooij, C. Regulation and essentiality of the StAR-related lipid transfer (START) domain-containing phospholipid transfer protein PFA0210c in malaria parasites. J. Biol. Chem. 2016, 291, 24280–24292. [Google Scholar] [CrossRef]
- Das, K.; Nozaki, T. Non-vesicular Lipid Transport Machinery in Entamoeba histolytica. Front. Cell. Infect. Microbiol. 2018, 8, 315. [Google Scholar] [CrossRef]
- Das, K.; Watanabe, N.; Nozaki, T. Two StAR-related lipid transfer proteins play specific roles in endocytosis, exocytosis, and motility in the parasitic protist Entamoeba histolytica. PLoS Pathog. 2021, 17, e1009551. [Google Scholar] [CrossRef]
- Ressurreição, M.; van Ooij, C. Lipid transport proteins in malaria, from Plasmodium parasites to their hosts. Biochim. Et Biophys. Acta (BBA)-Mol. Cell Biol. Lipids 2021, 1866, 159047. [Google Scholar] [CrossRef]
- Garten, M.; Beck, J.R.; Roth, R.; Tenkova-Heuser, T.; Heuser, J.; Istvan, E.S.; Bleck, C.K.E.; Goldberg, D.E.; Zimmerberg, J. Contacting domains segregate a lipid transporter from a solute transporter in the malarial host–parasite interface. Nat. Commun. 2020, 11, 1–10. [Google Scholar] [CrossRef]
- Cho, W. Membrane Targeting by C1 and C2 Domains. J. Biol. Chem. 2001, 276, 32407–32410. [Google Scholar] [CrossRef]
- Kasper, A.M.; Helmkamp, G.M., Jr. Intermembrane phospholipid fluxes catalyzed by bovine brain phospholipid exchange protein. Biochim. Biophys. Acta (BBA)-Lipids Lipid Metab. 1981, 664, 22–32. [Google Scholar] [CrossRef]
- Helmkamp, G.M. Phospholipid transfer proteins: Mechanism of action. J. Bioenerg. Biomembr. 1986, 18, 71–91. [Google Scholar] [CrossRef] [PubMed]
- Nichols, J.W. Kinetics of fluorescent-labeled phosphatidylcholine transfer between nonspecific lipid transfer protein and phospholipid vesicles. Biochemistry 1988, 27, 1889–1896. [Google Scholar] [CrossRef] [PubMed]
- Rueckert, D.G.; Schmidt, K. Lipid transfer proteins. Chem. Phys. Lipids 1990, 56, 1–20. [Google Scholar] [CrossRef] [PubMed]
- Wirtz, K.W. Phospholipid transfer proteins. Annu. Rev. Biochem. 1991, 60, 73–99. [Google Scholar] [CrossRef] [PubMed]
- Gadella, T.W., Jr.; Wirtz, K.W. Phospholipid binding and transfer by the nonspecific lipid-transfer protein (sterol carrier protein 2) A kinetic model. Eur. J. Biochem. 1994, 220, 1019–1028. [Google Scholar] [CrossRef][Green Version]
- Wirtz, K.W.; Schouten, A.; Gros, P. Phosphatidylinositol transfer proteins: From closed for transport to open for exchange. Adv. Enzym. Regul. 2005, 46, 301–311. [Google Scholar] [CrossRef]
- Voelker, D.R. Bridging gaps in phospholipid transport. Trends Biochem. Sci. 2005, 30, 396–404. [Google Scholar] [CrossRef]
- Levine, T.; Loewen, C. Inter-organelle membrane contact sites: Through a glass, darkly. Curr. Opin. Cell Biol. 2006, 18, 371–378. [Google Scholar] [CrossRef]
- Giorgi, C.; De Stefani, D.; Bononi, A.; Rizzuto, R.; Pinton, P. Structural and functional link between the mitochondrial network and the endoplasmic reticulum. Int. J. Biochem. Cell Biol. 2009, 41, 1817–1827. [Google Scholar] [CrossRef]
- Lebiedzinska, M.; Szabadkai, G.; Jones, A.W.; Duszynski, J.; Wieckowski, M.R. Interactions between the endoplasmic reticulum, mitochondria plasma membrane and other subcellular organelles. Int. J. Biochem. Cell Biol. 2009, 41, 1805–1816. [Google Scholar] [CrossRef] [PubMed]
- Curwin, A.; McMaster, C. Structure and function of the enigmatic Sec14 domain containing proteins and the etiology of human disease. Future Lipidol. 2008, 3, 399–410. [Google Scholar] [CrossRef]
- Bankaitis, V.A.; Aitken, J.R.; Cleves, A.E.; Dowhan, W. An essential role for a phospholipid transfer protein in yeast Golgi function. Nature 1990, 347, 561–562. [Google Scholar] [CrossRef] [PubMed]
- Bankaitis, V.A.; Malehorn, D.E.; Emr, S.D.; Greene, R. The Saccharomyces cerevisiae SEC14 gene encodes a cytosolic factor that is required for transport of secretory proteins from the yeast Golgi complex. J. Cell Biol. 1989, 108, 1271–1281. [Google Scholar] [CrossRef]
- Im, Y.J.; Raychaudhuri, S.; Prinz, W.A.; Hurley, J.H. Structural mechanism for sterol sensing and transport by OSBP-related proteins. Nature 2005, 437, 154–158. [Google Scholar] [CrossRef]
- Li, N.; Xu, C.; Li-Beisson, Y.; Philippar, K. Fatty Acid and Lipid Transport in Plant Cells. Trends Plant Sci. 2016, 21, 145–158. [Google Scholar] [CrossRef]
- Alpy, F.; Tomasetto, C. Give lipids a START: The StAR-related lipid transfer (START) domain in mammals. J. Cell Sci. 2005, 118, 2791–2801. [Google Scholar] [CrossRef]
- Yao, C.; Wilson, M.E. Dynamics of sterol synthesis during development of Leishmania spp. parasites to their virulent form. Parasites Vectors 2016, 9, 1–12. [Google Scholar] [CrossRef]
- Mukherjee, S.; Mishra, A.; Trenberth, K.E. Climate Change and Drought: A Perspective on Drought Indices. Curr. Clim. Chang. Rep. 2018, 4, 145–163. [Google Scholar] [CrossRef]
- Curwin, A.; Fairn, G.; McMaster, C.R. Phospholipid Transfer Protein Sec14 Is Required for Trafficking from Endosomes and Regulates Distinct trans-Golgi Export Pathways. J. Biol. Chem. 2009, 284, 7364–7375. [Google Scholar] [CrossRef]
- Mousley, C.; Tyeryar, K.; Ryan, M.; Bankaitis, V. Sec14p-like proteins regulate phosphoinositide homoeostasis and intracellular protein and lipid trafficking in yeast. Biochem. Soc. Trans. 2006, 34, 346–350. [Google Scholar] [CrossRef]
- Sirokmány, G.; Szidonya, L.; Káldi, K.; Gáborik, Z.; Ligeti, E.; Geiszt, M. Sec14 Homology Domain Targets p50RhoGAP to Endosomes and Provides a Link between Rab and Rho GTPases. J. Biol. Chem. 2006, 281, 6096–6105. [Google Scholar] [CrossRef] [PubMed]
- Nile, A.H.; Bankaitis, V.A.; Grabon, A. Mammalian diseases of phosphatidylinositol transfer proteins and their homologs. Clin. Lipidol. 2010, 5, 867–897. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Phillips, S.; Vincent, P.; Rizzieri, K.E.; Schaaf, G.; Bankaitis, V.A.; Gaucher, E.A. The Diverse Biological Functions of Phosphatidylinositol Transfer Proteins in Eukaryotes. Crit. Rev. Biochem. Mol. Biol. 2006, 41, 21–49. [Google Scholar] [CrossRef] [PubMed]
- Raychaudhuri, S.; Prinz, W.A. The diverse functions of oxysterol-binding proteins. Annu. Rev. Cell Dev. Biol. 2010, 26, 157–177. [Google Scholar] [CrossRef]
- Johansson, M.; Rocha, N.; Zwart, W.; Jordens, I.; Janssen, L.; Kuijl, C.; Olkkonen, V.M.; Neefjes, J. Activation of endosomal dynein motors by stepwise assembly of Rab7– RILP–p150Glued, ORP1L, and the receptor βlll spectrin. J. Cell Biol. 2007, 176, 459–471. [Google Scholar] [CrossRef]
- Rocha, N.; Kuijl, C.; van der Kant, R.; Janssen, L.; Houben, D.; Janssen, H.; Zwart, W.; Neefjes, J. Cholesterol sensor ORP1L contacts the ER protein VAP to control Rab7–RILP–p150Glued and late endosome positioning. J. Cell Biol. 2009, 185, 1209–1225. [Google Scholar] [CrossRef]
- Miliara, X.; Garnett, J.A.; Tatsuta, T.; Ali, F.A.; Baldie, H.; Pérez-Dorado, I.; Simpson, P.; Yague, E.; Langer, T.; Matthews, S. Structural insight into the TRIAP1/PRELI-like domain family of mitochondrial phospholipid transfer complexes. EMBO Rep. 2015, 16, 824–835. [Google Scholar] [CrossRef]
- Tatsuta, T.; Langer, T. Intramitochondrial phospholipid trafficking. Biochim. Biophys. Acta (BBA) Mol. Cell Biol. Lipids 2017, 1862, 81–89. [Google Scholar] [CrossRef]
- Jeffers, V.; Kamau, E.T.; Srinivasan, A.R.; Harper, J.; Sankaran, P.; Post, S.E.; Varberg, J.M.; Sullivan, W.J., Jr.; Boyle, J.P. TgPRELID, a Mitochondrial Protein Linked to Multidrug Resistance in the Parasite Toxoplasma gondii. mSphere 2017, 2, e00229-16. [Google Scholar] [CrossRef]
- Rojas, M.L.; Del Puerto, M.M.C.; Flores-Martín, J.; Racca, A.C.; Kourdova, L.T.; Miranda, A.L.; Panzetta-Dutari, G.M.; Genti-Raimondi, S. Role of the lipid transport protein StarD7 in mitochondrial dynamics. Biochim. Biophys. Acta (BBA) Mol. Cell Biol. Lipids 2021, 1866, 159029. [Google Scholar] [CrossRef] [PubMed]
- Reinisch, K.M.; Camilli, P.D. SMP-domain proteins at membrane contact sites: Structure and function. Biochim. Biophys. Acta 2016, 1861 Pt B, 924–927. [Google Scholar] [CrossRef]
- De Cádiz, A.E.; Jeelani, G.; Nakada-Tsukui, K.; Caler, E.; Nozaki, T. Transcriptome Analysis of Encystation in Entamoeba invadens. PLoS ONE 2013, 8, e74840. [Google Scholar] [CrossRef] [PubMed]
- Potocka, I.; Baldwin, T.C.; Kurczynska, E.U. Distribution of lipid transfer protein 1 (LTP1) epitopes associated with morphogenic events during somatic embryogenesis of Arabidopsis thaliana. Plant Cell Rep. 2012, 31, 2031–2045. [Google Scholar] [CrossRef] [PubMed]
- Kular, G.; Loubtchenkov, M.; Swigart, P.; Whatmore, J.; Ball, A.; Cockcroft, S.; Wetzker, R. Co-operation of phosphatidylinositol transfer protein with phosphoinositide 3-kinase γ in the formylmethionyl-leucylphenylalanine-dependent production of phosphatidylinositol 3, 4, 5-trisphosphate in human neutrophils. Biochem. J. 1997, 325, 299–301. [Google Scholar] [CrossRef] [PubMed]
- Kular, G.S.; Chaudhary, A.; Prestwich, G.; Swigart, P.; Wetzker, R.; Cockcroft, S. Co-operation of phosphatidylinositol transfer protein with phosphoinositide 3-kinase g in vitro. Adv. Enzym. Regul. 2002, 42, 53–62. [Google Scholar] [CrossRef]
- Cockcroft, S.; Garner, K. Potential role for phosphatidylinositol transfer protein (PITP) family in lipid transfer during phospholipase C signalling. Adv. Biol. Regul. 2013, 53, 280–291. [Google Scholar] [CrossRef]
- Fayngerts, S.A.; Wu, J.; Oxley, C.L.; Liu, X.; Vourekas, A.; Cathopoulis, T.; Wang, Z.; Cui, J.; Liu, S.; Sun, H.; et al. TIPE3 Is the Transfer Protein of Lipid Second Messengers that Promote Cancer. Cancer Cell 2014, 26, 465–478. [Google Scholar] [CrossRef]
- Clark, B.J. The mammalian START domain protein family in lipid transport in health and disease. J. Endocrinol. 2011, 212, 257–275. [Google Scholar] [CrossRef]
- Kumagai, K.; Kawano-Kawada, M.; Hanada, K. Phosphoregulation of the Ceramide Transport Protein CERT at Serine 315 in the Interaction with VAMP-associated Protein (VAP) for Inter-organelle Trafficking of Ceramide in Mammalian Cells. J. Biol. Chem. 2014, 289, 10748–10760. [Google Scholar] [CrossRef]
- Kölsch, V.; Charest, P.G.; Firtel, R.A. The regulation of cell motility and chemotaxis by phospholipid signaling. J. Cell Sci. 2008, 121, 551–559. [Google Scholar] [CrossRef] [PubMed]
- Thomas, F.J.M. Role of PI (4,5) P2 in vesicle exocytosis and membrane fusion. Subcell. Biochem. 2012, 59, 111–130. [Google Scholar]
- Haastert, B.; Mellanby, R.J.; Anderton, S.M.; O’Connor, R.A. T Cells at the Site of Autoimmune Inflammation Show Increased Potential for Trogocytosis. PLoS ONE 2013, 8, e81404. [Google Scholar] [CrossRef] [PubMed]
- Levin, R.; Grinstein, S.; Schlam, D. Phosphoinositides in phagocytosis and macropinocytosis. Biochim. Biophys. Acta (BBA) Mol. Cell Biol. Lipids 2015, 1851, 805–823. [Google Scholar] [CrossRef]
- Litvak, V.; Dahan, N.; Ramachandran, S.; Sabanay, H.; Lev, S. Maintenance of the diacylglycerol level in the Golgi apparatus by the Nir2 protein is critical for Golgi secretory function. Nature 2005, 7, 225–234. [Google Scholar] [CrossRef] [PubMed]
- Peretti, D.; Dahan, N.; Shimoni, E.; Hirschberg, K.; Lev, S. Coordinated Lipid Transfer between the Endoplasmic Reticulum and the Golgi Complex Requires the VAP Proteins and Is Essential for Golgi-mediated Transport. Mol. Biol. Cell 2008, 19, 3871–3884. [Google Scholar] [CrossRef]
- Mattjus, P. Glycolipid transfer proteins and membrane interaction. Biochim. Biophys. Acta (BBA) Biomembr. 2009, 1788, 267–272. [Google Scholar] [CrossRef]
- McGee, T.; Skinner, H.; Whitters, E.; Henry, S.; Bankaitis, V. A phosphatidylinositol transfer protein controls the phosphatidylcholine content of yeast Golgi membranes. J. Cell Biol. 1994, 124, 273–287. [Google Scholar] [CrossRef]
- Skinner, H.B.; McGee, T.P.; McMaster, C.R.; Fry, M.R.; Bell, R.M.; Bankaitis, V.A. The Saccharomyces cerevisiae phosphatidylinositol-transfer protein effects a ligand-dependent inhibition of choline-phosphate cytidylyltransferase activity. Proc. Natl. Acad. Sci. USA 1995, 92, 112–116. [Google Scholar] [CrossRef]
- Schaaf, G.; Ortlund, E.; Tyeryar, K.R.; Mousley, C.; Ile, K.E.; Garrett, T.A.; Ren, J.; Woolls, M.J.; Raetz, C.R.; Redinbo, M.R.; et al. Functional Anatomy of Phospholipid Binding and Regulation of Phosphoinositide Homeostasis by Proteins of the Sec14 Superfamily. Mol. Cell 2008, 29, 191–206. [Google Scholar] [CrossRef]
- Tanaka, K.; Horiguchi, K.; Yoshida, T.; Takeda, M.; Fujisawa, H.; Takeuchi, K.; Umeda, M.; Kato, S.; Ihara, S.; Nagata, S.; et al. Evidence that a phosphatidylinositol 3,4,5-triphosphate binding protein can function in nucleus. J. Biol. Chem. 1999, 274, 3919–3922. [Google Scholar] [CrossRef] [PubMed]
- Martelli, A.M.; Bortul, R.; Tabellini, G.; Bareggi, R.; Manzoli, L.; Narducci, P.; Cocco, L. Diacylglycerol kinases in nuclear lipid-dependent signal transduction pathways. Cell. Mol. Life Sci. 2002, 59, 1129–1137. [Google Scholar] [CrossRef] [PubMed]
- Audhya, A.; Scott, D. Regulation of PI4,5P2 synthesis by nuclearcytoplasmic shuttling of the Mss4 lipid kinase. EMBO J. 2003, 22, 4223–4236. [Google Scholar] [CrossRef] [PubMed]
- Irvine, R.F. Nuclear lipid signalling. Nat. Rev. Mol. Cell Biol. 2003, 4, 349–361. [Google Scholar] [CrossRef]
- Balla, A.; Balla, T. Phosphatidylinositol 4-kinases: Old enzymes with emerging functions. Trends Cell Biol. 2006, 16, 351–361. [Google Scholar] [CrossRef] [PubMed]
- Matsubara, T.; Shirai, Y.; Miyasaka, K.; Murakami, T.; Yamaguchi, Y.; Ueyama, T.; Kai, M.; Sakane, F.; Kanoh, H.; Hashimoto, T.; et al. Nuclear Transportation of Diacylglycerol Kinase γ and Its Possible Function in the Nucleus. J. Biol. Chem. 2006, 281, 6152–6164. [Google Scholar] [CrossRef]
- Carman, M.; Henry, S.A. Phosphatidic acid plays a central role in the transcriptional regulation of glycerophospholipid synthesis in Saccharomyces cerevisiae. J. Biol. Chem. 2007, 282, 37293–37297. [Google Scholar] [CrossRef]
- Demmel, L.; Beck, M.; Klose, C.; Schlaitz, A.-L.; Gloor, Y.; Hsu, P.P.; Havlis, J.; Shevchenko, A.; Krause, E.; Kalaidzidis, Y.; et al. Nucleocytoplasmic Shuttling of the Golgi Phosphatidylinositol 4-Kinase Pik1 Is Regulated by 14-3-3 Proteins and Coordinates Golgi Function with Cell Growth. Mol. Biol. Cell 2008, 19, 1046–1061. [Google Scholar] [CrossRef]
- Mishkind, M.; Vermeer, J.E.; Darwish, E.; Munnik, T. Heat stress activates phospholipase D and triggers PIP2accumulation at the plasma membrane and nucleus. Plant J. 2009, 60, 10–21. [Google Scholar] [CrossRef]
- Ren, H.; Federico, L.; Huang, H.; Sunkara, M.; Drennan, T.; Frohman, M.A.; Smyth, S.S.; Morris, A.J. A Phosphatidic Acid Binding/Nuclear Localization Motif Determines Lipin1 Function in Lipid Metabolism and Adipogenesis. Mol. Biol. Cell 2010, 21, 3171–3181. [Google Scholar] [CrossRef]
- Jang, Y.H.; Min, D.S. Nuclear Localization of Phospholipase D1 Mediates the Activation of Nuclear Protein Kinase Cα and Extracellular Signal-regulated Kinase Signaling Pathways. J. Biol. Chem. 2011, 286, 4680–4689. [Google Scholar] [CrossRef] [PubMed]
- Symeon, S. Phospholipid metabolism and nuclear function: Roles of the lipin family of phosphatidic acid phosphatase. Biochim. Biophys. Acta 2013, 1831, 575–581. [Google Scholar]
- Jülke, S.; Ludwig-Müller, J. Response of Arabidopsis thaliana Roots with Altered Lipid Transfer Protein (LTP) Gene Expression to the Clubroot Disease and Salt Stress. Plants 2015, 5, 2. [Google Scholar] [CrossRef]
- Karlsson, T.; Altankhuyag, A.; Dobrovolska, O.; Turcu, D.C.; Lewis, A.E. A polybasic motif in ErbB3-binding protein 1 (EBP1) has key functions in nucleolar localization and polyphosphoinositide interaction. Biochem. J. 2016, 473, 2033–2047. [Google Scholar] [CrossRef]
- Tsai, T.-Y.; Wang, W.-T.; Li, H.-K.; Chen, W.-J.; Tsai, Y.-H.; Chao, C.-H.; Lee, Y.-H.W. RNA helicase DDX3 maintains lipid homeostasis through upregulation of the microsomal triglyceride transfer protein by interacting with HNF4 and SHP. Sci. Rep. 2017, 7, srep41452. [Google Scholar] [CrossRef] [PubMed]
- Zhou, T.; Li, S.; Zhong, W.; Vihervaara, T.; Béaslas, O.; Perttilä, J.; Luo, W.; Jiang, Y.; Lehto, M.; Olkkonen, V.M.; et al. OSBP-Related Protein 8 (ORP8) Regulates Plasma and Liver Tissue Lipid Levels and Interacts with the Nucleoporin Nup62. PLoS ONE 2011, 6, e21078. [Google Scholar] [CrossRef]
- Béaslas, O.; Vihervaara, T.; Li, J.; Laurila, P.-P.; Yan, D.; Olkkonen, V.M. Silencing of OSBP-related protein 8 (ORP8) modifies the macrophage transcriptome, nucleoporin p62 distribution, and migration capacity. Exp. Cell Res. 2012, 318, 1933–1945. [Google Scholar] [CrossRef]
- Goldfinger, L.E.; Ptak, C.; Jeffery, E.D.; Shabanowitz, J.; Han, J.; Haling, J.R.; Sherman, N.E.; Fox, J.W.; Hunt, D.F.; Ginsberg, M.H. An Experimentally Derived Database of Candidate Ras-Interacting Proteins. J. Proteome Res. 2007, 6, 1806–1811. [Google Scholar] [CrossRef]
- Weber-Boyvat, M.; Zhong, W.; Yan, D.; Olkkonen, V.M. Oxysterol-binding proteins: Functions in cell regulation beyond lipid metabolism. Biochem. Pharmacol. 2013, 86, 89–95. [Google Scholar] [CrossRef]
- Messaoud, H.B.-B.; Guichard, M.; Lawton, P.; Delton, I.; Azzouz-Maache, S. Changes in Lipid and Fatty Acid Composition During Intramacrophagic Transformation of Leishmania donovani Complex Promastigotes into Amastigotes. Lipids 2017, 52, 433–441. [Google Scholar] [CrossRef]
- Weingärtner, A.; Drobot, B.; Herrmann, A.; Sa´nchez-Canete, M.P.; Gamarro, F.; Castanys, S.; Pomorski, T.G. Disruption of the Lipid-Transporting LdMT-LdRos3 Complex in Leishmania donovani Affects Membrane Lipid Asymmetry but Not Host Cell Invasion. PLoS ONE 2010, 5, e12443. [Google Scholar] [CrossRef] [PubMed]
- de Aquino, G.P.; Gomes, M.A.M.; Salinas, R.K.; Laranjeira-Silva, M.F. Lipid and fatty acid metabolism in trypanosomatids. Microb. Cell 2021, 8, 262–275. [Google Scholar] [CrossRef] [PubMed]
- Giri, S.; Shaha, C. Leishmania donovani parasite requires Atg8 protein for infectivity and survival under stress. Cell Death Dis. 2019, 10, 1–19. [Google Scholar] [CrossRef]
- Picazarri, K.; Nakada-Tsukui, K.; Nozaki, T. Autophagy during Proliferation and Encystation in the Protozoan Parasite Entamoeba invadens. Infect. Immun. 2008, 76, 278–288. [Google Scholar] [CrossRef] [PubMed]
- Picazarri, K.; Nakada-Tsukui, K.; Tsuboi, K.; Miyamoto, E.; Watanabe, N.; Kawakami, E.; Nozaki, T. Atg8 is involved in endosomal and phagosomal acidification in the parasitic protist Entamoeba histolytica. Cell Microbiol. 2015, 17, 1510–1522. [Google Scholar] [CrossRef]
- Nakada-Tsukui, K.; Watanabe, N.; Shibata, K.; Wahyuni, R.; Miyamoto, E.; Nozaki, T. Proteomic analysis of Atg8-dependent recruitment of phagosomal proteins in the enteric protozoan parasite Entamoeba histolytica. Front. Cell. Infect. Microbiol. 2022, 12, 961645. [Google Scholar] [CrossRef]
- Dong, G.; Filho, A.L.; Olivier, M. Modulation of Host-Pathogen Communication by Extracellular Vehicles (EVs) of the Protozoan Parasite Leishmania. Front. Cell. Infect. Microbiol. 2019, 9, 100. [Google Scholar] [CrossRef]
- Moyano, S.; Musso, J.; Feliziani, C.; Zamponi, N.; Frontera, L.S.; Ropolo, A.S.; Lanfredi-Rangel, A.; Lalle, M.; Touz, M.C. Exosome Biogenesis in the Protozoa Parasite Giardia lamblia: A Model of Reduced Interorganellar Crosstalk. Cells 2019, 8, 1600. [Google Scholar] [CrossRef]
- Urbanelli, L.; Magini, A.; Buratta, S.; Brozzi, A.; Sagini, K.; Polchi, A.; Tancini, B.; Emiliani, C. Signaling Pathways in Exosomes Biogenesis, Secretion and Fate. Genes 2013, 4, 152–170. [Google Scholar] [CrossRef]
- Donoso-Quezada, J.; Ayala-Mar, S.; González-Valdez, J. The role of lipids in exosome biology and intercellular communication: Function, analytics and applications. Traffic 2021, 22, 204–220. [Google Scholar] [CrossRef]
- Gupta, R.; Rathi, P.; Gupta, N.; Bradoo, S. Review: Lipase assays for conventional and molecular screening: An overview. Biotechnol. Appl. Biochem. 2003, 37, 63–71. [Google Scholar] [CrossRef] [PubMed]
- Chappuis, F.; Sundar, S.; Hailu, A.; Ghalib, H.; Rijal, S.; Peeling, R.W.; Alvar, J.; Boelaert, M. Visceral leishmaniasis: What are the needs for diagnosis, treatment and control? Nat. Rev. Microbiol. 2007, 5, 873–882. [Google Scholar] [CrossRef] [PubMed]
- Ramakrishnan, S.; Serricchio, M.; Striepen, B.; Bütikofer, P. Lipid synthesis in protozoan parasites: A comparison between kinetoplastids and apicomplexans. Prog. Lipid Res. 2013, 52, 488–512. [Google Scholar] [CrossRef] [PubMed]
- Ghosh, J.; Das, S.; Guha, R.; Ghosh, D.; Naskar, K.; Das, A.; Roy, S. Hyperlipidemia offers protection against Leishmania donovani infection: Role of membrane cholesterol. J. Lipid Res. 2012, 53, 2560–2572. [Google Scholar] [CrossRef] [PubMed]
- Lal, C.S.; Kumar, A.; Kumar, S.; Pandey, K.; Kumar, N.; Bimal, S.; Sinha, P.K.; Das, P. Hypocholesterolemia and increased triglyceride in pediatric visceral leishmaniasis. Clin. Chim. Acta 2007, 382, 151–153. [Google Scholar] [CrossRef]
- Ghosh, J.; Bose, M.; Roy, S.; Bhattacharyya, S.N. Leishmania donovani Targets Dicer1 to Downregulate miR-122, Lower Serum Cholesterol, and Facilitate Murine Liver Infection. Cell Host Microbe 2013, 13, 277–288. [Google Scholar] [CrossRef]
- Kima, P.E. PI3K signaling in Leishmania infections. Cell. Immunol. 2016, 309, 19–22. [Google Scholar] [CrossRef]
- Cummings, H.E.; Barbi, J.; Reville, P.; Oghumu, S.; Zorko, N.; Sarkar, A.; Keiser, T.L.; Lu, B.; Rückle, T.; Varikuti, S.; et al. Critical role for phosphoinositide 3-kinase gamma in parasite invasion and disease progression of cutaneous leishmaniasis. Proc. Natl. Acad. Sci. USA 2012, 109, 1251–1256. [Google Scholar] [CrossRef]
- Silverman, J.M.; Chan, S.K.; Robinson, D.P.; Dwyer, D.M.; Nandan, D.; Foster, L.J.; Reiner, N.E. Proteomic analysis of the secretome of Leishmania donovani. Genome Biol. 2008, 9, R35. [Google Scholar] [CrossRef]
- Corrales, R.M.; Sereno, D.; Mathieu-Daudé, F. Deciphering the Leishmania exoproteome: What we know and what we can learn. FEMS Immunol. Med. Microbiol. 2010, 58, 27–38. [Google Scholar] [CrossRef]
- Das, S.; Saha, T.; Yadav, S.; Shaha, C. A Novel Role of Secretory Cytosolic Tryparedoxin Peroxidase in Delaying Apoptosis of Leishmania-Infected Macrophages. Mol. Cell. Biol. 2022, 42, e00081-22. [Google Scholar] [CrossRef] [PubMed]
- Mottram, J.C.; Coombs, G.H.; Alexander, J. Cysteine peptidases as virulence factors of Leishmania. Curr. Opin. Microbiol. 2004, 7, 375–381. [Google Scholar] [CrossRef] [PubMed]
- Olivier, M.; Gregory, D.J.; Forget, G. Subversion Mechanisms by Which Leishmania Parasites Can Escape the Host Immune Response: A Signaling Point of View. Clin. Microbiol. Rev. 2005, 18, 293–305. [Google Scholar] [CrossRef]
- Santarém, N.; Silvestre, R.; Tavares, J.; Silva, M.; Cabral, S.; Maciel, J.; Cordeiro-Da-Silva, A. Immune Response Regulation by Leishmania Secreted and Nonsecreted Antigens. J. Biomed. Biotechnol. 2007, 2007, 85154. [Google Scholar] [CrossRef] [PubMed]
- Joshi, M.B.; Dwyer, D.M. Molecular and Functional Analyses of a Novel Class I Secretory Nuclease from the Human Pathogen, Leishmania donovani. J. Biol. Chem. 2007, 282, 10079–10095. [Google Scholar] [CrossRef]
- Peters, C.; Stierhof, Y.D.; Ilg, T. Proteophosphoglycan secreted by Leishmania mexicana amastigotes causes vacuole formation in macrophages. Infect. Immun. 1997, 65, 783–786. [Google Scholar] [CrossRef]
- Peters, C.; Kawakami, M.; Kaul, M.; Ilg, T.; Overath, P.; Aebischer, T. Secreted proteophosphoglycan ofLeishmania mexicana amastigotes activates complement by triggering the mannan binding lectin pathway. Eur. J. Immunol. 1997, 27, 2666–2672. [Google Scholar] [CrossRef]
- Joshi, M.B.; Rogers, M.E.; Shakarian, A.M.; Yamage, M.; Al-Harthi, S.A.; Bates, P.A.; Dwyer, D.M. Molecular characterization, expression, and in vivo analysis of LmexCht1: The chitinase of the human pathogen, Leishmania mexicana. J. Biol. Chem. 2005, 280, 3847–3861. [Google Scholar] [CrossRef]
- Rogers, M.E.; Hajmová, M.; Joshi, M.B.; Sadlova, J.; Dwyer, D.M.; Volf, P.; Bates, P.A. Leishmania chitinase facilitates colonization of sand fly vectors and enhances transmission to mice. Cell. Microbiol. 2008, 10, 1363–1372. [Google Scholar] [CrossRef]
- Kulkarni, M.M.; McMaster, W.R.; Kamysz, E.; Kamysz, W.; Engman, D.M.; McGwire, B.S. The major surface-metalloprotease of the parasitic protozoan, Leishmania, protects against antimicrobial peptide-induced apoptotic killing. Mol. Microbiol. 2006, 62, 1484–1497. [Google Scholar] [CrossRef]
- Santos, A.L.; Branquinha, M.H.; D’Avila-Levy, C.M. The ubiquitous gp63-like metalloprotease from lower trypanosomatids: In the search for a function. An. Da Acad. Bras. De Ciências 2006, 78, 687–714. [Google Scholar] [CrossRef] [PubMed]
- Atayde, V.D.; Aslan, H.; Townsend, S.; Hassani, K.; Kamhawi, S.; Olivier, M. Faculty Opinions recommendation of Exosome Secretion by the Parasitic Protozoan Leishmania within the Sand Fly Midgut. Cell Rep. 2015, 13, 957–967. [Google Scholar] [CrossRef] [PubMed]
- Forrest, D.M.; Batista, M.; Marchini, F.K.; Tempone, A.J.; Traub-Csekö, Y.M. Proteomic analysis of exosomes derived from procyclic and metacyclic-like cultured Leishmania infantum chagasi. J. Proteom. 2020, 227, 103902. [Google Scholar] [CrossRef]
- Gabriel, M.; Galué-Parra, A.; Pereira, W.L.A.; Pedersen, K.W.; da Silva, E.O. Leishmania 360°: Guidelines for Exosomal Research. Microorganisms 2021, 9, 2081. [Google Scholar] [CrossRef] [PubMed]
- Douanne, N.; Dong, G.; Douanne, M.; Olivier, M.; Fernandez-Prada, C. Unravelling the proteomic signature of extracellular vesicles released by drug-resistant Leishmania infantum parasites. PLoS Negl. Trop. Dis. 2020, 14, e0008439. [Google Scholar] [CrossRef] [PubMed]
- Holič, R.; Šťastný, D.; Griač, P. Sec14 family of lipid transfer proteins in yeasts. Biochim. Biophys. Acta (BBA) Mol. Cell Biol. Lipids 2021, 1866, 158990. [Google Scholar] [CrossRef]
- Yakir-Tamang, L.; Gerst, J.E. A Phosphatidylinositol-Transfer Protein and Phosphatidylinositol-4-phosphate 5-Kinase Control Cdc42 to Regulate the Actin Cytoskeleton and Secretory Pathway in Yeast. Mol. Biol. Cell 2009, 20, 3583–3597. [Google Scholar] [CrossRef]
- Cremona, O.; De Camilli, P. Phosphoinositides in membrane traffic at the synapse. J. Cell Sci. 2001, 114, 1041–1052. [Google Scholar] [CrossRef]
- Das, K.; Debnath, U.; Rahaman, S.B.; Upadhyay, S.; Das, D. Non-vesicular lipid transport machinery in Trichomonas vaginalis: Novel drug targets against Trichomoniasis. Curr. Top. Med. Chem. 2022, 23, 540–550. [Google Scholar] [CrossRef] [PubMed]
- Kelly, F.D.; Yates, P.A.; Landfear, S.M. Nutrient sensing in Leishmania: Flagellum and cytosol. Mol. Microbiol. 2020, 115, 849–859. [Google Scholar] [CrossRef]
- Clayton, C. Regulation of gene expression in trypanosomatids: Living with polycistronic transcription. Open Biol. 2019, 9, 190072. [Google Scholar] [CrossRef] [PubMed]
- Mandal, G.; Sharma, M.; Kruse, M.; Sander-Juelch, C.; Munro, L.A.; Wang, Y.; Vilg, J.V.; Tamás, M.J.; Bhattacharjee, H.; Wiese, M.; et al. Modulation of Leishmania major aquaglyceroporin activity by a mitogen-activated protein kinase. Mol. Microbiol. 2012, 85, 1204–1218. [Google Scholar] [CrossRef]
- Goldman-Pinkovich, A.; Balno, C.; Strasser, R.; Zeituni-Molad, M.; Bendelak, K.; Rentsch, D.; Ephros, M.; Wiese, M.; Jardim, A.; Myler, P.J.; et al. An Arginine Deprivation Response Pathway Is Induced in Leishmania during Macrophage Invasion. PLOS Pathog. 2016, 12, e1005494. [Google Scholar] [CrossRef]
- Salmon, D.; Vanwalleghem, G.; Morias, Y.; Denoeud, J.; Krumbholz, C.; Lhommé, F.; Bachmaier, S.; Kador, M.; Gossmann, J.; Dias, F.B.S.; et al. Adenylate Cyclases of Trypanosoma brucei Inhibit the Innate Immune Response of the Host. Science 2012, 337, 463–466. [Google Scholar] [CrossRef] [PubMed]
- Grabon, A.; Khan, D.; Bankaitis, V.A. Phosphatidylinositol transfer proteins and instructive regulation of lipid kinase biology. Biochim. Biophys. Acta (BBA) Mol. Cell Biol. Lipids 2015, 1851, 724–735. [Google Scholar] [CrossRef] [PubMed]
- Petropolis, D.B.; Rodrigues, J.C.F.; Viana, N.B.; Pontes, B.; Pereira, C.F.A.; Silva-Filho, F.C. Leishmania amazonensis promastigotes in 3D collagen I culture: An in vitro physiological environment for the study of extracellular matrix and host cell interactions. PeerJ 2014, 2, e317. [Google Scholar] [CrossRef]
- Sunter, J.; Gull, K. Shape, form, function and Leishmania pathogenicity: From textbook descriptions to biological understanding. Open Biol. 2017, 7, 170165. [Google Scholar] [CrossRef]
- Saada, E.A.; Kabututu, Z.P.; Lopez, M.; Shimogawa, M.M.; Langousis, G.; Oberholzer, M.; Riestra, A.; Jónsson, Z.O.; Wohlschlegel, J.A.; Hill, K.L. Insect Stage-Specific Receptor Adenylate Cyclases Are Localized to Distinct Subdomains of the Trypanosoma brucei Flagellar Membrane. Eukaryot. Cell 2014, 13, 1064–1076. [Google Scholar] [CrossRef]
- Varga, V.; Moreira-Leite, F.; Portman, N.; Gull, K. Protein diversity in discrete structures at the distal tip of the trypanosome flagellum. Proc. Natl. Acad. Sci. USA 2017, 114, E6546–E6555. [Google Scholar] [CrossRef]
- Forestier, C.-L.; Machu, C.; Loussert, C.; Pescher, P.; Späth, G.F. Imaging host cell –Leishmania interaction dynamics implicates parasite motility, lysosome recruitment, and host cell wounding in the infection process. Cell Host Microbe 2011, 9, 319–330. [Google Scholar] [CrossRef]
- Peretti, D.; Kim, S.; Tufi, R.; Lev, S. Lipid Transfer Proteins and Membrane Contact Sites in Human Cancer. Front. Cell Dev. Biol. 2020, 7, 371. [Google Scholar] [CrossRef]
- Bankaitis, V.A.; Mousley, C.J.; Schaaf, G. The Sec14 superfamily and mechanisms for crosstalk between lipid metabolism and lipid signaling. Trends Biochem. Sci. 2010, 35, 150–160. [Google Scholar] [CrossRef] [PubMed]
- Holm, A.; Tejle, K.; Magnusson, K.E.; Descoteaux, A.; Rasmusson, B. Leishmania donovani lipophosphoglycan causes periphagosomal actin accumulation: Correlation with impaired translocation of PKC alpha and defective phagosome maturation. Cell. Microbiol. 2001, 3, 439–447. [Google Scholar] [CrossRef]
- Lodge, R.; Descoteaux, A. Leishmania donovani promastigotes induce periphagosomal F-actin accumulation through retention of the GTPase Cdc42. Cell. Microbiol. 2005, 7, 1647–1658. [Google Scholar] [CrossRef] [PubMed]
- Schnabl, M.; Oskolkova, O.; Holic, R.; Brezna, B.; Pichler, H.; Zagorsek, M.; Kohlwein, S.D.; Paltauf, F.; Daum, G.; Griac, P. Subcellular localization of yeast Sec14 homologues and their involvement in regulation of phospholipid turnover. JBIC J. Biol. Inorg. Chem. 2003, 270, 3133–3145. [Google Scholar] [CrossRef] [PubMed]
- Nile, A.; Tripathi, A.; Yuan, P.; Mousley, C.; Suresh, S.; Wallace, I.; Shah, S.D.; Pohlhaus, D.T.; Temple, B.; Nislow, C.; et al. PITPs as targets for selectively interfering with phosphoinositide signaling in cells. Nat. Chem. Biol. 2014, 10, 76–84. [Google Scholar] [CrossRef]
- Khan, D.; McGrath, K.R.; Dorosheva, O.; Bankaitis, V.A.; Tripathi, A. Structural elements that govern Sec14-like PITP sensitivities to potent small molecule inhibitors. J. Lipid Res. 2016, 57, 650–662. [Google Scholar] [CrossRef]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Das, K.; Nozaki, T. Non-Vesicular Lipid Transport Machinery in Leishmania donovani: Functional Implications in Host-Parasite Interaction. Int. J. Mol. Sci. 2023, 24, 10637. https://doi.org/10.3390/ijms241310637
Das K, Nozaki T. Non-Vesicular Lipid Transport Machinery in Leishmania donovani: Functional Implications in Host-Parasite Interaction. International Journal of Molecular Sciences. 2023; 24(13):10637. https://doi.org/10.3390/ijms241310637
Chicago/Turabian StyleDas, Koushik, and Tomoyoshi Nozaki. 2023. "Non-Vesicular Lipid Transport Machinery in Leishmania donovani: Functional Implications in Host-Parasite Interaction" International Journal of Molecular Sciences 24, no. 13: 10637. https://doi.org/10.3390/ijms241310637
APA StyleDas, K., & Nozaki, T. (2023). Non-Vesicular Lipid Transport Machinery in Leishmania donovani: Functional Implications in Host-Parasite Interaction. International Journal of Molecular Sciences, 24(13), 10637. https://doi.org/10.3390/ijms241310637




