Characterization of Two Na+(K+, Li+)/H+ Antiporters from Natronorubrum daqingense
Abstract
1. Introduction
2. Results
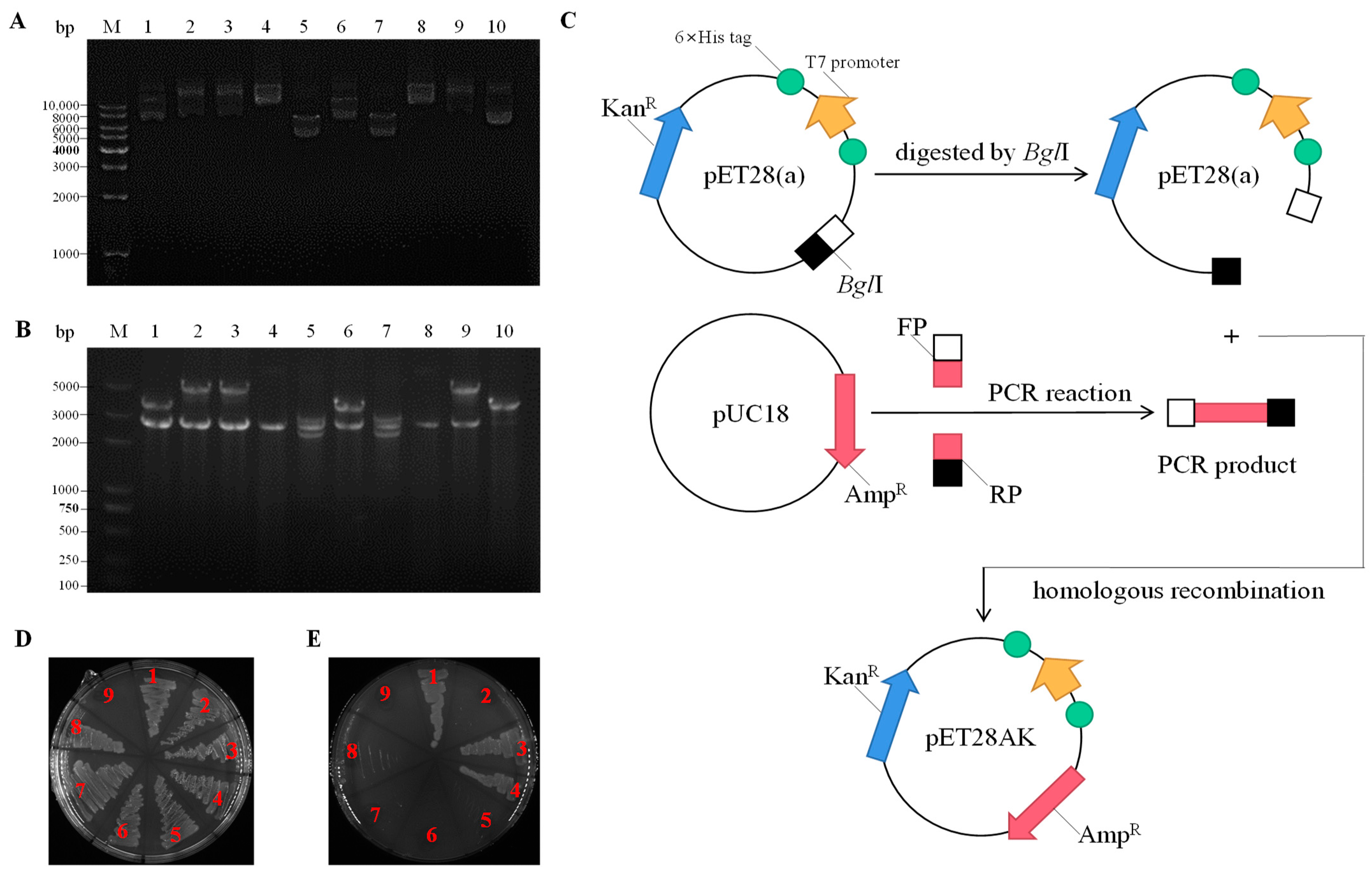
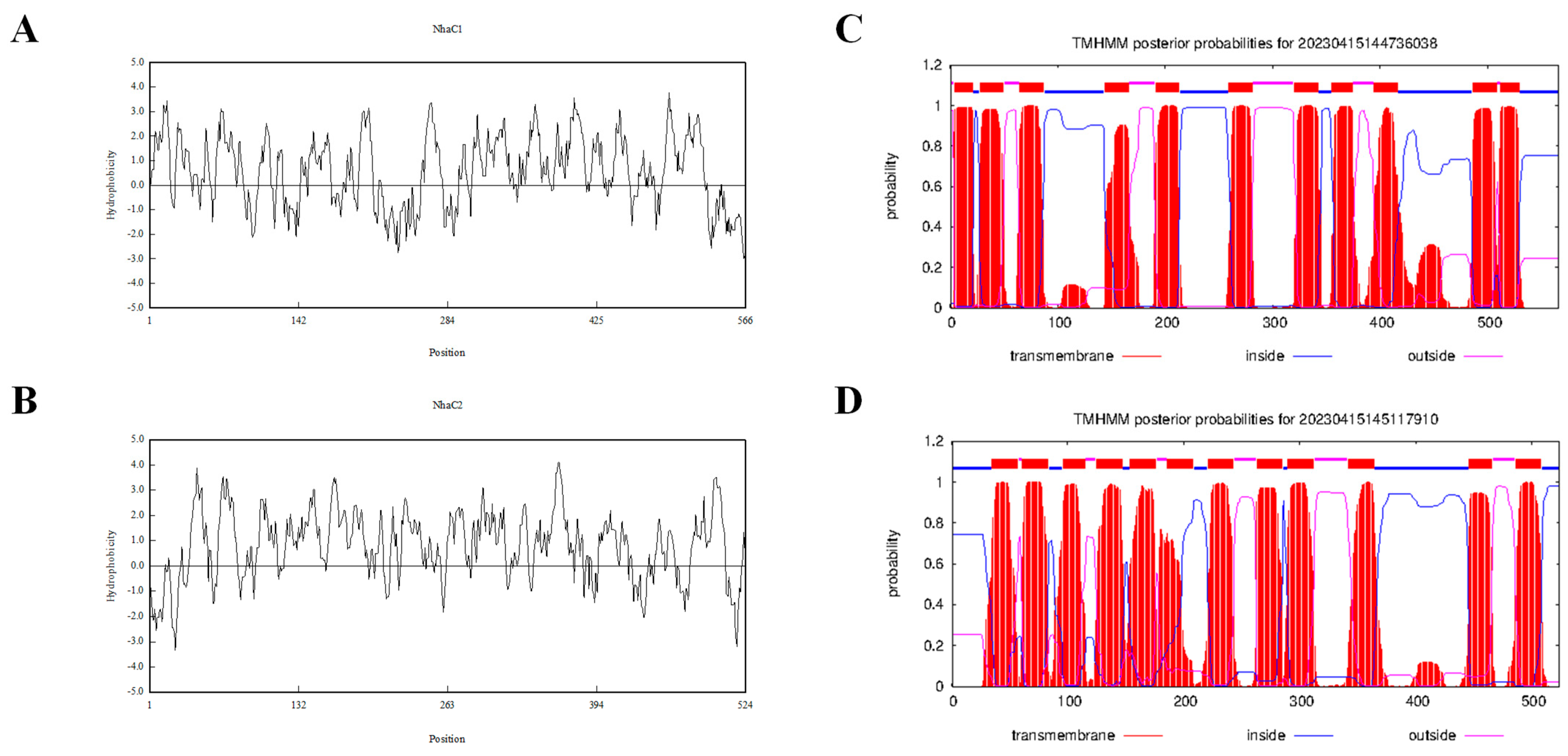
2.1. Cloning and Sequence Analysis of Na+(K+, Li+)/H+ Antiporter Genes
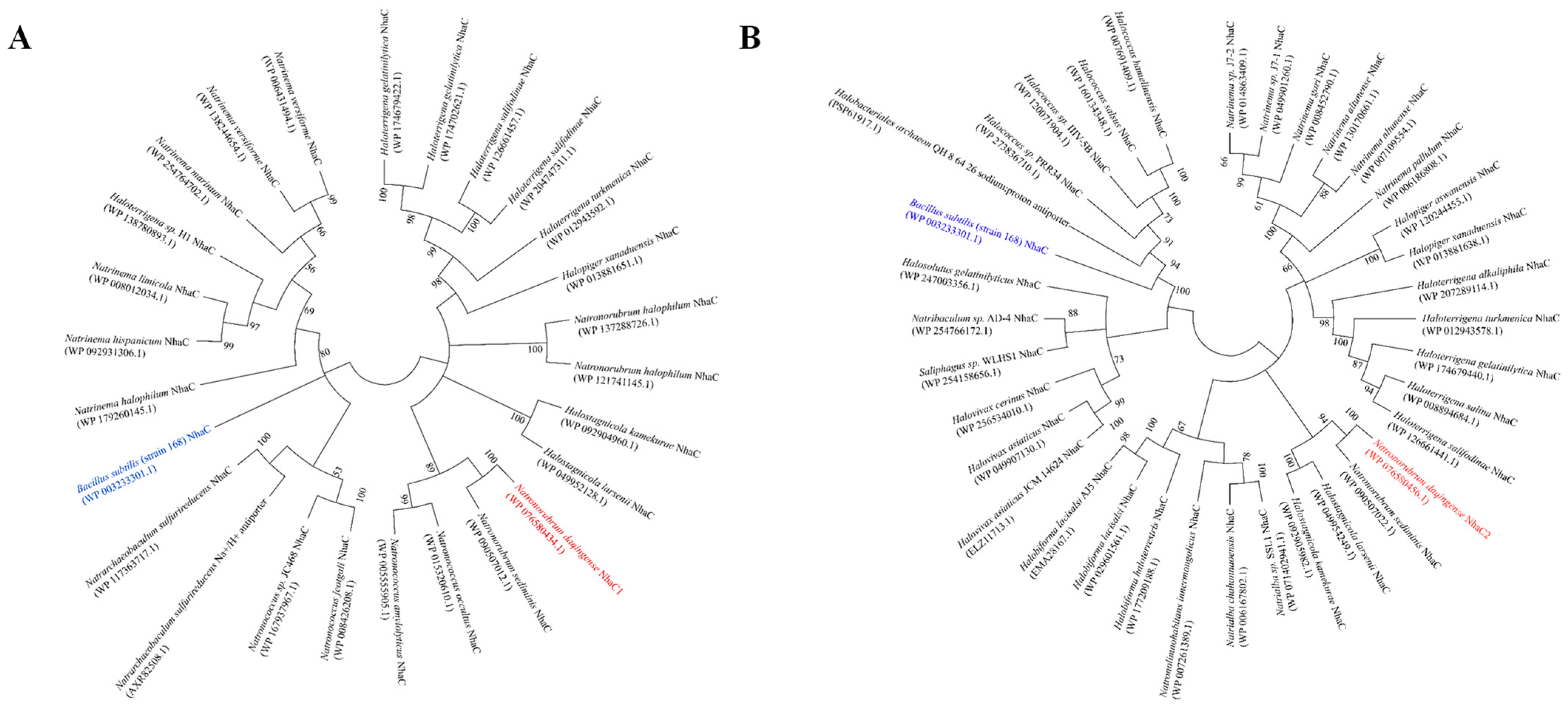
2.2. Phylogenetic Analysis Based on the Neighbour-Joining Algorithm
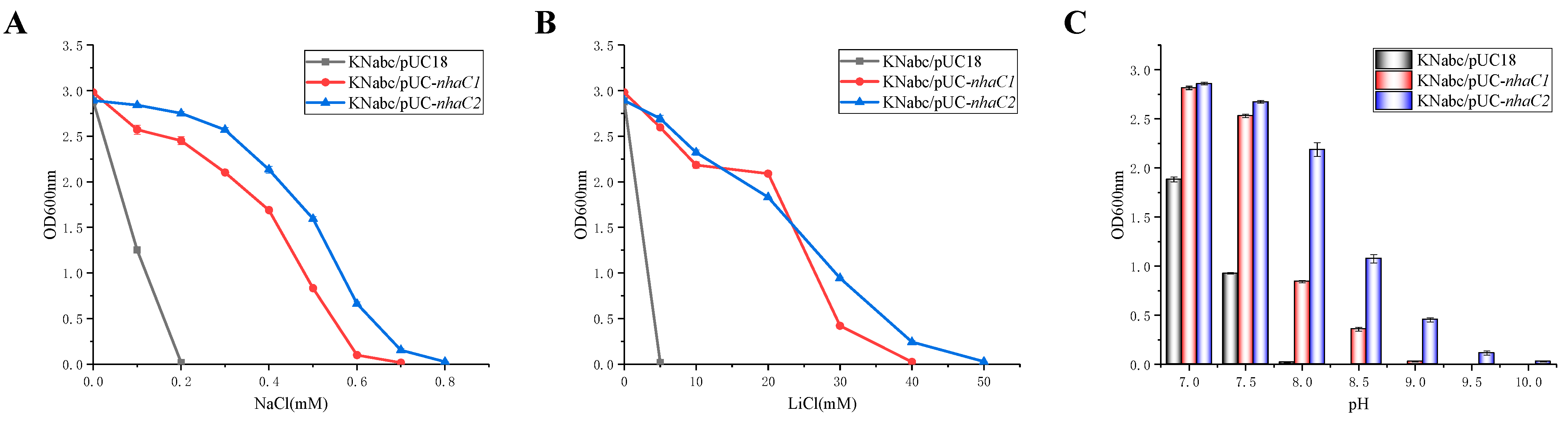
2.3. Test for Salt Tolerance and Alkaline pH Resistance
2.4. SDS-PAGE and Western Blot Analysis of NhaC1 and NhaC2
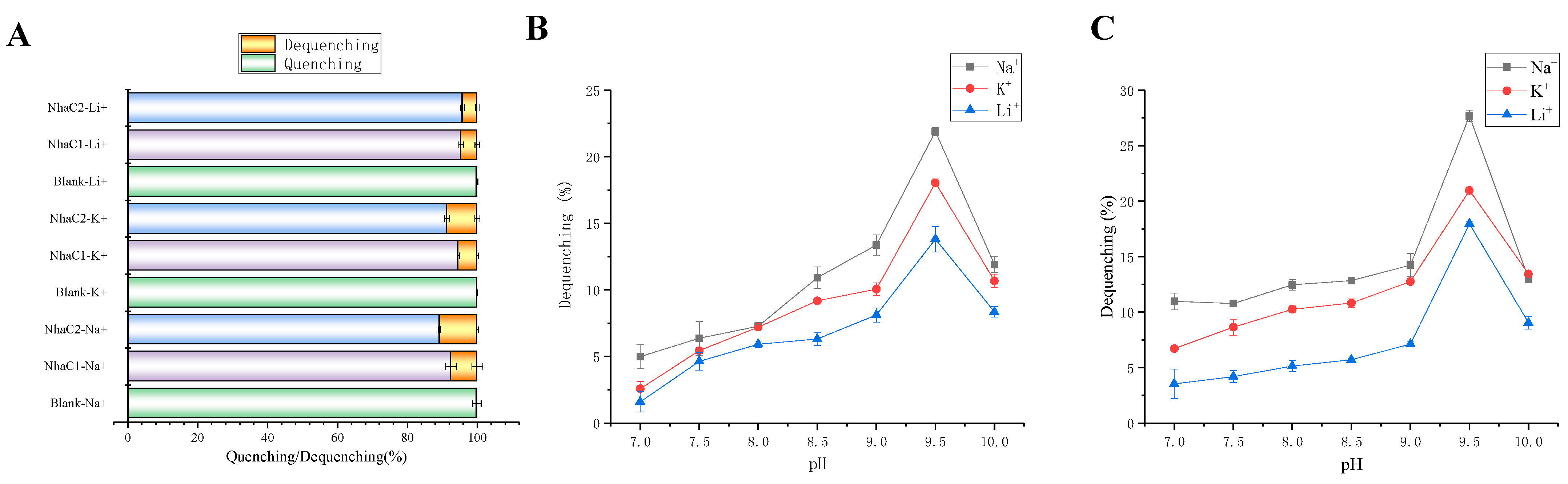
2.5. Detection of Na+(K+, Li+)/H+ Antiport Activity
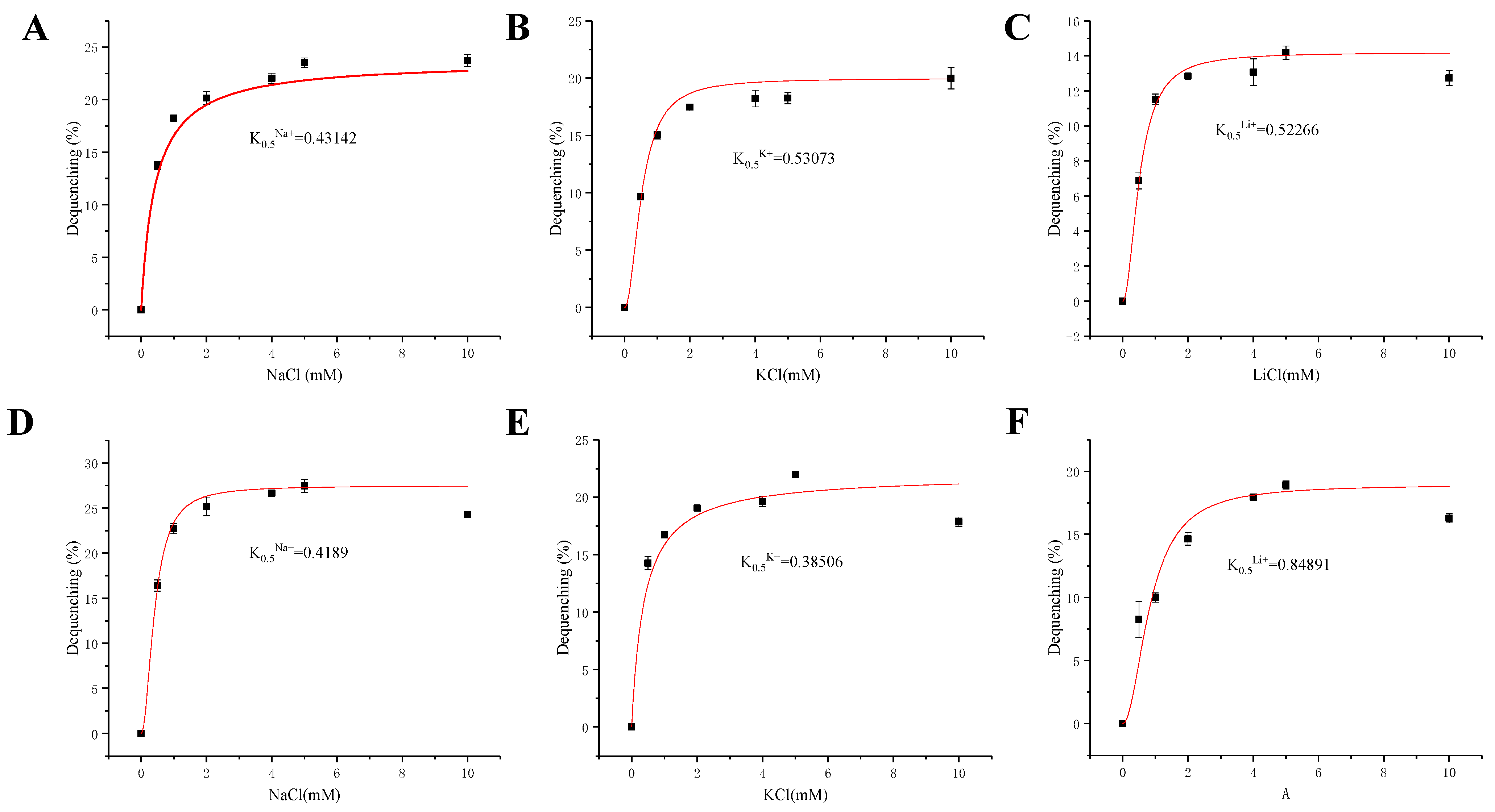
2.6. Calculation of K0.5 Values for Monovalent Cations
3. Discussion
4. Materials and Methods
4.1. Strains, Plasmids, and Growth Conditions
4.2. Screening of Na+(K+, Li+)/H+ Antiporter Genes
4.3. Preparation of Everted Membrane Vesicles
4.4. Preparation and Purification of Proteins
4.5. SDS-PAGE and Western Blot
4.6. Detection of Na+(K+, Li+)/H+ Antiport Activity
4.7. Calculation of pH Profile and K0.5 Values for Monovalent Cations
4.8. DNA Manipulation and Bioinformatics Analyses
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Chong, P.L.; Chang, A.; Yu, A.; Mammedova, A. Vesicular and Planar Membranes of Archaea Lipids: Unusual Physical Properties and Biomedical Applications. Int. J. Mol. Sci. 2022, 23, 7616. [Google Scholar] [CrossRef] [PubMed]
- Rao, A.; de Kok, N.A.W.; Driessen, A.J.M. Membrane Adaptations and Cellular Responses of Sulfolobus acidocaldarius to the Allylamine Terbinafine. Int. J. Mol. Sci. 2023, 24, 7328. [Google Scholar] [CrossRef] [PubMed]
- Margesin, R.; Schinner, F. Potential of halotolerant and halophilic microorganisms for biotechnology. Extremophiles 2001, 5, 73–83. [Google Scholar] [CrossRef]
- Oren, A. Diversity of halophilic microorganisms: Environments, phylogeny, physiology, and applications. J. Ind. Microbiol. Biotechnol. 2002, 28, 56–63. [Google Scholar] [CrossRef]
- Brett, C.L.; Donowitz, M.; Rao, R. Evolutionary origins of eukaryotic sodium/proton exchangers. Am. J. Physiol. 2005, 288, C223–C239. [Google Scholar] [CrossRef] [PubMed]
- Dimroth, P. Na(+)-coupled alternative to H(+)-coupled primary transport systems in bacteria. Bioessays 1991, 13, 463–468. [Google Scholar] [CrossRef]
- Krulwich, T.A.; Guffanti, A.A. The Na+ cycle of extreme alkalophiles: A secondary Na+/H+ antiporter and Na+/solute symporters. J. Bioenerg. Biomembr. 1989, 21, 663–677. [Google Scholar] [CrossRef]
- Patiño-Ruiz, M.; Ganea, C.; Călinescu, O. Prokaryotic Na+/H+ Exchangers-Transport Mechanism and Essential Residues. Int. J. Mol. Sci. 2022, 23, 9156. [Google Scholar] [CrossRef]
- Kumari, J.; Rathore, M.S. Na+/K+-ATPase a Primary Membrane Transporter: An Overview and Recent Advances with Special Reference to Alage. J. Membr. Biol. 2020, 253, 191–204. [Google Scholar] [CrossRef]
- Bakker, E.P.; Borchard, A.; Michels, M.; Altendorf, K.; Siebers, A. High-affinity potassium uptake system in Bacillus acidocaldarius showing immunological cross-reactivity with the Kdp system from Escherichia coli. J. Bacteriol. 1987, 169, 4342–4348. [Google Scholar] [CrossRef]
- Woehlke, G.; Dimroth, P. Anaerobic growth of Salmonella typhimurium on L(+)- and D(-)-tartrate involves an oxaloacetate decarboxylase Na+ pump. Arch. Microbiol. 1994, 162, 233–237. [Google Scholar] [CrossRef]
- Wifling, K.; Dimroth, P. Isolation and characterization of oxaloacetate decarboxylase of Salmonella typhimurium, a sodium ion pump. Arch. Microbiol. 1989, 152, 584–588. [Google Scholar] [CrossRef]
- Tokuda, H.; Unemoto, T. A respiration-dependent primary sodium extrusion system functioning at alkaline pH in the marine bacterium Vibrio alginolyticus. Biochem. Biophys. Res. Commun. 1981, 102, 265–271. [Google Scholar] [CrossRef]
- Tokuda, H.; Unemoto, T. Growth of a marine Vibrio alginolyticus and moderately halophilic V. costicola becomes uncoupler resistant when the respiration-dependent Na+ pump functions. J. Bacteriol. 1983, 156, 636–643. [Google Scholar] [CrossRef] [PubMed]
- Gärtner, P.; Weiss, D.S.; Harms, U.; Thauer, R.K. N5-methyltetrahydromethanopterin:coenzyme M methyltransferase from Methanobacterium thermoautotrophicum. Catalytic mechanism and sodium ion dependence. Eur. J. Biochem. 1994, 226, 465–472. [Google Scholar] [CrossRef]
- Weiss, D.S.; Gärtner, P.; Thauer, R.K. The energetics and sodium-ion dependence of N5-methyltetrahydromethanopterin:coenzyme M methyltransferase studied with cob(I)alamin as methyl acceptor and methylcob(III)alamin as methyl donor. Eur. J. Biochem. 1994, 226, 799–809. [Google Scholar] [CrossRef] [PubMed]
- Lienard, T.; Becher, B.; Marschall, M.; Bowien, S.; Gottschalk, G. Sodium ion translocation by N5-methyltetrahydromethanopterin: Coenzyme M methyltransferase from Methanosarcina mazei Gö1 reconstituted in ether lipid liposomes. Eur. J. Biochem. 1996, 239, 857–864. [Google Scholar] [CrossRef] [PubMed]
- Krulwich, T.A.; Hicks, D.B.; Ito, M. Cation/proton antiporter complements of bacteria: Why so large and diverse? Mol. Microbiol. 2009, 74, 257–260. [Google Scholar] [CrossRef]
- Prakash, S.; Cooper, G.; Singhi, S.; Saier, M.H., Jr. The ion transporter superfamily. Biochim. Biophys. Acta 2003, 1618, 79–92. [Google Scholar] [CrossRef]
- Neverisky, D.L.; Abbott, G.W. Ion channel-transporter interactions. Crit. Rev. Biochem. Mol. Biol. 2015, 51, 257–267. [Google Scholar] [CrossRef]
- Saier, M.H., Jr.; Reddy, V.S.; Tsu, B.V.; Ahmed, M.S.; Li, C.; Moreno-Hagelsieb, G. The Transporter Classification Database (TCDB): Recent advances. Nucleic Acids Res. 2016, 44, D372–D379. [Google Scholar] [CrossRef] [PubMed]
- Tsujii, M.; Tanudjaja, E.; Uozumi, N. Diverse Physiological Functions of Cation Proton Antiporters across Bacteria and Plant Cells. Int. J. Mol. Sci. 2020, 21, 4566. [Google Scholar] [CrossRef]
- Gardner, C.C.; James, P.F. The SLC9C2 Gene Product (Na+/H+Exchanger Isoform 11; NHE11) Is a Testis-Specific Protein Localized to the Head of Mature Mammalian Sperm. Int. J. Mol. Sci. 2023, 24, 5329. [Google Scholar] [CrossRef] [PubMed]
- Ito, M.; Guffanti, A.A.; Zemsky, J.; Ivey, D.M.; Krulwich, T.A. Role of the nhaC-encoded Na+/H+ antiporter of alkaliphilic Bacillus firmus OF4. J. Bacteriol. 1997, 179, 3851–3857. [Google Scholar] [CrossRef] [PubMed]
- Liew, C.W.; Illias, R.M.; Mahadi, N.M.; Najimudin, N. Expression of the Na+/H+ antiporter gene (g1-nhaC) of alkaliphilic Bacillus sp. G1 in Escherichia coli. FEMS Microbiol. Lett. 2007, 276, 114–122. [Google Scholar] [CrossRef]
- Haja, D.K.; Adams, M.W.W. pH Homeostasis and Sodium Ion Pumping by Multiple Resistance and pH Antiporters in Pyrococcus furiosus. Front. Microbiol. 2021, 12, 712104. [Google Scholar] [CrossRef]
- Wang, S.; Yang, Q.; Liu, Z.H.; Sun, L.; Wei, D.; Zhang, J.Z.; Song, J.Z.; Yuan, H.F. Haloterrigena daqingense sp. nov., an extremely haloalkaliphilic archaeon isolated from a saline-alkaline soil. Int. J. Syst. Evol. Microbiol. 2010, 60, 2267–2271. [Google Scholar] [CrossRef]
- de la Haba, R.R.; Minegishi, H.; Kamekura, M.; Shimane, Y.; Ventosa, A. Phylogenomics of Haloarchaea: The Controversy of the Genera Natrinema-Haloterrigena. Front. Microbiol. 2021, 12, 740909. [Google Scholar] [CrossRef]
- Wang, S.; Narsing Rao, M.P.; Wei, D.; Sun, L.; Fang, B.Z.; Li, W.Q.; Yu, L.H.; Li, W.J. Complete genome sequencing and comparative genome analysis of the extremely halophilic archea, Haloteriigena daqingense. Biotechnol. Appl. Biochem. 2022, 69, 1482–1488. [Google Scholar] [CrossRef]
- Meng, L.; Meng, F.; Zhang, R.; Zhang, Z.; Dong, P.; Sun, K.; Chen, J.; Zhang, W.; Yan, M.; Li, J.; et al. Characterization of a novel two-component Na+(Li+, K+)/H+ antiporter from Halomonas zhaodongensis. Sci. Rep. 2017, 7, 4221. [Google Scholar] [CrossRef]
- Nozaki, K.; Inaba, K.; Kuroda, T.; Tsuda, M.; Tsuchiya, T. Cloning and sequencing of the gene for Na+/H+ antiporter of Vibrio parahaemolyticus. Biochem. Biophys. Res. Commun. 1996, 222, 774–779. [Google Scholar] [CrossRef] [PubMed]
- Padan, E.; Schuldiner, S. Molecular physiology of Na+/H+ antiporters, key transporters in circulation of Na+ and H+ in cells. Biochim. Biophys. Acta. 1994, 1185, 129–151. [Google Scholar] [CrossRef] [PubMed]
- Ito, M.; Guffanti, A.A.; Oudega, B.; Krulwich, T.A. mrp, a multigene, multifunctional locus in Bacillus subtilis with roles in resistance to cholate and to Na+ and in pH homeostasis. J. Bacteriol. 1999, 181, 2394–2402. [Google Scholar] [CrossRef] [PubMed]
- Quinn, M.J.; Resch, C.T.; Sun, J.; Lind, E.J.; Dibrov, P.; Häse, C.C. NhaP1 is a K+(Na+)/H+ antiporter required for growth and internal pH homeostasis of Vibrio cholerae at low extracellular pH. Microbiology 2012, 158, 1094–1105. [Google Scholar] [CrossRef]
- Padan, E.; Bibi, E.; Ito, M.; Krulwich, T.A. Alkaline pH homeostasis in bacteria: New insights. Biochim. Biophys. Acta 2005, 1717, 67–88. [Google Scholar] [CrossRef]
- Altschul, S.F.; Gish, W.; Miller, W.; Myers, E.W.; Lipman, D.J. Basic local alignment search tool. J. Mol. Biol. 1990, 215, 403–410. [Google Scholar] [CrossRef] [PubMed]
- Wimmer, F.; Oberwinkler, T.; Bisle, B.; Tittor, J.; Oesterhelt, D. Identification of the arginine/ornithine antiporter ArcD from Halobacterium salinarum. FEBS Lett. 2008, 582, 3771–3775. [Google Scholar] [CrossRef]
- Karpel, R.; Olami, Y.; Taglicht, D.; Schuldiner, S.; Padan, E. Sequencing of the gene ant which affects the Na+/H+ antiporter activity in Escherichia coli. J. Biol. Chem. 1988, 263, 10408–10414. [Google Scholar] [CrossRef]
- Radchenko, M.V.; Tanaka, K.; Waditee, R.; Oshimi, S.; Matsuzaki, Y.; Fukuhara, M.; Kobayashi, H.; Takabe, T.; Nakamura, T. Potassium/proton antiport system of Escherichia coli. J. Biol. Chem. 2006, 281, 19822–19829. [Google Scholar] [CrossRef]
- Cheng, J.; Guffanti, A.A.; Krulwich, T.A. The chromosomal tetracycline resistance locus of Bacillus subtilis encodes a Na+/H+ antiporter that is physiologically important at elevated pH. J. Biol. Chem. 1994, 269, 27365–27371. [Google Scholar] [CrossRef]
- Saier, M.H.; Reddy, V.S.; Moreno-Hagelsieb, G.; Hendargo, K.J.; Zhang, Y.; Iddamsetty, V.; Lam, K.J.K.; Tian, N.; Russum, S.; Wang, J.; et al. The Transporter Classification Database (TCDB): 2021 update. Nucleic Acids Res. 2021, 49, D461–D467. [Google Scholar] [CrossRef] [PubMed]
- Jiang, J.; Wang, L.; Zhang, H.; Wu, H.; Huang, H.; Yang, L. Putative paired small multidrug resistance family proteins PsmrAB, the homolog of YvdSR, actually function as a novel two-component Na(+)/H(+) antiporter. FEMS Microbiol. Lett. 2013, 338, 31–38. [Google Scholar] [CrossRef] [PubMed]
- Green, M.R.; Sambrook, J. Molecular Cloning: A Laboratory Manual, 4th ed.; Cold Spring Harbor Laboratory Press: Long Island, NY, USA, 2012; pp. 1011–1024. [Google Scholar]
- Rosen, B.P. Ion extrusion systems in Escherichia coli. Methods Enzymol. 1986, 125, 328–336. [Google Scholar] [CrossRef]
- Saitou, N.; Nei, M. The neighbor-joining method: A new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 1987, 4, 406–425. [Google Scholar] [CrossRef]
- Xu, X.W.; Liu, S.J.; Tohty, D.; Oren, A.; Wu, M.; Zhou, P.J. Haloterrigena saccharevitans sp. nov., an extremely halophilic archaeon from Xin-Jiang, China. Int. J. Syst. Evol. Microbiol. 2005, 55, 2539–2542. [Google Scholar] [CrossRef] [PubMed]


| Characteristics | 1028-1-ORF3 | 1028-9-ORF2 |
|---|---|---|
| Gene ID | BB347_RS08225 | BB347_RS08285 |
| Gene abbreviated name | nhaC1 | nhaC2 |
| Protein abbreviated name | NhaC1 | NhaC2 |
| Accession | WP_076580434.1 | WP_076580456.1 |
| Amino acid sequence length(aa) | 566 | 524 |
| Number of hydrophobic amino acids (aa) | 302 | 301 |
| Predicted molecular weight (Da) | 60,722 | 54,786 |
| Number of TMSs | 11 | 12 |
| Definition | Na+/H+ antiporter NhaC family protein | Na+/H+ antiporter NhaC family protein |
| Strain or Plasmid | Description | Source or Reference |
|---|---|---|
| Strains | ||
| N.daqingense JX313T | Original strain, an extremely halophilic archaea | Isolated and identified by our lab [27] |
| E. coli DH5α | Host strain for cloning | Vazyme Biotech Co., Ltd. |
| E. coli KNabc | Na+/H+ antiporter-deficient strain, nhaA::KmR, nhaB::EmR, chaA::CmR | Donated by Prof. Juquan Jiang [31] |
| Plasmids | ||
| pUC18 | Cloning vector, AmpR | Comate Biosciences Co., Ltd. (Changchun, China) |
| pET-28a | Prokaryotic expression vector, KanR | Comate Biosciences Co., Ltd. |
| pET-28AK | Prokaryotic expression vector, KanR and AmpR | This study |
| pUC1028-1 | pUC18 carrying 3.5 kb DNA fragment with Na+/H+ antiport activity | This study |
| pUC1028-9 | pUC18 carrying 4.9 kb DNA fragment with Na+/H+ antiport activity | This study |
| pET28AK-nhaC1 | Heterologous expression vector of nhaC1 | This study |
| pET28AK-nhaC2 | Heterologous expression vector of nhaC2 | This study |
| Component | Soluble Film Buffer | Binding Buffer | Washing Buffer | Elution Buffer |
|---|---|---|---|---|
| Choline chloride | 140 mM | 140 mM | 140 mM | 140 mM |
| Tris | 25 mM | 25 mM | 25 mM | 25 mM |
| Glycerol | 10% | 10% | 10% | 10% |
| N-Dodecyl-β-D-maltoside | 0.02% | 0.02% | 0.02% | 0.02% |
| Imidazole | - | 10 M | 25/45/50/55/60/65/85 mM | 300 mM |
| Primers | Description | Sequence(from 5′ to 3′) | Source or Reference |
|---|---|---|---|
| 22F | Archaea 16S rDNA | ATTCCGGTTGATCCTGC | X.W.Xu, et al. [46] |
| 1540R | AGGAGGTGATCCAGCCGCAG | ||
| M13-47F | Sequencing primers of pUC18 | CGCCAGGGTTTTCCCAGTCACGAC | This study |
| M13R | CACACAGGAAACAGCTATGAC | This study | |
| T7 | Sequencing primers of pET-28a and pET28AK | TAATACGACTCACTATAGGG | This study |
| T7t | GCTAGTTATTGCTCAGCGG | This study | |
| Amp-F | To insertAmpR into pET-28a | CTGCHCGTTGGTGCGGATATCCGCGGAACCCCTATTTGTT | This study |
| Amp-R | GTATCCCACTACCGAGATATCTTACCAATGCTTAATCAGTGAGGC | This study | |
| 1-3FP | To insert DNA sequence of pUC1028-1-ORF3 into pUC18 | TATGACCATGATTACGAATTCATGTCTGACTTTGGAGCGCTTT | This study |
| 1-3RP | CAGGTCGACTCTAGAGGATCCTTACTCCTCAGGGTCCGTCCC | This study | |
| 9-2FP | To insert DNA sequence of pUC1028-9-ORF2 into pUC18 | TATGACCATGATTACGAATTCATGAGTGAAGCCAACGATAATTCA | This study |
| 9-2RP | CAGGTCGACTCTAGAGGATCCTCATAGTCGTGCCACCTCCTCG | This study | |
| NhaC1-EF | To insert nhaC1 into pET28AK | CAGCAAATGGGTCGCGGATCCATGTCTGACTTTGGAGCGCTTT | This study |
| NhaC1-ER | TTGTCGACGGAGCTCGAATTCTTACTCCTCAGGGTCCGTCCC | This study | |
| NhaC1-EF | To insert nhaC2 into pET28AK | CAGCAAATGGGTCGCGGATCCATGAGTGAAGCCAACGATAATTCA | This study |
| NhaC1-ER | TTGTCGACGGAGCTCGAATTCTCACACCCCCCAGAAGAACG | This study |
| NhaC1 Homologs | Accession | NhaC2 Homologs | Accession |
|---|---|---|---|
| Nd_NhaC1(this study) | WP_076580434.1 | Nd_NhaC2(this study) | WP_076580456.1 |
| Ns_NhaC | WP_090507012.1 | Ns_NhaC | WP_090507022.1 |
| NsA_NhaC | WP_278304797.1 | Nt_NhaC | WP_076607379.1 |
| Nti_NhaC | WP_006090653.1 | Np_spa | TYL39413.1 |
| Nte_NhaC | WP_090303449.1 | NsuJ_NhaC | ELY48302.1 |
| No_NhaC | WP_015320610.1 | Nh_NhaC1 | WP_170972344.1 |
| Na_NhaC | WP_005555905.1 | Nh_NhaC2 | WP_162989723.1 |
| Hk_NhaC | WP_092904960.1 | Nsu_NhaC | WP_049890100.1 |
| Nh_NhaC | WP_137288726.1 | Np_NhaC | WP_187432893.1 |
| Ht_NhaC | WP_012943592.1 | Hi_NhaC | WP_049954249.1 |
| Ha_NhaC | WP_120244443.1 | Na_NhaC | WP_152938558.1 |
| Hx_NhaC | WP_013881651.1 | Nt_NhaC | WP_006090630.1 |
| NsA_NhaC | WP_278304817.1 | ||
| HiA_NhaC | EMA28167.1 | ||
| NsC_NhaC | WP_252487510.1 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, Q.; Qiao, M.; Song, J. Characterization of Two Na+(K+, Li+)/H+ Antiporters from Natronorubrum daqingense. Int. J. Mol. Sci. 2023, 24, 10786. https://doi.org/10.3390/ijms241310786
Wang Q, Qiao M, Song J. Characterization of Two Na+(K+, Li+)/H+ Antiporters from Natronorubrum daqingense. International Journal of Molecular Sciences. 2023; 24(13):10786. https://doi.org/10.3390/ijms241310786
Chicago/Turabian StyleWang, Qi, Mengwei Qiao, and Jinzhu Song. 2023. "Characterization of Two Na+(K+, Li+)/H+ Antiporters from Natronorubrum daqingense" International Journal of Molecular Sciences 24, no. 13: 10786. https://doi.org/10.3390/ijms241310786
APA StyleWang, Q., Qiao, M., & Song, J. (2023). Characterization of Two Na+(K+, Li+)/H+ Antiporters from Natronorubrum daqingense. International Journal of Molecular Sciences, 24(13), 10786. https://doi.org/10.3390/ijms241310786






