Liquid Biopsy in NSCLC: An Investigation with Multiple Clinical Implications
Abstract
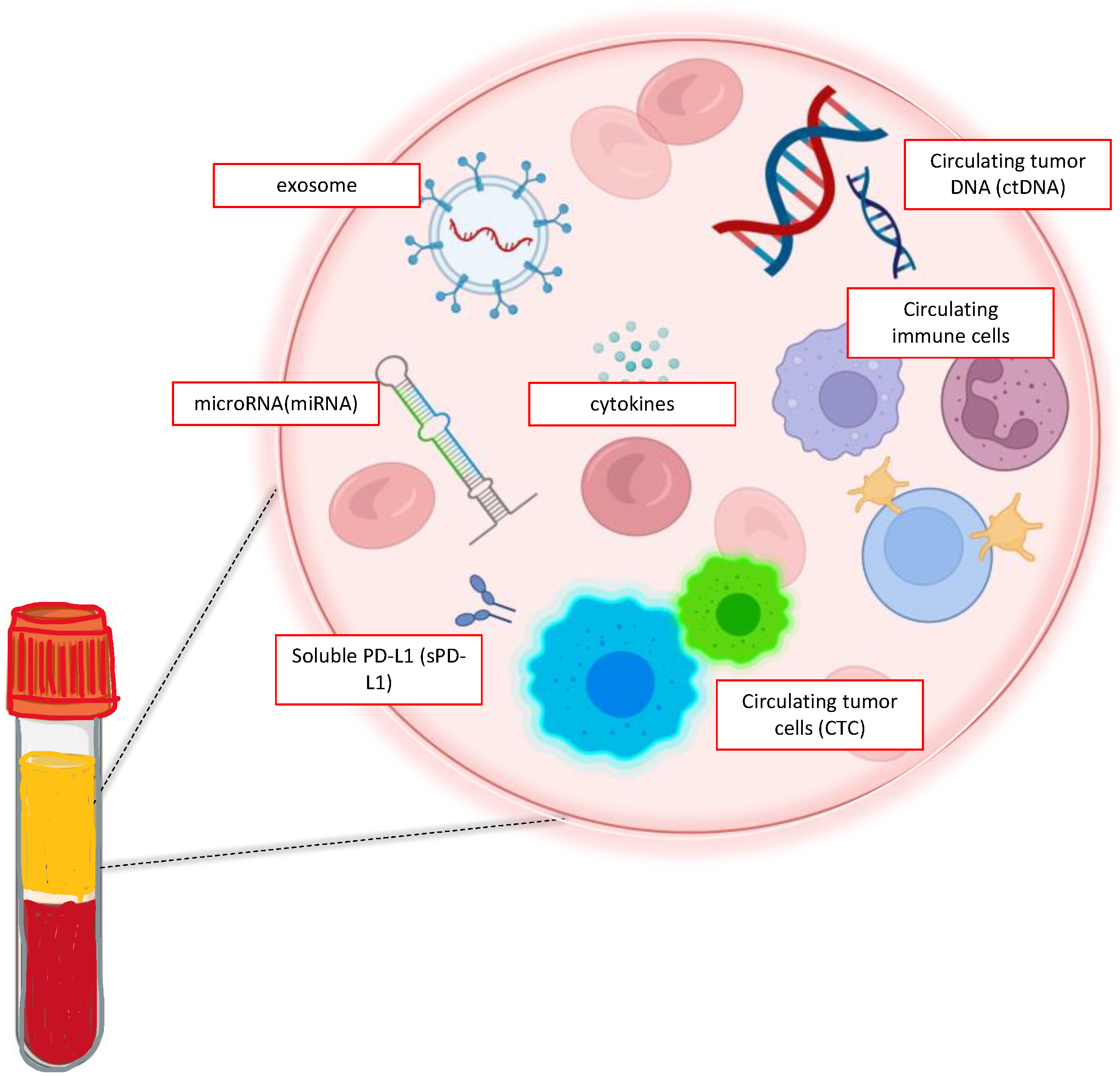
1. Introduction
2. Liquid Biopsy: In the Lab
2.1. CTCs
2.2. ctDNA
2.3. Exosomes and miRNA
2.4. TEPs
3. Liquid Biopsy Diagnostic’s Application When the Tissue Is Unavailable
4. Liquid Biopsy in Screening and in Early Nsclc Disease Stage
4.1. Screening
4.2. Neoadjuvant Setting
4.3. Minimal Residual Disease (MRD) and Adjuvant Setting
5. Liquid Biopsy in Oncogene Addicted Advanced Nsclc: A Focus on Target Therapies
6. Liquid Biopsy in Non-Oncogene Addicted Advanced Nsclc: A Focus on Immunotherapy
6.1. ICI Response or Resistance Prediction
6.2. Treatment Response Monitoring
7. Discussion and Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- American Cancer Society. Facts & Figures 2019. Available online: Https://Www.Cancer.Org/Content/Dam/Cancer-Org/Research/Cancer-Facts-and-Statistics/Annual-Cancer-Facts-and-Figures/2019/Cancer-Facts-and-Figures-2019.Pdf (accessed on 13 June 2020).
- Yang, S.-R.; Schultheis, A.M.; Yu, H.; Mandelker, D.; Ladanyi, M.; Büttner, R. Precision Medicine in Non-Small Cell Lung Cancer: Current Applications and Future Directions. Semin. Cancer Biol. 2022, 84, 184–198. [Google Scholar] [CrossRef] [PubMed]
- Tan, A.C.; Tan, D.S.W. Targeted Therapies for Lung Cancer Patients with Oncogenic Driver Molecular Alterations. J. Clin. Oncol. 2022, 40, 611–625. [Google Scholar] [CrossRef] [PubMed]
- Hendriks, L.E.; Kerr, K.M.; Menis, J.; Mok, T.S.; Nestle, U.; Passaro, A.; Peters, S.; Planchard, D.; Smit, E.F.; Solomon, B.J.; et al. Oncogene-Addicted Metastatic Non-Small-Cell Lung Cancer: ESMO Clinical Practice Guideline for Diagnosis, Treatment and Follow-Up. Ann. Oncol. 2023, 34, 339–357. [Google Scholar] [CrossRef] [PubMed]
- Hendriks, L.E.; Kerr, K.M.; Menis, J.; Mok, T.S.; Nestle, U.; Passaro, A.; Peters, S.; Planchard, D.; Smit, E.F.; Solomon, B.J.; et al. Non-Oncogene-Addicted Metastatic Non-Small-Cell Lung Cancer: ESMO Clinical Practice Guideline for Diagnosis, Treatment and Follow-Up. Ann. Oncol. 2023, 34, 358–376. [Google Scholar] [CrossRef]
- Rolfo, C.; Mack, P.; Scagliotti, G.V.; Aggarwal, C.; Arcila, M.E.; Barlesi, F.; Bivona, T.; Diehn, M.; Dive, C.; Dziadziuszko, R.; et al. Liquid Biopsy for Advanced NSCLC: A Consensus Statement from the International Association for the Study of Lung Cancer. J. Thorac. Oncol. 2021, 16, 1647–1662. [Google Scholar] [CrossRef]
- Mosele, F.; Remon, J.; Mateo, J.; Westphalen, C.B.; Barlesi, F.; Lolkema, M.P.; Normanno, N.; Scarpa, A.; Robson, M.; Meric-Bernstam, F.; et al. Recommendations for the Use of Next-Generation Sequencing (NGS) for Patients with Metastatic Cancers: A Report from the ESMO Precision Medicine Working Group. Ann. Oncol. 2020, 31, 1491–1505. [Google Scholar] [CrossRef]
- Esagian, S.M.; Grigoriadou, G.Ι.; Nikas, I.P.; Boikou, V.; Sadow, P.M.; Won, J.-K.; Economopoulos, K.P. Comparison of Liquid-Based to Tissue-Based Biopsy Analysis by Targeted next Generation Sequencing in Advanced Non-Small Cell Lung Cancer: A Comprehensive Systematic Review. J. Cancer Res. Clin. Oncol. 2020, 146, 2051–2066. [Google Scholar] [CrossRef]
- Reck, M.; Remon, J.; Hellmann, M.D. First-Line Immunotherapy for Non–Small-Cell Lung Cancer. J. Clin. Oncol. 2022, 40, 586–597. [Google Scholar] [CrossRef]
- MacManus, M.; Hegi-Johnson, F. Overcoming Immunotherapy Resistance in NSCLC. Lancet Oncol. 2022, 23, 191–193. [Google Scholar] [CrossRef]
- Fernández-Lázaro, D.; García Hernández, J.L.; García, A.C.; Córdova Martínez, A.; Mielgo-Ayuso, J.; Cruz-Hernández, J.J. Liquid Biopsy as Novel Tool in Precision Medicine: Origins, Properties, Identification and Clinical Perspective of Cancer’s Biomarkers. Diagnostics 2020, 10, 215. [Google Scholar] [CrossRef]
- Diaz, L.A.; Bardelli, A. Liquid Biopsies: Genotyping Circulating Tumor DNA. J. Clin. Oncol. 2014, 32, 579–586. [Google Scholar] [CrossRef]
- Siravegna, G.; Mussolin, B.; Venesio, T.; Marsoni, S.; Seoane, J.; Dive, C.; Papadopoulos, N.; Kopetz, S.; Corcoran, R.B.; Siu, L.L.; et al. How Liquid Biopsies Can Change Clinical Practice in Oncology. Ann. Oncol. 2019, 30, 1580–1590. [Google Scholar] [CrossRef]
- Li, W.; Liu, J.-B.; Hou, L.-K.; Yu, F.; Zhang, J.; Wu, W.; Tang, X.-M.; Sun, F.; Lu, H.-M.; Deng, J.; et al. Liquid Biopsy in Lung Cancer: Significance in Diagnostics, Prediction, and Treatment Monitoring. Mol. Cancer 2022, 21, 25. [Google Scholar] [CrossRef]
- Pantel, K.; Alix-Panabières, C. Liquid Biopsy and Minimal Residual Disease—Latest Advances and Implications for Cure. Nat. Rev. Clin. Oncol. 2019, 16, 409–424. [Google Scholar] [CrossRef]
- Ferreira, M.M.; Ramani, V.C.; Jeffrey, S.S. Circulating Tumor Cell Technologies. Mol. Oncol. 2016, 10, 374–394. [Google Scholar] [CrossRef]
- Nikolaos, G.G.; Konstantinos, A.K.; Melpomeni, G.A.; Lefki-Pavlina, N.G.; Ioannis, S.V. A Novel Combined Methodology for Isolation and Detection of Circulating Tumor Cells Based on Flow Cytometry and Cellular Filtration Technologies. Int. J. Cancer Clin. Res. 2020, 7, 132. [Google Scholar] [CrossRef]
- Andree, K.C.; Van Dalum, G.; Terstappen, L.W.M.M. Challenges in Circulating Tumor Cell Detection by the CellSearch System. Mol. Oncol. 2016, 10, 395–407. [Google Scholar] [CrossRef]
- Hofman, V.; Bonnetaud, C.; Ilie, M.I.; Vielh, P.; Vignaud, J.M.; Fléjou, J.F.; Lantuejoul, S.; Piaton, E.; Mourad, N.; Butori, C.; et al. Preoperative Circulating Tumor Cell Detection Using the Isolation by Size of Epithelial Tumor Cell Method for Patients with Lung Cancer Is a New Prognostic Biomarker. Clin. Cancer Res. 2011, 17, 827–835. [Google Scholar] [CrossRef]
- Wei, T.; Zhu, D.; Yang, Y.; Yuan, G.; Xie, H.; Shen, R. The Application of Nano-Enrichment in CTC Detection and the Clinical Significance of CTCs in Non-Small Cell Lung Cancer (NSCLC) Treatment. PLoS ONE 2019, 14, e0219129. [Google Scholar] [CrossRef]
- Bankó, P.; Lee, S.Y.; Nagygyörgy, V.; Zrínyi, M.; Chae, C.H.; Cho, D.H.; Telekes, A. Technologies for Circulating Tumor Cell Separation from Whole Blood. J. Hematol. Oncol. 2019, 12, 48. [Google Scholar] [CrossRef]
- Yang, Y.; Liu, H.; Chen, Y.; Xiao, N.; Zheng, Z.; Liu, H.; Wan, J. Liquid Biopsy on the Horizon in Immunotherapy of Non-Small Cell Lung Cancer: Current Status, Challenges, and Perspectives. Cell Death Dis. 2023, 14, 230. [Google Scholar] [CrossRef] [PubMed]
- Oliveira, K.C.S.; Ramos, I.B.; Silva, J.M.C.; Barra, W.F.; Riggins, G.J.; Palande, V.; Pinho, C.T.; Frenkel-Morgenstern, M.; Santos, S.E.B.; Assumpcao, P.P.; et al. Current Perspectives on Circulating Tumor DNA, Precision Medicine, and Personalized Clinical Management of Cancer. Mol. Cancer Res. 2020, 18, 517–528. [Google Scholar] [CrossRef] [PubMed]
- Sacher, A.G.; Komatsubara, K.M.; Oxnard, G.R. Application of Plasma Genotyping Technologies in Non–Small Cell Lung Cancer: A Practical Review. J. Thorac. Oncol. 2017, 12, 1344–1356. [Google Scholar] [CrossRef] [PubMed]
- Germano, G.; Mauri, G.; Siravegna, G.; Dive, C.; Pierce, J.; Di Nicolantonio, F.; D’Incalci, M.; Bardelli, A.; Siena, S.; Sartore-Bianchi, A. Parallel Evaluation of Circulating Tumor DNA and Circulating Tumor Cells in Metastatic Colorectal Cancer. Clin. Color. Cancer 2018, 17, 80–83. [Google Scholar] [CrossRef] [PubMed]
- Guibert, N.; Pradines, A.; Favre, G.; Mazieres, J. Current and Future Applications of Liquid Biopsy in Nonsmall Cell Lung Cancer from Early to Advanced Stages. Eur. Respir. Rev. 2020, 29, 190052. [Google Scholar] [CrossRef] [PubMed]
- Normanno, N.; Denis, M.G.; Thress, K.S.; Ratcliffe, M.; Reck, M. Guide to Detecting Epidermal Growth Factor Receptor (EGFR) Mutations in CtDNA of Patients with Advanced Non-Small-Cell Lung Cancer. Oncotarget 2017, 8, 12501–12516. [Google Scholar] [CrossRef]
- Uchida, J.; Kato, K.; Kukita, Y.; Kumagai, T.; Nishino, K.; Daga, H.; Nagatomo, I.; Inoue, T.; Kimura, M.; Oba, S.; et al. Diagnostic Accuracy of Noninvasive Genotyping of EGFR in Lung Cancer Patients by Deep Sequencing of Plasma Cell-Free DNA. Clin. Chem. 2015, 61, 1191–1196. [Google Scholar] [CrossRef]
- Rolfo, C.; Russo, A. Liquid Biopsy for Early Stage Lung Cancer Moves Ever Closer. Nat. Rev. Clin. Oncol. 2020, 17, 523–524. [Google Scholar] [CrossRef]
- Abbosh, C.; Frankell, A.M.; Harrison, T.; Kisistok, J.; Garnett, A.; Johnson, L.; Veeriah, S.; Moreau, M.; Chesh, A.; Chaunzwa, T.L.; et al. Tracking Early Lung Cancer Metastatic Dissemination in TRACERx Using CtDNA. Nature 2023, 616, 553–562. [Google Scholar] [CrossRef]
- Assaf, Z.J.F.; Zou, W.; Fine, A.D.; Socinski, M.A.; Young, A.; Lipson, D.; Freidin, J.F.; Kennedy, M.; Polisecki, E.; Nishio, M.; et al. A Longitudinal Circulating Tumor DNA-Based Model Associated with Survival in Metastatic Non-Small-Cell Lung Cancer. Nat. Med. 2023, 29, 859–868. [Google Scholar] [CrossRef]
- Kustanovich, A.; Schwartz, R.; Peretz, T.; Grinshpun, A. Life and Death of Circulating Cell-Free DNA. Cancer Biol. Ther. 2019, 20, 1057–1067. [Google Scholar] [CrossRef]
- Yáñez-Mó, M.; Siljander, P.R.-M.; Andreu, Z.; Bedina Zavec, A.; Borràs, F.E.; Buzas, E.I.; Buzas, K.; Casal, E.; Cappello, F.; Carvalho, J.; et al. Biological Properties of Extracellular Vesicles and Their Physiological Functions. J. Extracell. Vesicles 2015, 4, 27066. [Google Scholar] [CrossRef]
- Labani-Motlagh, A.; Naseri, S.; Wenthe, J.; Eriksson, E.; Loskog, A. Systemic Immunity upon Local Oncolytic Virotherapy Armed with Immunostimulatory Genes May Be Supported by Tumor-Derived Exosomes. Mol. Ther. Oncolytics 2021, 20, 508–518. [Google Scholar] [CrossRef]
- Lin, S.; Gregory, R.I. MicroRNA Biogenesis Pathways in Cancer. Nat. Rev. Cancer 2015, 15, 321–333. [Google Scholar] [CrossRef]
- Wang, J.; Chen, J.; Sen, S. MicroRNA as Biomarkers and Diagnostics: MicroRNAs as Biomarkers for Diagnostics. J. Cell. Physiol. 2016, 231, 25–30. [Google Scholar] [CrossRef]
- Di Leva, G.; Croce, C.M. MiRNA Profiling of Cancer. Curr. Opin. Genet. Dev. 2013, 23, 3–11. [Google Scholar] [CrossRef]
- Wu, Y.; Zhang, Y.; Zhang, X.; Luo, S.; Yan, X.; Qiu, Y.; Zheng, L.; Li, L. Research Advances for Exosomal MiRNAs Detection in Biosensing: From the Massive Study to the Individual Study. Biosens. Bioelectron. 2021, 177, 112962. [Google Scholar] [CrossRef]
- Wang, H.; Peng, R.; Wang, J.; Qin, Z.; Xue, L. Circulating MicroRNAs as Potential Cancer Biomarkers: The Advantage and Disadvantage. Clin. Epigenetics 2018, 10, 59. [Google Scholar] [CrossRef]
- Bushman, F.D.; Cantu, A.; Everett, J.; Sabatino, D.; Berry, C. Challenges in Estimating Numbers of Vectors Integrated in Gene-Modified Cells Using DNA Sequence Information. Mol. Ther. 2021, 29, 3328–3331. [Google Scholar] [CrossRef]
- Nilsson, R.J.; Balaj, L.; Hulleman, E.; van Rijn, S.; Pegtel, D.M.; Walraven, M.; Widmark, A.; Gerritsen, W.R.; Verheul, H.M.; Vandertop, W.P.; et al. Blood Platelets Contain Tumor-Derived RNA Biomarkers. Blood 2011, 118, 3680–3683. [Google Scholar] [CrossRef]
- Myron, G. Best, Pieter Wesseling, Thomas Wurdinger; Tumor-Educated Platelets as a Noninvasive Biomarker Source for Cancer Detection and Progression Monitoring. Cancer Res. 2018, 78, 3407–3412. [Google Scholar] [CrossRef]
- Dagogo-Jack, I.; Shaw, A.T. Tumour Heterogeneity and Resistance to Cancer Therapies. Nat. Rev. Clin. Oncol. 2018, 15, 81–94. [Google Scholar] [CrossRef]
- De Mattos-Arruda, L.; Weigelt, B.; Cortes, J.; Won, H.H.; Ng, C.K.Y.; Nuciforo, P.; Bidard, F.-C.; Aura, C.; Saura, C.; Peg, V.; et al. Capturing Intra-Tumor Genetic Heterogeneity by de Novo Mutation Profiling of Circulating Cell-Free Tumor DNA: A Proof-of-Principle. Ann. Oncol. 2014, 25, 1729–1735. [Google Scholar] [CrossRef] [PubMed]
- Luo, J.; Shen, L.; Zheng, D. Diagnostic Value of Circulating Free DNA for the Detection of EGFR Mutation Status in NSCLC: A Systematic Review and Meta-Analysis. Sci. Rep. 2014, 4, 6269. [Google Scholar] [CrossRef] [PubMed]
- Qiu, M.; Wang, J.; Xu, Y.; Ding, X.; Li, M.; Jiang, F.; Xu, L.; Yin, R. Circulating Tumor DNA Is Effective for the Detection of EGFR Mutation in Non–Small Cell Lung Cancer: A Meta-Analysis. Cancer Epidemiol. Biomark. Prev. 2015, 24, 206–212. [Google Scholar] [CrossRef]
- Ettinger, D.S.; Wood, D.E.; Aisner, D.L.; Akerley, W.; Bauman, J.R.; Bharat, A.; Bruno, D.S.; Chang, J.Y.; Chirieac, L.R.; D’amico, T.A.; et al. Non-Small Cell Lung Cancer, Version 3.2022, NCCN Clinical Practice Guidelines in Oncology. J. Natl. Compr. Cancer Netw. 2022, 20, 497–530. [Google Scholar] [CrossRef]
- Goldman, J.W.; Noor, Z.S.; Remon, J.; Besse, B.; Rosenfeld, N. Are Liquid Biopsies a Surrogate for Tissue EGFR Testing? Ann. Oncol. 2018, 29, i38–i46. [Google Scholar] [CrossRef]
- Reck, M.; Hagiwara, K.; Han, B.; Tjulandin, S.; Grohé, C.; Yokoi, T.; Morabito, A.; Novello, S.; Arriola, E.; Molinier, O.; et al. CtDNA Determination of EGFR Mutation Status in European and Japanese Patients with Advanced NSCLC: The ASSESS Study. J. Thorac. Oncol. 2016, 11, 1682–1689. [Google Scholar] [CrossRef]
- Remon, J.; Soria, J.C.; Planchard, D.; Jovelet, C.; Pannet, C.; Lacroix, L.; Gazzah, A.; Lawson, A.; Smalley, S.; Howarth, K.; et al. Abstract 3192: Liquid Biopsies for Molecular Profiling of Mutations in Non-Small Cell Lung Cancer Patients Lacking Tissue Samples. Cancer Res. 2016, 76, 3192. [Google Scholar] [CrossRef]
- Li, Y.-Z.; Kong, S.-N.; Liu, Y.-P.; Yang, Y.; Zhang, H.-M. Can Liquid Biopsy Based on CtDNA/CfDNA Replace Tissue Biopsy for the Precision Treatment of EGFR-Mutated NSCLC? J. Clin. Med. 2023, 12, 1438. [Google Scholar] [CrossRef]
- Douillard, J.-Y.; Ostoros, G.; Cobo, M.; Ciuleanu, T.; Cole, R.; McWalter, G.; Walker, J.; Dearden, S.; Webster, A.; Milenkova, T.; et al. Gefitinib Treatment in EGFR Mutated Caucasian NSCLC: Circulating-Free Tumor DNA as a Surrogate for Determination of EGFR Status. J. Thorac. Oncol. 2014, 9, 1345–1353. [Google Scholar] [CrossRef]
- Deng, Q.; Fang, Q.; Sun, H.; Singh, A.P.; Alexander, M.; Li, S.; Cheng, H.; Zhou, S. Detection of Plasma EGFR Mutations for Personalized Treatment of Lung Cancer Patients without Pathologic Diagnosis. Cancer Med. 2020, 9, 2085–2095. [Google Scholar] [CrossRef]
- Xu, J.; Liu, Z.; Bai, H.; Dong, G.; Zhong, J.; Wan, R.; Zang, A.; Li, X.; Li, Q.; Guo, J.; et al. Evaluation of Clinical Outcomes of Icotinib in Patients with Clinically Diagnosed Advanced Lung Cancer With EGFR -Sensitizing Variants Assessed by Circulating Tumor DNA Testing: A Phase 2 Nonrandomized Clinical Trial. JAMA Oncol. 2022, 8, 1328. [Google Scholar] [CrossRef]
- Raez, L.E.; Brice, K.; Dumais, K.; Lopez-Cohen, A.; Wietecha, D.; Izquierdo, P.A.; Santos, E.S.; Powery, H.W. Liquid Biopsy Versus Tissue Biopsy to Determine Front Line Therapy in Metastatic Non-Small Cell Lung Cancer (NSCLC). Clin. Lung Cancer 2023, 24, 120–129. [Google Scholar] [CrossRef]
- Mack, P.C.; Banks, K.C.; Espenschied, C.R.; Burich, R.A.; Zill, O.A.; Lee, C.E.; Riess, J.W.; Mortimer, S.A.; Talasaz, A.; Lanman, R.B.; et al. Spectrum of Driver Mutations and Clinical Impact of Circulating Tumor DNA Analysis in Non–Small Cell Lung Cancer: Analysis of over 8000 Cases. Cancer 2020, 126, 3219–3228. [Google Scholar] [CrossRef]
- Zugazagoitia, J.; Ramos, I.; Trigo, J.M.; Palka, M.; Gómez-Rueda, A.; Jantus-Lewintre, E.; Camps, C.; Isla, D.; Iranzo, P.; Ponce-Aix, S.; et al. Clinical Utility of Plasma-Based Digital next-Generation Sequencing in Patients with Advance-Stage Lung Adenocarcinomas with Insufficient Tumor Samples for Tissue Genotyping. Ann. Oncol. 2019, 30, 290–296. [Google Scholar] [CrossRef]
- Dziadziuszko, R.; Mok, T.; Peters, S.; Han, J.-Y.; Alatorre-Alexander, J.; Leighl, N.; Sriuranpong, V.; Pérol, M.; De Castro Junior, G.; Nadal, E.; et al. Blood First Assay Screening Trial (BFAST) in Treatment-Naive Advanced or Metastatic NSCLC: Initial Results of the Phase 2 ALK-Positive Cohort. J. Thorac. Oncol. 2021, 16, 2040–2050. [Google Scholar] [CrossRef]
- Paik, P.K.; Veillon, R.; Cortot, A.B.; Felip, E.; Sakai, H.; Mazieres, J.; Griesinger, F.; Horn, L.; Senellart, H.; Van Meerbeeck, J.P.; et al. Phase II Study of Tepotinib in NSCLC Patients with METex14 Mutations. J. Clin. Oncol. 2019, 37, 9005. [Google Scholar] [CrossRef]
- The National Lung Screening Trial Research Team Reduced Lung-Cancer Mortality with Low-Dose Computed Tomographic Screening. N. Engl. J. Med. 2011, 365, 395–409. [CrossRef]
- Sozzi, G.; Conte, D.; Leon, M.; Cirincione, R.; Roz, L.; Ratcliffe, C.; Roz, E.; Cirenei, N.; Bellomi, M.; Pelosi, G.; et al. Quantification of Free Circulating DNA As a Diagnostic Marker in Lung Cancer. J. Clin. Oncol. 2003, 21, 3902–3908. [Google Scholar] [CrossRef]
- Sozzi, G.; Roz, L.; Conte, D.; Mariani, L.; Andriani, F.; Lo Vullo, S.; Verri, C.; Pastorino, U. Plasma DNA Quantification in Lung Cancer Computed Tomography Screening: Five-Year Results of a Prospective Study. Am. J. Respir. Crit. Care Med. 2009, 179, 69–74. [Google Scholar] [CrossRef] [PubMed]
- Paci, M.; Maramotti, S.; Bellesia, E.; Formisano, D.; Albertazzi, L.; Ricchetti, T.; Ferrari, G.; Annessi, V.; Lasagni, D.; Carbonelli, C.; et al. Circulating Plasma DNA as Diagnostic Biomarker in Non-Small Cell Lung Cancer. Lung Cancer 2009, 64, 92–97. [Google Scholar] [CrossRef] [PubMed]
- Catarino, R.; Coelho, A.; Araújo, A.; Gomes, M.; Nogueira, A.; Lopes, C.; Medeiros, R. Circulating DNA: Diagnostic Tool and Predictive Marker for Overall Survival of NSCLC Patients. PLoS ONE 2012, 7, e38559. [Google Scholar] [CrossRef] [PubMed]
- Newman, A.M.; Bratman, S.V.; To, J.; Wynne, J.F.; Eclov, N.C.W.; Modlin, L.A.; Liu, C.L.; Neal, J.W.; Wakelee, H.A.; Merritt, R.E.; et al. An Ultrasensitive Method for Quantitating Circulating Tumor DNA with Broad Patient Coverage. Nat. Med. 2014, 20, 548–554. [Google Scholar] [CrossRef]
- Phallen, J.; Sausen, M.; Adleff, V.; Leal, A.; Hruban, C.; White, J.; Anagnostou, V.; Fiksel, J.; Cristiano, S.; Papp, E.; et al. Direct Detection of Early-Stage Cancers Using Circulating Tumor DNA. Sci. Transl. Med. 2017, 9, eaan2415. [Google Scholar] [CrossRef]
- Ooki, A.; Maleki, Z.; Tsay, J.-C.J.; Goparaju, C.; Brait, M.; Turaga, N.; Nam, H.-S.; Rom, W.N.; Pass, H.I.; Sidransky, D.; et al. A Panel of Novel Detection and Prognostic Methylated DNA Markers in Primary Non–Small Cell Lung Cancer and Serum DNA. Clin. Cancer Res. 2017, 23, 7141–7152. [Google Scholar] [CrossRef]
- Hulbert, A.; Jusue-Torres, I.; Stark, A.; Chen, C.; Rodgers, K.; Lee, B.; Griffin, C.; Yang, A.; Huang, P.; Wrangle, J.; et al. Early Detection of Lung Cancer Using DNA Promoter Hypermethylation in Plasma and Sputum. Clin. Cancer Res. 2017, 23, 1998–2005. [Google Scholar] [CrossRef]
- Klein, E.A.; Richards, D.; Cohn, A.; Tummala, M.; Lapham, R.; Cosgrove, D.; Chung, G.; Clement, J.; Gao, J.; Hunkapiller, N.; et al. Clinical Validation of a Targeted Methylation-Based Multi-Cancer Early Detection Test Using an Independent Validation Set. Ann. Oncol. 2021, 32, 1167–1177. [Google Scholar] [CrossRef]
- Hubbell, E.; Clarke, C.A.; Aravanis, A.M.; Berg, C.D. Modeled Reductions in Late-Stage Cancer with a Multi-Cancer Early Detection Test. Cancer Epidemiol. Biomark. Prev. 2021, 30, 460–468. [Google Scholar] [CrossRef]
- Neal, R.D.; Johnson, P.; Clarke, C.A.; Hamilton, S.A.; Zhang, N.; Kumar, H.; Swanton, C.; Sasieni, P. Cell-Free DNA–Based Multi-Cancer Early Detection Test in an Asymptomatic Screening Population (NHS-Galleri): Design of a Pragmatic, Prospective Randomised Controlled Trial. Cancers 2022, 14, 4818. [Google Scholar] [CrossRef]
- Reis, P.P.; Drigo, S.A.; Carvalho, R.F.; Lopez Lapa, R.M.; Felix, T.F.; Patel, D.; Cheng, D.; Pintilie, M.; Liu, G.; Tsao, M.-S. Circulating MiR-16-5p, MiR-92a-3p, and MiR-451a in Plasma from Lung Cancer Patients: Potential Application in Early Detection and a Regulatory Role in Tumorigenesis Pathways. Cancers 2020, 12, 2071. [Google Scholar] [CrossRef]
- Abdollahi, A.; Rahmati, S.; Ghaderi, B.; Sigari, N.; Nikkhoo, B.; Sharifi, K.; Abdi, M. A Combined Panel of Circulating MicroRNA as a Diagnostic Tool for Detection of the Non-Small Cell Lung Cancer. QJM Int. J. Med. 2019, 112, 779–785. [Google Scholar] [CrossRef]
- Chen, K.; Zhao, H.; Shi, Y.; Yang, F.; Wang, L.T.; Kang, G.; Nie, Y.; Wang, J. Perioperative Dynamic Changes in Circulating Tumor DNA in Patients with Lung Cancer (DYNAMIC). Clin. Cancer Res. 2019, 25, 7058–7067. [Google Scholar] [CrossRef]
- Sozzi, G.; Boeri, M.; Rossi, M.; Verri, C.; Suatoni, P.; Bravi, F.; Roz, L.; Conte, D.; Grassi, M.; Sverzellati, N.; et al. Clinical Utility of a Plasma-Based MiRNA Signature Classifier Within Computed Tomography Lung Cancer Screening: A Correlative MILD Trial Study. J. Clin. Oncol. 2014, 32, 768–773. [Google Scholar] [CrossRef]
- Pastorino, U.; Boeri, M.; Sestini, S.; Sabia, F.; Milanese, G.; Silva, M.; Suatoni, P.; Verri, C.; Cantarutti, A.; Sverzellati, N.; et al. Baseline Computed Tomography Screening and Blood MicroRNA Predict Lung Cancer Risk and Define Adequate Intervals in the BioMILD Trial. Ann. Oncol. 2022, 33, 395–405. [Google Scholar] [CrossRef]
- Montani, F.; Marzi, M.J.; Dezi, F.; Dama, E.; Carletti, R.M.; Bonizzi, G.; Bertolotti, R.; Bellomi, M.; Rampinelli, C.; Maisonneuve, P.; et al. MiR-Test: A Blood Test for Lung Cancer Early Detection. JNCI J. Natl. Cancer Inst. 2015, 107, djv063. [Google Scholar] [CrossRef]
- Yang, Y.; Hu, Z.; Zhou, Y.; Zhao, G.; Lei, Y.; Li, G.; Chen, S.; Chen, K.; Shen, Z.; Chen, X.; et al. The Clinical Use of Circulating MicroRNAs as Non-Invasive Diagnostic Biomarkers for Lung Cancers. Oncotarget 2017, 8, 90197–90214. [Google Scholar] [CrossRef]
- Shen, J.; Liao, J.; Guarnera, M.A.; Fang, H.; Cai, L.; Stass, S.A.; Jiang, F. Analysis of MicroRNAs in Sputum to Improve Computed Tomography for Lung Cancer Diagnosis. J. Thorac. Oncol. 2014, 9, 33–40. [Google Scholar] [CrossRef]
- Su, Y.; Guarnera, M.A.; Fang, H.; Jiang, F. Small Non-Coding RNA Biomarkers in Sputum for Lung Cancer Diagnosis. Mol. Cancer 2016, 15, 36. [Google Scholar] [CrossRef]
- Cazzoli, R.; Buttitta, F.; Di Nicola, M.; Malatesta, S.; Marchetti, A.; Rom, W.N.; Pass, H.I. MicroRNAs Derived from Circulating Exosomes as Noninvasive Biomarkers for Screening and Diagnosing Lung Cancer. J. Thorac. Oncol. 2013, 8, 1156–1162. [Google Scholar] [CrossRef]
- Rabinowits, G.; Gerçel-Taylor, C.; Day, J.M.; Taylor, D.D.; Kloecker, G.H. Exosomal MicroRNA: A Diagnostic Marker for Lung Cancer. Clin. Lung Cancer 2009, 10, 42–46. [Google Scholar] [CrossRef] [PubMed]
- Yu, Y.; Chen, Z.; Dong, J.; Wei, P.; Hu, R.; Zhou, C.; Sun, N.; Luo, M.; Yang, W.; Yao, R.; et al. Folate Receptor-Positive Circulating Tumor Cells as a Novel Diagnostic Biomarker in Non-Small Cell Lung Cancer. Transl. Oncol. 2013, 6, 697–702. [Google Scholar] [CrossRef] [PubMed]
- Fiorelli, A.; Accardo, M.; Carelli, E.; Angioletti, D.; Santini, M.; Di Domenico, M. Circulating Tumor Cells in Diagnosing Lung Cancer: Clinical and Morphologic Analysis. Ann. Thorac. Surg. 2015, 99, 1899–1905. [Google Scholar] [CrossRef] [PubMed]
- Ilie, M.; Hofman, V.; Long-Mira, E.; Selva, E.; Vignaud, J.-M.; Padovani, B.; Mouroux, J.; Marquette, C.-H.; Hofman, P. “Sentinel” Circulating Tumor Cells Allow Early Diagnosis of Lung Cancer in Patients with Chronic Obstructive Pulmonary Disease. PLoS ONE 2014, 9, e111597. [Google Scholar] [CrossRef]
- Goswami, C.; Chawla, S.; Thakral, D.; Pant, H.; Verma, P.; Malik, P.S.; Jayadeva; Gupta, R.; Ahuja, G.; Sengupta, D. Molecular Signature Comprising 11 Platelet-Genes Enables Accurate Blood-Based Diagnosis of NSCLC. BMC Genom. 2020, 21, 744. [Google Scholar] [CrossRef]
- Chabon, J.J.; Hamilton, E.G.; Kurtz, D.M.; Esfahani, M.S.; Moding, E.J.; Stehr, H.; Schroers-Martin, J.; Nabet, B.Y.; Chen, B.; Chaudhuri, A.A.; et al. Integrating Genomic Features for Non-Invasive Early Lung Cancer Detection. Nature 2020, 580, 245–251. [Google Scholar] [CrossRef]
- Robbins, H.A.; Alcala, K.; Moez, E.K.; Guida, F.; Thomas, S.; Zahed, H.; Warkentin, M.T.; Smith-Byrne, K.; Brhane, Y.; Muller, D.; et al. Design and Methodological Considerations for Biomarker Discovery and Validation in the Integrative Analysis of Lung Cancer Etiology and Risk (INTEGRAL) Program. Ann. Epidemiol. 2023, 77, 1–12. [Google Scholar] [CrossRef]
- Ponomaryova, A.A.; Rykova, E.Y.; Cherdyntseva, N.V.; Skvortsova, T.E.; Dobrodeev, A.Y.; Zav’yalov, A.A.; Bryzgalov, L.O.; Tuzikov, S.A.; Vlassov, V.V.; Laktionov, P.P. Potentialities of Aberrantly Methylated Circulating DNA for Diagnostics and Post-Treatment Follow-up of Lung Cancer Patients. Lung Cancer 2013, 81, 397–403. [Google Scholar] [CrossRef]
- Provencio, M.; Serna-Blasco, R.; Nadal, E.; Insa, A.; García-Campelo, M.R.; Casal Rubio, J.; Dómine, M.; Majem, M.; Rodríguez-Abreu, D.; Martínez-Martí, A.; et al. Overall Survival and Biomarker Analysis of Neoadjuvant Nivolumab Plus Chemotherapy in Operable Stage IIIA Non–Small-Cell Lung Cancer (NADIM Phase II Trial). J. Clin. Oncol. 2022, 40, 2924–2933. [Google Scholar] [CrossRef]
- Forde, P.M.; Spicer, J.; Lu, S.; Provencio, M.; Mitsudomi, T.; Awad, M.M.; Felip, E.; Broderick, S.R.; Brahmer, J.R.; Swanson, S.J.; et al. Neoadjuvant Nivolumab plus Chemotherapy in Resectable Lung Cancer. N. Engl. J. Med. 2022, 386, 1973–1985. [Google Scholar] [CrossRef]
- Kris, M.G.; Grindheim, J.M.; Chaft, J.E.; Lee, J.M.; Johnson, B.E.; Rusch, V.W.; Bunn, P.A.; Pass, H.; Schum, E.; Carlisle, J.; et al. 1O Dynamic Circulating Tumour DNA (CtDNA) Response to Neoadjuvant (NA) Atezolizumab (Atezo) and Surgery (Surg) and Association with Outcomes in Patients (Pts) with NSCLC. Ann. Oncol. 2021, 32, S1373. [Google Scholar] [CrossRef]
- Chaft, J.E.; Oezkan, F.; Kris, M.G.; Bunn, P.A.; Wistuba, I.I.; Kwiatkowski, D.J.; Owen, D.H.; Tang, Y.; Johnson, B.E.; Lee, J.M.; et al. Neoadjuvant Atezolizumab for Resectable Non-Small Cell Lung Cancer: An Open-Label, Single-Arm Phase II Trial. Nat. Med. 2022, 28, 2155–2161. [Google Scholar] [CrossRef]
- Yue, D.; Liu, W.; Chen, C.; Zhang, T.; Ma, Y.; Cui, L.; Gu, Y.; Bei, T.; Zhao, X.; Zhang, B.; et al. Circulating Tumor DNA Predicts Neoadjuvant Immunotherapy Efficacy and Recurrence-Free Survival in Surgical Non-Small Cell Lung Cancer Patients. Transl. Lung Cancer Res. 2022, 11, 263–276. [Google Scholar] [CrossRef]
- Shen, H.; Jin, Y.; Zhao, H.; Wu, M.; Zhang, K.; Wei, Z.; Wang, X.; Wang, Z.; Li, Y.; Yang, F.; et al. Potential Clinical Utility of Liquid Biopsy in Early-Stage Non-Small Cell Lung Cancer. BMC Med. 2022, 20, 480. [Google Scholar] [CrossRef]
- Detterbeck, F.C. The Eighth Edition TNM Stage Classification for Lung Cancer: What Does It Mean on Main Street? J. Thorac. Cardiovasc. Surg. 2018, 155, 356–359. [Google Scholar] [CrossRef]
- Pignon, J.-P.; Tribodet, H.; Scagliotti, G.V.; Douillard, J.-Y.; Shepherd, F.A.; Stephens, R.J.; Dunant, A.; Torri, V.; Rosell, R.; Seymour, L.; et al. Lung Adjuvant Cisplatin Evaluation: A Pooled Analysis by the LACE Collaborative Group. J. Clin. Oncol. 2008, 26, 3552–3559. [Google Scholar] [CrossRef]
- Spigel, D.R.; Faivre-Finn, C.; Gray, J.E.; Vicente, D.; Planchard, D.; Paz-Ares, L.; Vansteenkiste, J.F.; Garassino, M.C.; Hui, R.; Quantin, X.; et al. Five-Year Survival Outcomes from the PACIFIC Trial: Durvalumab After Chemoradiotherapy in Stage III Non–Small-Cell Lung Cancer. J. Clin. Oncol. 2022, 40, 1301–1311. [Google Scholar] [CrossRef]
- Nagasaka, M.; Uddin, M.H.; Al-Hallak, M.N.; Rahman, S.; Balasubramanian, S.; Sukari, A.; Azmi, A.S. Liquid Biopsy for Therapy Monitoring in Early-Stage Non-Small Cell Lung Cancer. Mol. Cancer 2021, 20, 82. [Google Scholar] [CrossRef]
- Guo, N.; Lou, F.; Ma, Y.; Li, J.; Yang, B.; Chen, W.; Ye, H.; Zhang, J.-B.; Zhao, M.-Y.; Wu, W.-J.; et al. Circulating Tumor DNA Detection in Lung Cancer Patients before and after Surgery. Sci. Rep. 2016, 6, 33519. [Google Scholar] [CrossRef]
- Chen, K.; Kang, G.; Zhao, H.; Zhang, K.; Zhang, J.; Yang, F.; Wang, J. Liquid Biopsy in Newly Diagnosed Patients with Locoregional (I-IIIA) Non-Small Cell Lung Cancer. Expert Rev. Mol. Diagn. 2019, 19, 419–427. [Google Scholar] [CrossRef]
- Bratman, S.V.; Newman, A.M.; Alizadeh, A.A.; Diehn, M. Potential Clinical Utility of Ultrasensitive Circulating Tumor DNA Detection with CAPP-Seq. Expert Rev. Mol. Diagn. 2015, 15, 715–719. [Google Scholar] [CrossRef] [PubMed]
- Chaudhuri, A.A.; Chabon, J.J.; Lovejoy, A.F.; Newman, A.M.; Stehr, H.; Azad, T.D.; Khodadoust, M.S.; Esfahani, M.S.; Liu, C.L.; Zhou, L.; et al. Early Detection of Molecular Residual Disease in Localized Lung Cancer by Circulating Tumor DNA Profiling. Cancer Discov. 2017, 7, 1394–1403. [Google Scholar] [CrossRef] [PubMed]
- Moding, E.J.; Liu, Y.; Nabet, B.Y.; Chabon, J.J.; Chaudhuri, A.A.; Hui, A.B.; Bonilla, R.F.; Ko, R.B.; Yoo, C.H.; Gojenola, L.; et al. Circulating Tumor DNA Dynamics Predict Benefit from Consolidation Immunotherapy in Locally Advanced Non-Small-Cell Lung Cancer. Nat. Cancer 2020, 1, 176–183. [Google Scholar] [CrossRef] [PubMed]
- Parikh, A.R.; Leshchiner, I.; Elagina, L.; Goyal, L.; Levovitz, C.; Siravegna, G.; Livitz, D.; Rhrissorrakrai, K.; Martin, E.E.; Van Seventer, E.E.; et al. Liquid versus Tissue Biopsy for Detecting Acquired Resistance and Tumor Heterogeneity in Gastrointestinal Cancers. Nat. Med. 2019, 25, 1415–1421. [Google Scholar] [CrossRef] [PubMed]
- Nigro, M.C.; Marchese, P.V.; Deiana, C.; Casadio, C.; Galvani, L.; Di Federico, A.; De Giglio, A. Clinical Utility and Application of Liquid Biopsy Genotyping in Lung Cancer: A Comprehensive Review. Lung Cancer Targets Ther. 2023, 14, 11–25. [Google Scholar] [CrossRef]
- Siravegna, G.; Mussolin, B.; Buscarino, M.; Corti, G.; Cassingena, A.; Crisafulli, G.; Ponzetti, A.; Cremolini, C.; Amatu, A.; Lauricella, C.; et al. Clonal Evolution and Resistance to EGFR Blockade in the Blood of Colorectal Cancer Patients. Nat. Med. 2015, 21, 795–801. [Google Scholar] [CrossRef]
- VanderLaan, P.A.; Yamaguchi, N.; Folch, E.; Boucher, D.H.; Kent, M.S.; Gangadharan, S.P.; Majid, A.; Goldstein, M.A.; Huberman, M.S.; Kocher, O.N.; et al. Success and Failure Rates of Tumor Genotyping Techniques in Routine Pathological Samples with Non-Small-Cell Lung Cancer. Lung Cancer 2014, 84, 39–44. [Google Scholar] [CrossRef]
- Bonanno, L.; Dal Maso, A.; Pavan, A.; Zulato, E.; Calvetti, L.; Pasello, G.; Guarneri, V.; Conte, P.; Indraccolo, S. Liquid Biopsy and Non-Small Cell Lung Cancer: Are We Looking at the Tip of the Iceberg? Br. J. Cancer 2022, 127, 383–393. [Google Scholar] [CrossRef]
- Wu, Y.-L.; Sequist, L.V.; Hu, C.-P.; Feng, J.; Lu, S.; Huang, Y.; Li, W.; Hou, M.; Schuler, M.; Mok, T.; et al. EGFR Mutation Detection in Circulating Cell-Free DNA of Lung Adenocarcinoma Patients: Analysis of LUX-Lung 3 and 6. Br. J. Cancer 2017, 116, 175–185. [Google Scholar] [CrossRef]
- Han, B.; Tjulandin, S.; Hagiwara, K.; Normanno, N.; Wulandari, L.; Laktionov, K.; Hudoyo, A.; He, Y.; Zhang, Y.-P.; Wang, M.-Z.; et al. EGFR Mutation Prevalence in Asia-Pacific and Russian Patients with Advanced NSCLC of Adenocarcinoma and Non-Adenocarcinoma Histology: The IGNITE Study. Lung Cancer 2017, 113, 37–44. [Google Scholar] [CrossRef]
- Karlovich, C.; Goldman, J.W.; Sun, J.-M.; Mann, E.; Sequist, L.V.; Konopa, K.; Wen, W.; Angenendt, P.; Horn, L.; Spigel, D.; et al. Assessment of EGFR Mutation Status in Matched Plasma and Tumor Tissue of NSCLC Patients from a Phase I Study of Rociletinib (CO-1686). Clin. Cancer Res. 2016, 22, 2386–2395. [Google Scholar] [CrossRef]
- Sacher, A.G.; Paweletz, C.; Dahlberg, S.E.; Alden, R.S.; O’Connell, A.; Feeney, N.; Mach, S.L.; Jänne, P.A.; Oxnard, G.R. Prospective Validation of Rapid Plasma Genotyping for the Detection of EGFR and KRAS Mutations in Advanced Lung Cancer. JAMA Oncol. 2016, 2, 1014. [Google Scholar] [CrossRef]
- Oxnard, G.R.; Thress, K.S.; Alden, R.S.; Lawrance, R.; Paweletz, C.P.; Cantarini, M.; Yang, J.C.-H.; Barrett, J.C.; Jänne, P.A. Association Between Plasma Genotyping and Outcomes of Treatment with Osimertinib (AZD9291) in Advanced Non–Small-Cell Lung Cancer. J. Clin. Oncol. 2016, 34, 3375–3382. [Google Scholar] [CrossRef]
- Zheng, D.; Ye, X.; Zhang, M.Z.; Sun, Y.; Wang, J.Y.; Ni, J.; Zhang, H.P.; Zhang, L.; Luo, J.; Zhang, J.; et al. Plasma EGFR T790M CtDNA Status Is Associated with Clinical Outcome in Advanced NSCLC Patients with Acquired EGFR-TKI Resistance. Sci. Rep. 2016, 6, 20913. [Google Scholar] [CrossRef]
- Liu, Z.; Xie, Z.; Zhao, S.; Ye, D.; Cai, X.; Cheng, B.; Li, C.; Xiong, S.; Li, J.; Liang, H.; et al. Presence of Allele Frequency Heterogeneity Defined by CtDNA Profiling Predicts Unfavorable Overall Survival of NSCLC. Transl. Lung Cancer Res. 2019, 8, 1045–1050. [Google Scholar] [CrossRef]
- Mok, T.; Wu, Y.-L.; Lee, J.S.; Yu, C.-J.; Sriuranpong, V.; Sandoval-Tan, J.; Ladrera, G.; Thongprasert, S.; Srimuninnimit, V.; Liao, M.; et al. Detection and Dynamic Changes of EGFR Mutations from Circulating Tumor DNA as a Predictor of Survival Outcomes in NSCLC Patients Treated with First-Line Intercalated Erlotinib and Chemotherapy. Clin. Cancer Res. 2015, 21, 3196–3203. [Google Scholar] [CrossRef]
- Karachaliou, N.; Mayo-de Las Casas, C.; Queralt, C.; De Aguirre, I.; Melloni, B.; Cardenal, F.; Garcia-Gomez, R.; Massuti, B.; Sánchez, J.M.; Porta, R.; et al. Association of EGFR L858R Mutation in Circulating Free DNA With Survival in the EURTAC Trial. JAMA Oncol. 2015, 1, 149. [Google Scholar] [CrossRef]
- Yu, R.; Bai, H.; Li, T.; Gao, B.; Han, J.; Chang, G.; Zhang, P.; Fei, K.; He, X.; Wang, J. TP53 Mutations in Circulating Tumor DNA in Advanced Epidermal Growth Factor Receptor-Mutant Lung Adenocarcinoma Patients Treated with Gefitinib. Transl. Oncol. 2021, 14, 101163. [Google Scholar] [CrossRef]
- Nygaard, A.D.; Garm Spindler, K.-L.; Pallisgaard, N.; Andersen, R.F.; Jakobsen, A. The Prognostic Value of KRAS Mutated Plasma DNA in Advanced Non-Small Cell Lung Cancer. Lung Cancer 2013, 79, 312–317. [Google Scholar] [CrossRef]
- Gautschi, O.; Huegli, B.; Ziegler, A.; Gugger, M.; Heighway, J.; Ratschiller, D.; Mack, P.C.; Gumerlock, P.H.; Kung, H.J.; Stahel, R.A.; et al. Origin and Prognostic Value of Circulating KRAS Mutations in Lung Cancer Patients. Cancer Lett. 2007, 254, 265–273. [Google Scholar] [CrossRef]
- Li, j.; Dong, W.; Liu, L.N.; Huang, Y.J.; Xiao, M.F. Liquid Biopsy for ALK-Positive Early Non-Small-Cell Lung Cancer Predicts Disease Relapse. Future Oncol. 2021, 17, 81–90. [Google Scholar] [CrossRef] [PubMed]
- Kwon, M.; Ku, B.M.; Olsen, S.; Park, S.; Lefterova, M.; Odegaard, J.; Jung, H.; Sun, J.; Lee, S.; Ahn, J.S.; et al. Longitudinal Monitoring by Next-generation Sequencing of Plasma Cell-free DNA in ALK Rearranged NSCLC Patients Treated with ALK Tyrosine Kinase Inhibitors. Cancer Med. 2022, 11, 2944–2956. [Google Scholar] [CrossRef] [PubMed]
- Li, M.; Zhang, X. BRAF Mutations and Resistance of Non-Small Cell Lung Cancer to BRAF-Targeted Therapies Using Liquid Biopsy. Asia-Pac. J. Oncol. Nurs. 2021, 8, 110–111. [Google Scholar] [CrossRef] [PubMed]
- Bean, J.; Brennan, C.; Shih, J.-Y.; Riely, G.; Viale, A.; Wang, L.; Chitale, D.; Motoi, N.; Szoke, J.; Broderick, S.; et al. MET Amplification Occurs with or without T790M Mutations in EGFR Mutant Lung Tumors with Acquired Resistance to Gefitinib or Erlotinib. Proc. Natl. Acad. Sci. USA 2007, 104, 20932–20937. [Google Scholar] [CrossRef]
- Della Corte, C.M.; Bellevicine, C.; Vicidomini, G.; Vitagliano, D.; Malapelle, U.; Accardo, M.; Fabozzi, A.; Fiorelli, A.; Fasano, M.; Papaccio, F.; et al. SMO Gene Amplification and Activation of the Hedgehog Pathway as Novel Mechanisms of Resistance to Anti-Epidermal Growth Factor Receptor Drugs in Human Lung Cancer. Clin. Cancer Res. 2015, 21, 4686–4697. [Google Scholar] [CrossRef]
- Della Corte, C.M.; Malapelle, U.; Vigliar, E.; Pepe, F.; Troncone, G.; Ciaramella, V.; Troiani, T.; Martinelli, E.; Belli, V.; Ciardiello, F.; et al. Efficacy of Continuous EGFR-Inhibition and Role of Hedgehog in EGFR Acquired Resistance in Human Lung Cancer Cells with Activating Mutation of EGFR. Oncotarget 2017, 8, 23020–23032. [Google Scholar] [CrossRef]
- Morgillo, F.; Amendola, G.; Della Corte, C.M.; Giacomelli, C.; Botta, L.; Di Maro, S.; Messere, A.; Ciaramella, V.; Taliani, S.; Marinelli, L.; et al. Dual MET and SMO Negative Modulators Overcome Resistance to EGFR Inhibitors in Human Nonsmall Cell Lung Cancer. J. Med. Chem. 2017, 60, 7447–7458. [Google Scholar] [CrossRef]
- Papadimitrakopoulou, V.A.; Wu, Y.-L.; Han, J.-Y.; Ahn, M.-J.; Ramalingam, S.S.; John, T.; Okamoto, I.; Yang, J.C.-H.; Bulusu, K.C.; Laus, G.; et al. Analysis of Resistance Mechanisms to Osimertinib in Patients with EGFR T790M Advanced NSCLC from the AURA3 Study. Ann. Oncol. 2018, 29, viii741. [Google Scholar] [CrossRef]
- Buder, A.; Hochmair, M.J.; Schwab, S.; Bundalo, T.; Schenk, P.; Errhalt, P.; Mikes, R.E.; Absenger, G.; Patocka, K.; Baumgartner, B.; et al. Cell-Free Plasma DNA-Guided Treatment with Osimertinib in Patients With Advanced EGFR-Mutated NSCLC. J. Thorac. Oncol. 2018, 13, 821–830. [Google Scholar] [CrossRef]
- Del Re, M.; Bordi, P.; Rofi, E.; Restante, G.; Valleggi, S.; Minari, R.; Crucitta, S.; Arrigoni, E.; Chella, A.; Morganti, R.; et al. The Amount of Activating EGFR Mutations in Circulating Cell-Free DNA Is a Marker to Monitor Osimertinib Response. Br. J. Cancer 2018, 119, 1252–1258. [Google Scholar] [CrossRef]
- Ariyasu, R.; Nishikawa, S.; Uchibori, K.; Oh-hara, T.; Yoshizawa, T.; Dotsu, Y.; Koyama, J.; Saiki, M.; Sonoda, T.; Kitazono, S.; et al. High Ratio of T790M to EGFR Activating Mutations Correlate with the Osimertinib Response in Non-Small-Cell Lung Cancer. Lung Cancer 2018, 117, 1–6. [Google Scholar] [CrossRef]
- Zhou, C.; Imamura, F.; Cheng, Y.; Okamoto, I.; Chul Cho, B.; Chih Lin, M.; Majem, M.; Gautschi, O.; Gray, J.E.; Boyer, M.J.; et al. Early Clearance of Plasma EGFR Mutations as a Predictor of Response to Osimertinib and Comparator EGFR-TKIs in the FLAURA Trial. J. Clin. Oncol. 2019, 37, 9020. [Google Scholar] [CrossRef]
- Mack, P.C.; Redman, M.W.; Moon, J.; Goldberg, S.B.; Herbst, R.S.; Melnick, M.A.C.; Walther, Z.; Hirsch, F.R.; Politi, K.A.; Kelly, K.; et al. Residual Circu- Lating Tumor DNA (CtDNA) after Two Months of Therapy to Predict Progression-Free and Overall Survival in Pa- Tients Treated on S1403 with Afatinib þ/_ Cetuximab. J. Clin. Oncol. 2020, 38, 9532. [Google Scholar] [CrossRef]
- Piotrowska, Z.; Ahn, M.-J.; Pang, Y.K.; How, S.H.; Kim, S.-W.; Voon, P.J.; Cortinovis, D.L.; De Castro Carpeno, J.; Tiseo, M.; Abreu, D.R.; et al. LBA53 ELIOS: A Multicentre, Molecular Profiling Study of Patients (Pts) with Epidermal Growth Factor Receptor-Mutated (EGFRm) Advanced NSCLC Treated with First-Line (1L) Osimertinib. Ann. Oncol. 2022, 33, S1420–S1421. [Google Scholar] [CrossRef]
- Schoenfeld, A.J.; Chan, J.M.; Kubota, D.; Sato, H.; Rizvi, H.; Daneshbod, Y.; Chang, J.C.; Paik, P.K.; Offin, M.; Arcila, M.E.; et al. Tumor Analyses Reveal Squamous Transformation and Off-Target Alterations as Early Resistance Mechanisms to First-Line Osimertinib in EGFR-Mutant Lung Cancer. Clin. Cancer Res. 2020, 26, 2654–2663. [Google Scholar] [CrossRef]
- Bauml, J.; Cho, B.C.; Park, K.; Lee, K.H.; Cho, E.K.; Kim, D.-W.; Kim, S.W.; Haura, E.B.; Sabari, J.K.; Sanborn, R.E.; et al. Amivantamab in Combination with Lazertinib for the Treatment of Osimertinib-Relapsed, Chemotherapy-Naïve EGFR Mutant (EGFRm) Non-Small Cell Lung Cancer (NSCLC) and Potential Biomarkers for Response. J. Clin. Oncol. 2021, 39, 9006. [Google Scholar] [CrossRef]
- Oxnard, G.R.; Yang, J.C.-H.; Yu, H.; Kim, S.-W.; Saka, H.; Horn, L.; Goto, K.; Ohe, Y.; Mann, H.; Thress, K.S.; et al. TATTON: A Multi-Arm, Phase Ib Trial of Osimertinib Combined with Selumetinib, Savolitinib, or Durvalumab in EGFR-Mutant Lung Cancer. Ann. Oncol. 2020, 31, 507–516. [Google Scholar] [CrossRef]
- Oxnard, G.R.; Cantarini, M.; Frewer, P.; Hawkins, G.; Peters, J.; Howarth, P.; Ahmed, G.F.; Sahota, T.; Hartmaier, R.; Li-Sucholeiki, X.; et al. SAVANNAH: A Phase II Trial of Osimertinib plus Savolitinib for Patients (Pts) with EGFR-Mutant, MET-Driven (MET+), Locally Advanced or Metastatic Non-Small Cell Lung Cancer (NSCLC), Following Disease Progression on Osimertinib. J. Clin. Oncol. 2019, 37, TPS9119. [Google Scholar] [CrossRef]
- Rehman, M.; Kim, C.; Reuss, J.E.; Kiedrowski, L.A.; Garg, R.J.; Liu, S.V. Divergent RET- and BRAF-Mediated Resistance to Osimertinib in EGFR -Mutant NSCLC: A Case Report. JCO Precis. Oncol. 2021, 5, 939–942. [Google Scholar] [CrossRef]
- Hou, H.; Sun, D.; Zhang, C.; Liu, D.; Zhang, X. ALK Rearrangements as Mechanisms of Acquired Resistance to Osimertinib in EGFR Mutant Non-small Cell Lung Cancer. Thorac. Cancer 2021, 12, 962–969. [Google Scholar] [CrossRef]
- Gadgeel, S.M.; Mok, T.S.K.; Peters, S.; Alexander, J.A.A.; Leighl, N.B.; Sriuranpong, V.; Perol, M.; De Castro, G.; Nadal, E.; De Marinis, F.; et al. Phase II/III Blood First Assay Screening Trial (BFAST) in Patients (Pts) with Treatment-Naïve NSCLC: Initial Results from the ALK+ Cohort. Ann. Oncol. 2019, 30, v918. [Google Scholar] [CrossRef]
- Horn, L.; Whisenant, J.G.; Wakelee, H.; Reckamp, K.L.; Qiao, H.; Leal, T.A.; Du, L.; Hernandez, J.; Huang, V.; Blumenschein, G.R.; et al. Monitoring Therapeutic Response and Resistance: Analysis of Circulating Tumor DNA in Patients With ALK+ Lung Cancer. J. Thorac. Oncol. 2019, 14, 1901–1911. [Google Scholar] [CrossRef] [PubMed]
- Al-Obeidi, E.; Riess, J.W.; Malapelle, U.; Rolfo, C.; Gandara, D.R. Convergence of Precision Oncology and Liquid Biopsy in Non-Small Cell Lung Cancer. Hematol. Oncol. Clin. N. Am. 2023, 37, 475–487. [Google Scholar] [CrossRef] [PubMed]
- Hasegawa, N.; Kohsaka, S.; Kurokawa, K.; Shinno, Y.; Takeda Nakamura, I.; Ueno, T.; Kojima, S.; Kawazu, M.; Suehara, Y.; Ishijima, M.; et al. Highly Sensitive Fusion Detection Using Plasma Cell-free RNA in Non-small-cell Lung Cancers. Cancer Sci. 2021, 112, 4393–4403. [Google Scholar] [CrossRef] [PubMed]
- Lantuejoul, S.; Sound-Tsao, M.; Cooper, W.A.; Girard, N.; Hirsch, F.R.; Roden, A.C.; Lopez-Rios, F.; Jain, D.; Chou, T.-Y.; Motoi, N.; et al. PD-L1 Testing for Lung Cancer in 2019: Perspective from the IASLC Pathology Committee. J. Thorac. Oncol. 2020, 15, 499–519. [Google Scholar] [CrossRef]
- Herbst, R.; Jassem, J.; Abogunrin, S.; James, D.; McCool, R.; Belleli, R.; Giaccone, G.; De Marinis, F. A Network Meta-Analysis of Cancer Immunotherapies Versus Chemotherapy for First-Line Treatment of Patients with Non-Small Cell Lung Cancer and High Programmed Death-Ligand 1 Expression. Front. Oncol. 2021, 11, 676732. [Google Scholar] [CrossRef]
- Ferrara, R.; Mezquita, L.; Texier, M.; Lahmar, J.; Audigier-Valette, C.; Tessonnier, L.; Mazieres, J.; Zalcman, G.; Brosseau, S.; Le Moulec, S.; et al. Comparison of Fast-Progression, Hyperprogressive Disease, and Early Deaths in Advanced Non–Small-Cell Lung Cancer Treated With PD-1/PD-L1 Inhibitors or Chemotherapy. JCO Precis. Oncol. 2020, 4, 829–840. [Google Scholar] [CrossRef]
- Mino-Kenudson, M.; Schalper, K.; Cooper, W.; Dacic, S.; Hirsch, F.R.; Jain, D.; Lopez-Rios, F.; Tsao, M.S.; Yatabe, Y.; Beasley, M.B.; et al. Predictive Biomarkers for Immunotherapy in Lung Cancer: Perspective from the International Association for the Study of Lung Cancer Pathology Committee. J. Thorac. Oncol. 2022, 17, 1335–1354. [Google Scholar] [CrossRef]
- U.S. Food and Drug Administration. FDA Approves Pembrolizumab for Adults and Children with TMB-H Solid Tumors. News Release. FDA. Available online: Https://Bit.Ly/30QEt40 (accessed on 17 June 2020).
- Bravaccini, S.; Bronte, G.; Ulivi, P. TMB in NSCLC: A Broken Dream? Int. J. Mol. Sci. 2021, 22, 6536. [Google Scholar] [CrossRef]
- Peters, S.; Dziadziuszko, R.; Morabito, A.; Felip, E.; Gadgeel, S.M.; Cheema, P.; Cobo, M.; Andric, Z.; Barrios, C.H.; Yamaguchi, M.; et al. Atezolizumab versus Chemotherapy in Advanced or Metastatic NSCLC with High Blood-Based Tumor Mutational Burden: Primary Analysis of BFAST Cohort C Randomized Phase 3 Trial. Nat. Med. 2022, 28, 1831–1839. [Google Scholar] [CrossRef]
- Gandara, D.R.; Paul, S.M.; Kowanetz, M.; Schleifman, E.; Zou, W.; Li, Y.; Rittmeyer, A.; Fehrenbacher, L.; Otto, G.; Malboeuf, C.; et al. Blood-Based Tumor Mutational Burden as a Predictor of Clinical Benefit in Non-Small-Cell Lung Cancer Patients Treated with Atezolizumab. Nat. Med. 2018, 24, 1441–1448. [Google Scholar] [CrossRef]
- Wang, Z.; Duan, J.; Cai, S.; Han, M.; Dong, H.; Zhao, J.; Zhu, B.; Wang, S.; Zhuo, M.; Sun, J.; et al. Assessment of Blood Tumor Mutational Burden as a Potential Biomarker for Immunotherapy in Patients with Non–Small Cell Lung Cancer With Use of a Next-Generation Sequencing Cancer Gene Panel. JAMA Oncol. 2019, 5, 696. [Google Scholar] [CrossRef]
- Herbst, R.S.; Giaccone, G.; de Marinis, F.; Reinmuth, N.; Vergnenegre, A.; Barrios, C.H.; Morise, M.; Felip, E.; Andric, Z.; Geater, S.; et al. Atezolizumab for First-Line Treatment of PD-L1–Selected Patients with NSCLC. N. Engl. J. Med. 2020, 383, 1328–1339. [Google Scholar] [CrossRef]
- Paz-Ares, L.; Ciuleanu, T.-E.; Cobo, M.; Schenker, M.; Zurawski, B.; Menezes, J.; Richardet, E.; Bennouna, J.; Felip, E.; Juan-Vidal, O.; et al. 98O First-Line Nivolumab (NIVO) + Ipilimumab (IPI) + 2 Cycles Chemotherapy (Chemo) vs 4 Cycles Chemo in Advanced Non-Small Cell Lung Cancer (ANSCLC): Association of Blood and Tissue Tumor Mutational Burden (TMB) with Efficacy in CheckMate 9LA. J. Thorac. Oncol. 2021, 16, S750–S751. [Google Scholar] [CrossRef]
- Jiang, T.; Chen, J.; Xu, X.; Cheng, Y.; Chen, G.; Pan, Y.; Fang, Y.; Wang, Q.; Huang, Y.; Yao, W.; et al. On-Treatment Blood TMB as Predictors for Camrelizumab plus Chemotherapy in Advanced Lung Squamous Cell Carcinoma: Biomarker Analysis of a Phase III Trial. Mol. Cancer 2022, 21, 4. [Google Scholar] [CrossRef]
- Sinoquet, L.; Jacot, W.; Quantin, X.; Alix-Panabières, C. Liquid Biopsy and Immuno-Oncology for Advanced Nonsmall Cell Lung Cancer. Clin. Chem. 2023, 69, 23–40. [Google Scholar] [CrossRef] [PubMed]
- Kapeleris, J.; Kulasinghe, A.; Warkiani, M.E.; Vela, I.; Kenny, L.; O’Byrne, K.; Punyadeera, C. The Prognostic Role of Circulating Tumor Cells (CTCs) in Lung Cancer. Front. Oncol. 2018, 8, 311. [Google Scholar] [CrossRef]
- Manjunath, Y.; Upparahalli, S.V.; Suvilesh, K.N.; Avella, D.M.; Kimchi, E.T.; Staveley-O’Carroll, K.F.; Li, G.; Kaifi, J.T. Circulating Tumor Cell Clusters Are a Potential Biomarker for Detection of Non-Small Cell Lung Cancer. Lung Cancer 2019, 134, 147–150. [Google Scholar] [CrossRef]
- Koch, C.; Joosse, S.A.; Schneegans, S.; Wilken, O.J.W.; Janning, M.; Loreth, D.; Müller, V.; Prieske, K.; Banys-Paluchowski, M.; Horst, L.J.; et al. Pre-Analytical and Analytical Variables of Label-Independent Enrichment and Automated Detection of Circulating Tumor Cells in Cancer Patients. Cancers 2020, 12, 442. [Google Scholar] [CrossRef]
- Hofman, P.; Heeke, S.; Alix-Panabières, C.; Pantel, K. Liquid Biopsy in the Era of Immuno-Oncology: Is It Ready for Prime-Time Use for Cancer Patients? Ann. Oncol. 2019, 30, 1448–1459. [Google Scholar] [CrossRef]
- Guibert, N.; Delaunay, M.; Lusque, A.; Boubekeur, N.; Rouquette, I.; Clermont, E.; Mourlanette, J.; Gouin, S.; Dormoy, I.; Favre, G.; et al. PD-L1 Expression in Circulating Tumor Cells of Advanced Non-Small Cell Lung Cancer Patients Treated with Nivolumab. Lung Cancer 2018, 120, 108–112. [Google Scholar] [CrossRef] [PubMed]
- Ilié, M.; Szafer-Glusman, E.; Hofman, V.; Chamorey, E.; Lalvée, S.; Selva, E.; Leroy, S.; Marquette, C.-H.; Kowanetz, M.; Hedge, P.; et al. Detection of PD-L1 in Circulating Tumor Cells and White Blood Cells from Patients with Advanced Non-Small-Cell Lung Cancer. Ann. Oncol. 2018, 29, 193–199. [Google Scholar] [CrossRef] [PubMed]
- Yin, Z.; Yu, M.; Ma, T.; Zhang, C.; Huang, S.; Karimzadeh, M.R.; Momtazi-Borojeni, A.A.; Chen, S. Mechanisms Underlying Low-Clinical Responses to PD-1/PD-L1 Blocking Antibodies in Immunotherapy of Cancer: A Key Role of Exosomal PD-L1. J. Immunother. Cancer 2021, 9, e001698. [Google Scholar] [CrossRef] [PubMed]
- Yang, Q.; Chen, M.; Gu, J.; Niu, K.; Zhao, X.; Zheng, L.; Xu, Z.; Yu, Y.; Li, F.; Meng, L.; et al. Novel Biomarkers of Dynamic Blood PD-L1 Expression for Immune Checkpoint Inhibitors in Advanced Non-Small-Cell Lung Cancer Patients. Front. Immunol. 2021, 12, 665133. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Zeng, H.; Zhang, H.; Han, Y. The Role of Exosomal PD-L1 in Tumor Immunotherapy. Transl. Oncol. 2021, 14, 101047. [Google Scholar] [CrossRef] [PubMed]
- Chen, G.; Huang, A.C.; Zhang, W.; Zhang, G.; Wu, M.; Xu, W.; Yu, Z.; Yang, J.; Wang, B.; Sun, H.; et al. Exosomal PD-L1 Contributes to Immunosuppression and Is Associated with Anti-PD-1 Response. Nature 2018, 560, 382–386. [Google Scholar] [CrossRef]
- Zhu, X.; Lang, J. Soluble PD-1 and PD-L1: Predictive and Prognostic Significance in Cancer. Oncotarget 2017, 8, 97671–97682. [Google Scholar] [CrossRef]
- Scirocchi, F.; Strigari, L.; Di Filippo, A.; Napoletano, C.; Pace, A.; Rahimi, H.; Botticelli, A.; Rughetti, A.; Nuti, M.; Zizzari, I.G. Soluble PD-L1 as a Prognostic Factor for Immunotherapy Treatment in Solid Tumors: Systematic Review and Meta-Analysis. Int. J. Mol. Sci. 2022, 23, 14496. [Google Scholar] [CrossRef]
- Oh, S.Y.; Kim, S.; Keam, B.; Kim, T.M.; Kim, D.-W.; Heo, D.S. Soluble PD-L1 Is a Predictive and Prognostic Biomarker in Advanced Cancer Patients Who Receive Immune Checkpoint Blockade Treatment. Sci. Rep. 2021, 11, 19712. [Google Scholar] [CrossRef]
- Guibert, N.; Jones, G.; Beeler, J.F.; Plagnol, V.; Morris, C.; Mourlanette, J.; Delaunay, M.; Keller, L.; Rouquette, I.; Favre, G.; et al. Targeted Sequencing of Plasma Cell-Free DNA to Predict Response to PD1 Inhibitors in Advanced Non-Small Cell Lung Cancer. Lung Cancer 2019, 137, 1–6. [Google Scholar] [CrossRef]
- Marinelli, D.; Mazzotta, M.; Scalera, S.; Terrenato, I.; Sperati, F.; D’Ambrosio, L.; Pallocca, M.; Corleone, G.; Krasniqi, E.; Pizzuti, L.; et al. KEAP1-Driven Co-Mutations in Lung Adenocarcinoma Unresponsive to Immunotherapy despite High Tumor Mutational Burden. Ann. Oncol. 2020, 31, 1746–1754. [Google Scholar] [CrossRef]
- Pavan, A.; Boscolo Bragadin, A.; Calvetti, L.; Ferro, A.; Zulato, E.; Attili, I.; Nardo, G.; Dal Maso, A.; Frega, S.; Menin, A.G.; et al. Role of next Generation Sequencing-Based Liquid Biopsy in Advanced Non-Small Cell Lung Cancer Patients Treated with Immune Checkpoint Inhibitors: Impact of STK11, KRAS and TP53 Mutations and Co-Mutations on Outcome. Transl. Lung Cancer Res. 2021, 10, 202–220. [Google Scholar] [CrossRef]
- Rizvi, H.; Sanchez-Vega, F.; La, K.; Chatila, W.; Jonsson, P.; Halpenny, D.; Plodkowski, A.; Long, N.; Sauter, J.L.; Rekhtman, N.; et al. Molecular Determinants of Response to Anti–Programmed Cell Death (PD)-1 and Anti–Programmed Death-Ligand 1 (PD-L1) Blockade in Patients with Non–Small-Cell Lung Cancer Profiled with Targeted Next-Generation Sequencing. J. Clin. Oncol. 2018, 36, 633–641. [Google Scholar] [CrossRef]
- Peng, X.-X.; Yu, R.; Wu, X.; Wu, S.-Y.; Pi, C.; Chen, Z.-H.; Zhang, X.-C.; Gao, C.-Y.; Shao, Y.W.; Liu, L.; et al. Correlation of Plasma Exosomal MicroRNAs with the Efficacy of Immunotherapy in EGFR/ALK Wild-Type Advanced Non-Small Cell Lung Cancer. J. Immunother. Cancer 2020, 8, e000376. [Google Scholar] [CrossRef]
- Fan, J.; Yin, Z.; Xu, J.; Wu, F.; Huang, Q.; Yang, L.; Jin, Y.; Yang, G. Circulating MicroRNAs Predict the Response to Anti-PD-1 Therapy in Non-Small Cell Lung Cancer. Genomics 2020, 112, 2063–2071. [Google Scholar] [CrossRef]
- Halvorsen, A.R.; Sandhu, V.; Sprauten, M.; Flote, V.G.; Kure, E.H.; Brustugun, O.T.; Helland, Å. Circulating MicroRNAs Associated with Prolonged Overall Survival in Lung Cancer Patients Treated with Nivolumab. Acta Oncol. 2018, 57, 1225–1231. [Google Scholar] [CrossRef]
- Boeri, M.; Milione, M.; Proto, C.; Signorelli, D.; Lo Russo, G.; Galeone, C.; Verri, C.; Mensah, M.; Centonze, G.; Martinetti, A.; et al. Circulating MiRNAs and PD-L1 Tumor Expression Are Associated with Survival in Advanced NSCLC Patients Treated with Immunotherapy: A Prospective Study. Clin. Cancer Res. 2019, 25, 2166–2173. [Google Scholar] [CrossRef]
- Goldberg, S.B.; Narayan, A.; Kole, A.J.; Decker, R.H.; Teysir, J.; Carriero, N.J.; Lee, A.; Nemati, R.; Nath, S.K.; Mane, S.M.; et al. Early Assessment of Lung Cancer Immunotherapy Response via Circulating Tumor DNA. Clin. Cancer Res. 2018, 24, 1872–1880. [Google Scholar] [CrossRef]
- Ricciuti, B.; Jones, G.; Severgnini, M.; Alessi, J.V.; Recondo, G.; Lawrence, M.; Forshew, T.; Lydon, C.; Nishino, M.; Cheng, M.; et al. Early Plasma Circulating Tumor DNA (CtDNA) Changes Predict Response to First-Line Pembrolizumab-Based Therapy in Non-Small Cell Lung Cancer (NSCLC). J. Immunother. Cancer 2021, 9, e001504. [Google Scholar] [CrossRef]
- Cabel, L.; Riva, F.; Servois, V.; Livartowski, A.; Daniel, C.; Rampanou, A.; Lantz, O.; Romano, E.; Milder, M.; Buecher, B.; et al. Circulating Tumor DNA Changes for Early Monitoring of Anti-PD1 Immunotherapy: A Proof-of-Concept Study. Ann. Oncol. 2017, 28, 1996–2001. [Google Scholar] [CrossRef]
- Zhang, Q.; Luo, J.; Wu, S.; Si, H.; Gao, C.; Xu, W.; Abdullah, S.E.; Higgs, B.W.; Dennis, P.A.; van der Heijden, M.S.; et al. Prognostic and Predictive Impact of Circulating Tumor DNA in Patients with Advanced Cancers Treated with Immune Checkpoint Blockade. Cancer Discov. 2020, 10, 1842–1853. [Google Scholar] [CrossRef]
- Iijima, Y.; Hirotsu, Y.; Amemiya, K.; Ooka, Y.; Mochizuki, H.; Oyama, T.; Nakagomi, T.; Uchida, Y.; Kobayashi, Y.; Tsutsui, T.; et al. Very Early Response of Circulating Tumour–Derived DNA in Plasma Predicts Efficacy of Nivolumab Treatment in Patients with Non–Small Cell Lung Cancer. Eur. J. Cancer 2017, 86, 349–357. [Google Scholar] [CrossRef] [PubMed]
- van der Leest, P.; Hiddinga, B.; Miedema, A.; Aguirre Azpurua, M.L.; Rifaela, N.; ter Elst, A.; Timens, W.; Groen, H.J.M.; van Kempen, L.C.; Hiltermann, T.J.N.; et al. Circulating Tumor DNA as a Biomarker for Monitoring Early Treatment Responses of Patients with Advanced Lung Adenocarcinoma Receiving Immune Checkpoint Inhibitors. Mol. Oncol. 2021, 15, 2910–2922. [Google Scholar] [CrossRef] [PubMed]
- Thompson, J.C.; Carpenter, E.L.; Silva, B.A.; Rosenstein, J.; Chien, A.L.; Quinn, K.; Espenschied, C.R.; Mak, A.; Kiedrowski, L.A.; Lefterova, M.; et al. Serial Monitoring of Circulating Tumor DNA by Next-Generation Gene Sequencing as a Biomarker of Response and Survival in Patients with Advanced NSCLC Receiving Pembrolizumab-Based Therapy. JCO Precis. Oncol. 2021, 5, 510–524. [Google Scholar] [CrossRef]
- Bratman, S.V.; Yang, S.Y.C.; Iafolla, M.A.J.; Liu, Z.; Hansen, A.R.; Bedard, P.L.; Lheureux, S.; Spreafico, A.; Razak, A.A.; Shchegrova, S.; et al. Personalized Circulating Tumor DNA Analysis as a Predictive Biomarker in Solid Tumor Patients Treated with Pembrolizumab. Nat. Cancer 2020, 1, 873–881. [Google Scholar] [CrossRef]
- Ren, S.; Chen, J.; Xu, X.; Jiang, T.; Cheng, Y.; Chen, G.; Pan, Y.; Fang, Y.; Wang, Q.; Huang, Y.; et al. Camrelizumab Plus Carboplatin and Paclitaxel as First-Line Treatment for Advanced Squamous NSCLC (CameL-Sq): A Phase 3 Trial. J. Thorac. Oncol. 2022, 17, 544–557. [Google Scholar] [CrossRef]
- Mondelo-Macía, P.; García-González, J.; León-Mateos, L.; Anido, U.; Aguín, S.; Abdulkader, I.; Sánchez-Ares, M.; Abalo, A.; Rodríguez-Casanova, A.; Díaz-Lagares, Á.; et al. Clinical Potential of Circulating Free DNA and Circulating Tumour Cells in Patients with Metastatic Non-small-cell Lung Cancer Treated with Pembrolizumab. Mol. Oncol. 2021, 15, 2923–2940. [Google Scholar] [CrossRef]
- Anagnostou, V.; Forde, P.M.; White, J.R.; Niknafs, N.; Hruban, C.; Naidoo, J.; Marrone, K.; Sivakumar, I.K.A.; Bruhm, D.C.; Rosner, S.; et al. Dynamics of Tumor and Immune Responses during Immune Checkpoint Blockade in Non–Small Cell Lung Cancer. Cancer Res. 2019, 79, 1214–1225. [Google Scholar] [CrossRef]
- Park, H.J.; Kim, K.W.; Pyo, J.; Suh, C.H.; Yoon, S.; Hatabu, H.; Nishino, M. Incidence of Pseudoprogression during Immune Checkpoint Inhibitor Therapy for Solid Tumors: A Systematic Review and Meta-Analysis. Radiology 2020, 297, 87–96. [Google Scholar] [CrossRef]
- Alama, A.; Coco, S.; Genova, C.; Rossi, G.; Fontana, V.; Tagliamento, M.; Dal Bello, M.G.; Rosa, A.; Boccardo, S.; Rijavec, E.; et al. Prognostic Relevance of Circulating Tumor Cells and Circulating Cell-Free DNA Association in Metastatic Non-Small Cell Lung Cancer Treated with Nivolumab. J. Clin. Med. 2019, 8, 1011. [Google Scholar] [CrossRef]
- Hellmann, M.D.; Nabet, B.Y.; Rizvi, H.; Chaudhuri, A.A.; Wells, D.K.; Dunphy, M.P.S.; Chabon, J.J.; Liu, C.L.; Hui, A.B.; Arbour, K.C.; et al. Circulating Tumor DNA Analysis to Assess Risk of Progression after Long-Term Response to PD-(L)1 Blockade in NSCLC. Clin. Cancer Res. 2020, 26, 2849–2858. [Google Scholar] [CrossRef]
- Tamminga, M.; de Wit, S.; Hiltermann, T.J.N.; Timens, W.; Schuuring, E.; Terstappen, L.W.M.M.; Groen, H.J.M. Circulating Tumor Cells in Advanced Non-Small Cell Lung Cancer Patients Are Associated with Worse Tumor Response to Checkpoint Inhibitors. J. Immunother. Cancer 2019, 7, 173. [Google Scholar] [CrossRef]
- Janning, M.; Kobus, F.; Babayan, A.; Wikman, H.; Velthaus, J.-L.; Bergmann, S.; Schatz, S.; Falk, M.; Berger, L.-A.; Böttcher, L.-M.; et al. Determination of PD-L1 Expression in Circulating Tumor Cells of NSCLC Patients and Correlation with Response to PD-1/PD-L1 Inhibitors. Cancers 2019, 11, 835. [Google Scholar] [CrossRef]
- Castellano, G.M.; Aisner, J.; Burley, S.K.; Vallat, B.; Yu, H.A.; Pine, S.R.; Ganesan, S. A Novel Acquired Exon 20 EGFR M766Q Mutation in Lung Adenocarcinoma Mediates Osimertinib Resistance but Is Sensitive to Neratinib and Poziotinib. J. Thorac. Oncol. 2019, 14, 1982–1988. [Google Scholar] [CrossRef]
- Park, C.-K.; Jun, H.R.; Oh, H.-J.; Lee, J.-Y.; Cho, H.-J.; Kim, Y.-C.; Lee, J.E.; Yoon, S.H.; Choi, C.M.; Lee, J.C.; et al. Evaluation of Blood Tumor Mutation Burden for the Efficacy of Second-Line Atezolizumab Treatment in Non-Small Cell Lung Cancer: BUDDY Trial. Cells 2023, 12, 1246. [Google Scholar] [CrossRef]
- National Comprehensive Cancer Network: NCCN Clinical Practice Guidelines in Oncology: Non-Small Cell Lung Cancer. Version 5.2021. Available online: https://www.nccn.org/guidelines/guidelines-detail?category=1&id=1450 (accessed on 1 May 2023).

| Study | Number of Patients | Clinical Stage | ctDNA Methodology | Mutations Monitored | Treatment | ctDNA MRD Landmark | |
|---|---|---|---|---|---|---|---|
| Sensitivity | Specificity | ||||||
| Chaudhuri et al. [103] | 37 | IB–IIIB | CAPP-Seq | Multiple | CRT or RT and/or surgery ± chemotherapy | 94% | 100% |
| Chen et al. [74] | 25 | IIB–IIIB | cSMART | Multiple | Surgery ± chemotherapy | 44% | 88% |
| Moding et al. [104] | 12 | IIB–IIIB | CAPP-Seq | Multiple | CRT | 100% | 100% |
| Abbosh et al. [30] | 108 | IA–III | Anchored-multiplex PCR (AMP) | Multiple | Surgery ± chemotherapy | 49% | N.E. * |
| Study | Sample Size | Method of Detection | Type of Sample | Alteration Detected | Sensitivity |
|---|---|---|---|---|---|
| Douillard et al. [52] | 652 | QUIAGEN therascreen EGFR RGQ PCR Kit | cfDNA (plasma) | EGFR-sensitizing mutations | 65.7% |
| Reck et al. [49] | 1162 | QUIAGEN therascreen EGFR RGQ PCR Kit; Cobas EGFR mutations test version 2; Cycleave; PNA-LNA PCR Clamp; other | ctDNA | EGFR-sensitizing mutations | 46% |
| Wu et al. [110] | 334 (plasma) 287 (serum) | QUIAGEN therascreen EGFR RGQ PCR Kit | cfDNA | EGFR-sensitizing mutations | 60.5% (plasma) 28.6% (serum) |
| Han et al. [111] | 2561 | Cobas EGFR mutations test version 2 | ctDNA | EGFR-sensitizing mutations | 46.9% |
| Karlovich et al. [112] | 153 | Cobas EGFR mutations test version 2; BEAMing (Symex Inostics GmbH) | cfDNA (plasma) | EGFR-sensitizing mutations and T790M resistance mutation | Activating mutations: 73%/82% T790M: 64%/73% |
| Oxnard et al. [114] | 216 | BEAMing (Sysmex Inostics GmbH) | cfDNA (plasma) | EGFR-sensitizing mutations and T790M resistance mutation | T790M: 70% L858R: 86% Del 19: 82% |
| Sacher et al. [113] | 180 | Droplet digital PCR (ddPCR) | cfDNA (plasma) | EGFR-sensitizing mutations and T790M resistance mutation | L858R: 74% Del 19: 82% T790M: 77 |
| Zheng et al. [115] | 117 | Droplet digital PCR (ddPCR) | ctDNA | T790M resistance mutation | 81% |
| Study | Number of Patients/Study Type | Treatment | Outcomes |
|---|---|---|---|
| Cabel et al. [182] | 15 (NSCLC and other cancer types) patients/prospective, pilot study | ≥2nd line nivolumab or pembrolizumab | ctDNA clearance after 8 weeks associated with improved OS and PFS |
| Goldberg et al. [180] | 28 Stage IV NSCLC patients/prospective | Anti-PD-L1/PD-1 ± anti-CTLA4 | Reduction in ctDNA levels <50% from baseline correlated with improved PFS, OS and better ORR |
| Van der Leest et al. [185] | 100 Stage IV NSCLC patients/retrospective | Anti-PD-L1/PD-1, ≥1st line | ctDNA decrease at 4–6 weeks associated with improved PFS and OS |
| Thompson et al. [186] | 45 Stage IV NSCLC patients/prospective | Pembrolizumab ± chemotherapy, ≥1st line | A decrease >50% of ctDNA at 3 weeks correlated with better ORR, PFS and OS |
| Ricciuti et al. [181] | 45 Stage IV NSCLC patients/retrospective | Pembrolizumab ± chemotherapy, 1st line | Decrease of ctDNA at 3 weeks associated with improved PFS, OS and better ORR |
| Ren et al. [188] | 134 Stage III and IV NSCLC patients/exploratory analysis of phase III trial | Camrelizumab + chemotherapy, 1st line | ctDNA clearance or decrease after two cycles correlated with improved PFS and OS |
| Mondelo-Macia et al. [189] | 50 Stage advanced NSCLC/prospective | Pembrolizumab ± chemotherapy, 1st line | Decrease of ctDNA at 12 weeks associated with improved PFS |
| Anagnostou et al. [190] | 24 Stage IV NSCLC/retrospective | Anti-PD-1 ± anti-CTLA4/antiLAG3/chemotherapy, ≥1st line | ctDNA clearance associated with improved PFS and OS |
| Bratman et al. [187] | 106 Stage IV patients (NSCLC and other cancer types) | Pembrolizumab, ≥1st line | ctDNA clearance associated with improved PFS and OS |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bertoli, E.; De Carlo, E.; Basile, D.; Zara, D.; Stanzione, B.; Schiappacassi, M.; Del Conte, A.; Spina, M.; Bearz, A. Liquid Biopsy in NSCLC: An Investigation with Multiple Clinical Implications. Int. J. Mol. Sci. 2023, 24, 10803. https://doi.org/10.3390/ijms241310803
Bertoli E, De Carlo E, Basile D, Zara D, Stanzione B, Schiappacassi M, Del Conte A, Spina M, Bearz A. Liquid Biopsy in NSCLC: An Investigation with Multiple Clinical Implications. International Journal of Molecular Sciences. 2023; 24(13):10803. https://doi.org/10.3390/ijms241310803
Chicago/Turabian StyleBertoli, Elisa, Elisa De Carlo, Debora Basile, Diego Zara, Brigida Stanzione, Monica Schiappacassi, Alessandro Del Conte, Michele Spina, and Alessandra Bearz. 2023. "Liquid Biopsy in NSCLC: An Investigation with Multiple Clinical Implications" International Journal of Molecular Sciences 24, no. 13: 10803. https://doi.org/10.3390/ijms241310803
APA StyleBertoli, E., De Carlo, E., Basile, D., Zara, D., Stanzione, B., Schiappacassi, M., Del Conte, A., Spina, M., & Bearz, A. (2023). Liquid Biopsy in NSCLC: An Investigation with Multiple Clinical Implications. International Journal of Molecular Sciences, 24(13), 10803. https://doi.org/10.3390/ijms241310803






