PROTEIN PHOSPHATASE 2C08, a Negative Regulator of Abscisic Acid Signaling, Promotes Internode Elongation in Rice
Abstract
1. Introduction
2. Results
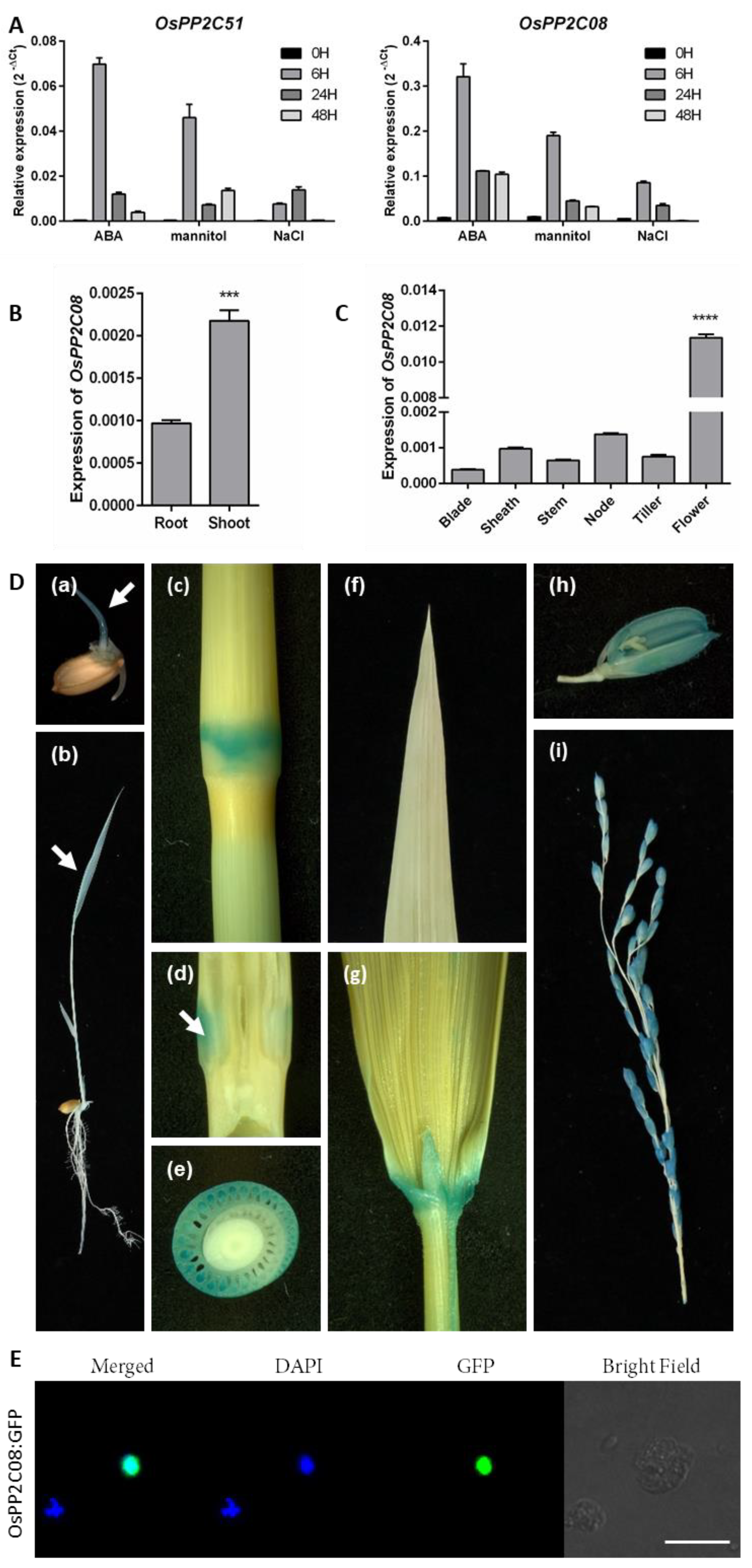
2.1. Expression Pattern of OsPP2C08 and Subcellular Localization of OsPP2C08
2.2. Overexpression of OsPP2C08 in Rice Causes ABA Insensitive Phenotypes
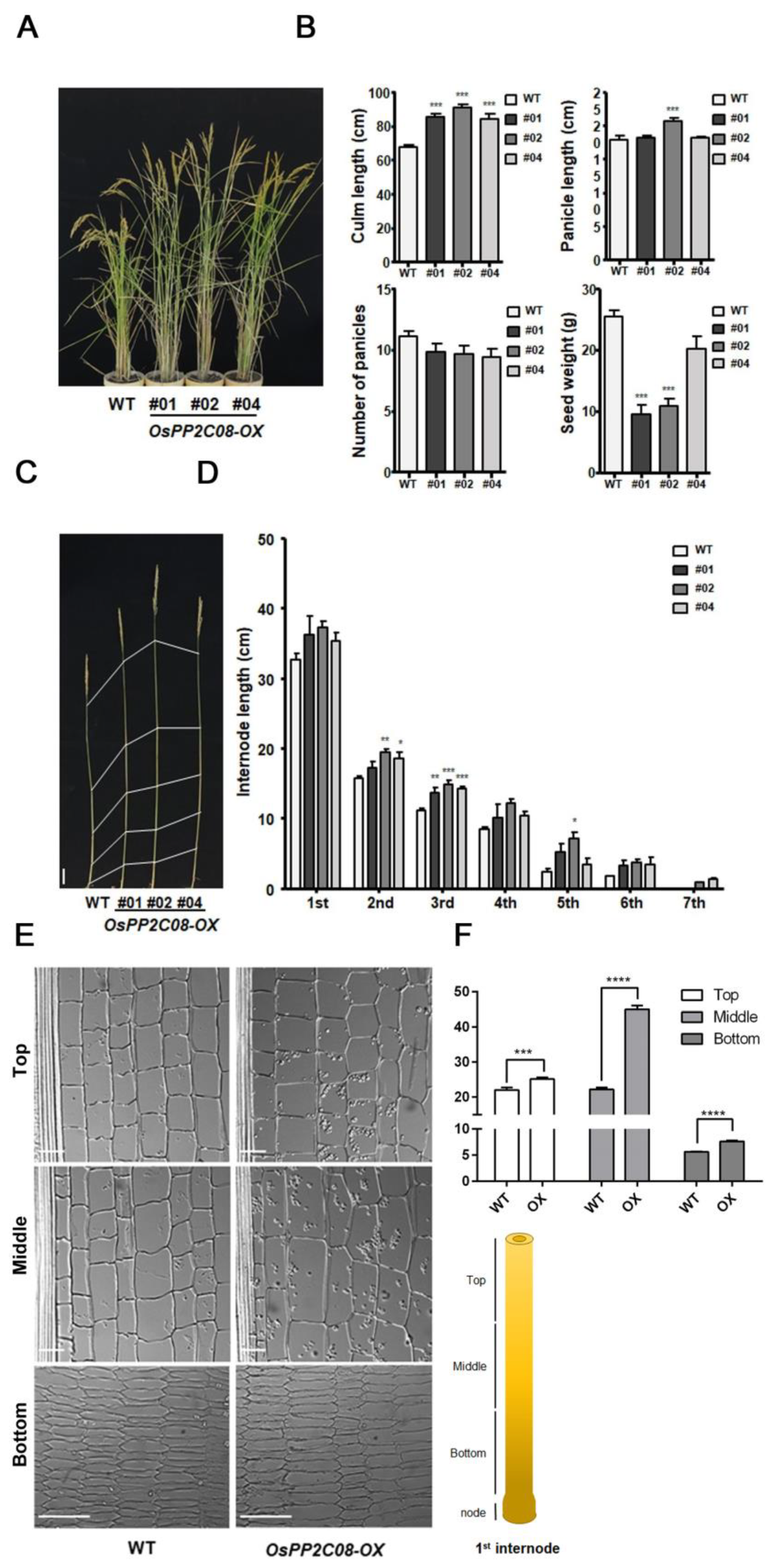
2.3. OsPP2C08 Promotes Internode Elongation in Rice
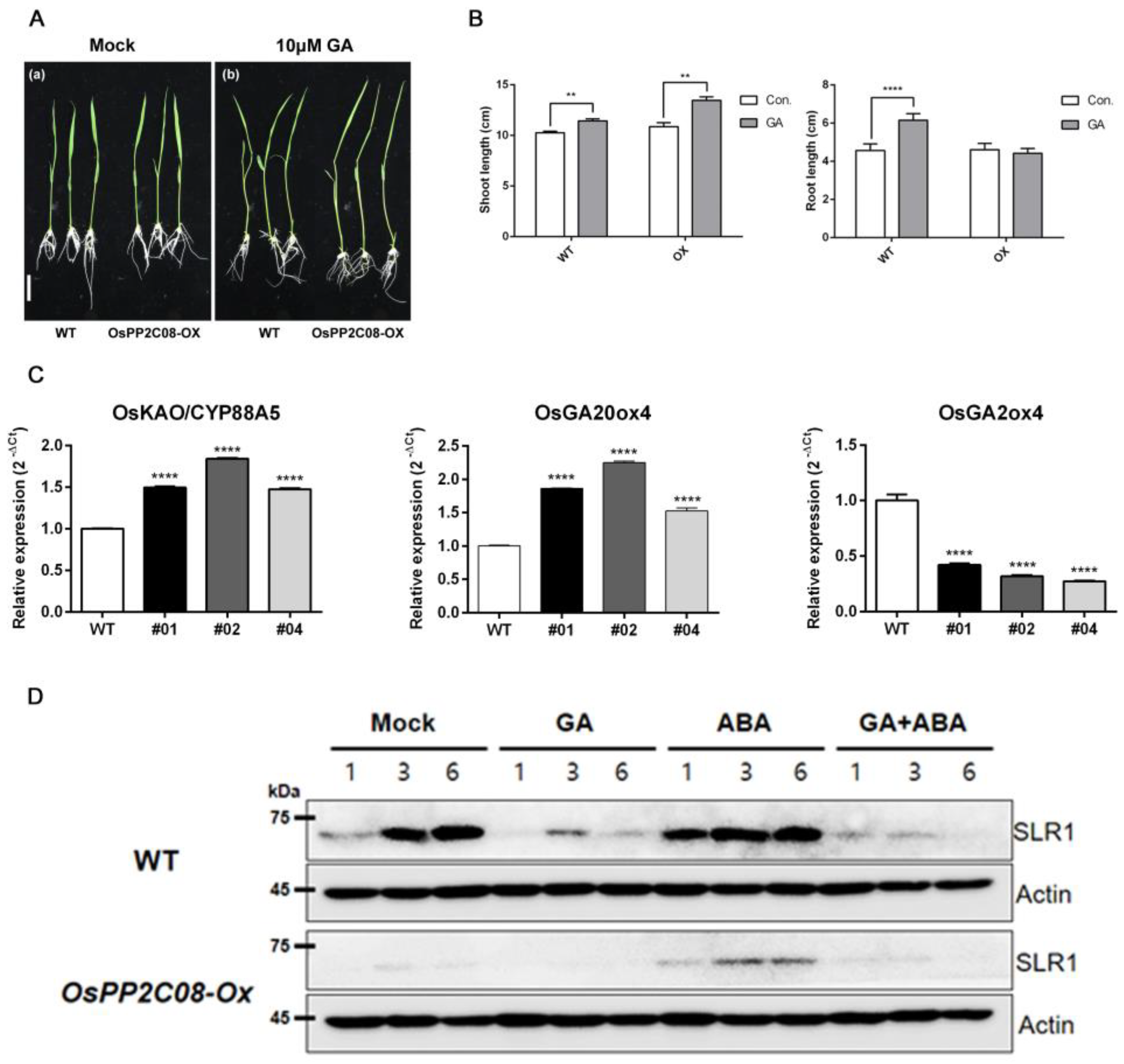
2.4. OsPP2C08 Overexpression Positively Regulates the Expression of GA Biosynthesis Genes
3. Discussion
4. Materials and Methods
4.1. Generation of Transgenic Rice
4.2. Total RNA Isolation and Reverse Transcription Quantitative Polymerase Chain Reaction (RT-qPCR)
4.3. Protoplast Isolation and Subcellular Localization
4.4. Post-Germination Assay
4.5. Growth Conditions for Analyzing Agricultural Traits
4.6. Histochemical GUS Assay and Measurement of Cell Size
4.7. Immunoblot Analysis of SLENDER RICE 1 (SLR1)
4.8. Transcriptome Analysis
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- González-García, M.P.; Rodríguez, D.; Nicolás, C.; Rodríguez, P.L.; Nicolás, G.; Lorenzo, O. Negative regulation of abscisic acid signaling by the Fagus sylvatica FsPP2C1 plays a role in seed dormancy regulation and promotion of seed germination. Plant Physiol. 2003, 133, 135–144. [Google Scholar] [CrossRef] [PubMed]
- Hasanuzzaman, M.; Nahar, K.; Alam, M.; Roychowdhury, R.; Fujita, M. Physiological, Biochemical, and Molecular Mecha-nisms of Heat Stress Tolerance in Plants. Int. J. Mol. Sci. 2013, 14, 9643–9684. [Google Scholar] [CrossRef] [PubMed]
- Sah, S.K.; Reddy, K.R.; Li, J. Abscisic Acid and Abiotic Stress Tolerance in Crop Plants. Front. Plant Sci. 2016, 7, 571. [Google Scholar] [CrossRef] [PubMed]
- Nambara, E.; Marion-Poll, A. Abscisic Acid Biosynthesis and Catabolism. Annu. Rev. Plant Biol. 2005, 56, 165–185. [Google Scholar] [CrossRef] [PubMed]
- Fujita, Y.; Fujita, M.; Shinozaki, K.; Yamaguchi-Shinozaki, K. ABA-mediated transcriptional regulation in response to osmotic stress in plants. J. Plant Res. 2011, 124, 509–525. [Google Scholar] [CrossRef] [PubMed]
- Endo, A.; Egawa, C.; Oohashi, M.; Meguro-Maoka, A.; Shimosaka, E.; Sato, Y. Ectopic expression of mutated type 2C protein phosphatase OsABI-LIKE2 decreases abscisic acid sensitivity in Arabidopsis and rice. Sci. Rep. 2018, 17, 12320. [Google Scholar] [CrossRef]
- Min, M.K.; Kim, R.; Hong, W.J.; Jung, K.H.; Lee, J.Y.; Kim, B.G. OsPP2C09 Is a Bifunctional Regulator in Both ABA-Dependent and Independent Abiotic Stress Signaling Pathways. Int. J. Mol. Sci. 2021, 22, 393. [Google Scholar] [CrossRef]
- Dugardeyn, J.; Vandenbussche, F.; Van Der Straeten, M. To grow or not to grow: What can we learn on ethylene-gibberellin cross-talk by in silico gene expression analysis. J. Exp. Bot. 2007, 59, 1–16. [Google Scholar] [CrossRef]
- Mutasa-Göttgens, E.; Hedden, P. Gibberellin as a factor in floral regulatory networks. J. Exp. Bot. 2009, 60, 1979–1989. [Google Scholar] [CrossRef]
- Hirano, K.; Kouketu, E.; Katoh, H.; Aya, K.; Ueguchi-Tanaka, M.; Matsuoka, M. The suppressive function of the rice DELLA protein SLR1 is dependent on its transcriptional activation activity. Plant J. 2012, 71, 443–453. [Google Scholar] [CrossRef]
- Hedden, P. The Current Status of Research on Gibberellin Biosynthesis. Plant Cell. Physiol. 2020, 61, 1832–1849. [Google Scholar] [CrossRef]
- Ito, T.; Fukazawa, J. SCARECROW-LIKE3 regulates the transcription of gibberellin-related genes by acting as a transcriptional co-repressor of GAI-ASSOCIATED FACTOR1. Plant Mol. Biol. 2021, 105, 463–482. [Google Scholar] [CrossRef]
- Han, C.; Yang, P.F. Studies on the molecular mechanisms of seed germination. Proteomics 2015, 15, 1671–1679. [Google Scholar] [CrossRef]
- Wang, C.; Liu, Y.; Li, S.-S.; Han, G.-Z. Insights into the Origin and Evolution of the Plant Hormone Signaling Machinery. Plant. Physiol. 2015, 167, 872–886. [Google Scholar] [CrossRef] [PubMed]
- Weiss, D.; Ori, N. Mechanisms of Cross Talk between Gibberellin and Other Hormones. Plant Physiol. 2007, 144, 1240–1246. [Google Scholar] [CrossRef] [PubMed]
- Javid, M.G.; Sorooshzadeh, A.; Moradi, F.; Sanavy, S.A.M.M.; Allahdadi, I. The Role of Phytohormones in Alleviating Salt Stress in Crop Plants. Aust. J. Crop Sci. 2011, 5, 726–734. [Google Scholar]
- Seo, M.; Hanada, A.; Kuwahara, A.; Endo, A.; Okamoto, M.; Yamauchi, Y.; North, H.; Marion-Poll, A.; Sun, T.-P.; Koshiba, T.; et al. Regulation of hormone metabolism in Arabidopsis seeds: Phytochrome regulation of abscisic acid metabolism and ab-scisic acid regulation of gibberellin metabolism. Plant J. 2006, 48, 354–366. [Google Scholar] [CrossRef]
- Liu, X.; Hu, P.; Huang, M.; Tang, Y.; Li, Y.; Li, L.; Hou, X. The NF-YC–RGL2 module integrates GA and ABA signalling to reg-ulate seed germination in Arabidopsis. Nat. Commun. 2016, 7, 12768. [Google Scholar] [CrossRef]
- Fan, Z.-Q.; Tan, X.-L.; Shan, W.; Kuang, J.-F.; Lu, W.-J.; Lin, H.-T.; Su, X.-G.; Lakshmanan, P.; Zhao, M.-L.; Chen, J.-Y. Involve-ment of BrNAC041 in ABA-GA antagonism in the leaf senescence of Chinese flowering cabbage. Postharvest Biol. Technol. 2020, 168, 111254. [Google Scholar] [CrossRef]
- Kim, W.; Lee, Y.; Park, J.; Lee, N.; Choi, G. HONSU, a protein phosphatase 2C, regulates seed dormancy by inhibiting ABA signaling in Arabidopsis. Plant Cell Physiol. 2013, 54, 555–572. [Google Scholar] [CrossRef]
- Cantoro, R.; Crocco, C.D.; Benech-Arnold, R.L.; Rodríguez, M.V. In vitro binding of Sorghum bicolor transcription factors ABI4 and ABI5 to a conserved region of a GA 2-OXIDASE promoter: Possible role of this interaction in the expression of seed dormancy. J. Exp. Bot. 2013, 64, 5721–5735. [Google Scholar] [CrossRef] [PubMed]
- Bhatnagar, N.; Min, M.-K.; Choi, E.-H.; Kim, N.; Moon, S.-J.; Yoon, I.; Kwon, T.; Jung, K.-H.; Kim, B.-G. The protein phosphatase 2C clade A protein OsPP2C51 positively regulates seed germination by directly inactivating OsbZIP10. Plant Mol. Biol. 2017, 93, 389–401. [Google Scholar] [CrossRef]
- Knöller, A.S.; Blakeslee, J.J.; Richards, E.L.; Peer, W.A.; Murphy, A.S. Brachytic2/ZmABCB1 functions in IAA export from in-tercalary meristems. J. Exp. Bot. 2010, 61, 3689–3696. [Google Scholar] [CrossRef]
- Kim, S.; Park, S.-I.; Kwon, H.; Cho, M.H.; Kim, B.-G.; Chung, J.H.; Nam, M.H.; Song, J.S.; Kim, K.-H.; Yoon, I.S. The Rice Ab-scisic Acid-Responsive RING Finger E3 Ligase OsRF1 Targets OsPP2C09 for Degradation and Confers Drought and Salinity Tolerance in Rice. IS1. Front. Plant Sci. 2021, 12, 797940. [Google Scholar] [CrossRef]
- Melcher, K.; Ng, L.-M.; Zhou, X.E.; Soon, F.-F.; Xu, Y.; Suino-Powell, K.M.; Park, S.-Y.; Weiner, J.J.; Fujii, H.; Chinnusamy, V.; et al. A gate-latch-lock mechanism for hormone signalling by abscisic acid receptors. Nature 2009, 462, 602–608. [Google Scholar] [CrossRef]
- Zhu, J.K. Abiotic Stress Signaling and Responses in Plants. Cell 2016, 167, 313–324. [Google Scholar] [CrossRef] [PubMed]
- Rigoulot, S.B.; Petzold, H.E.; Williams, S.P.; Brunner, A.M.; Beers, E.P. Populus trichocarpa clade A PP2C protein phosphatases: Their stress-induced expression patterns, interactions in core abscisic acid signaling, and potential for regulation of growth and development. Plant Mol. Biol. 2019, 100, 303–317. [Google Scholar] [CrossRef] [PubMed]
- Miao, J.; Li, X.; Li, X.; Tan, W.; You, A.; Wu, S.; Tao, Y.; Chen, C.; Wang, J.; Zhang, D.; et al. OsPP2C09, a negative regulatory factor in abscisic acid signalling, plays an essential role in balancing plant growth and drought tolerance in rice. New Phytol. 2020, 227, 1417–1433. [Google Scholar] [CrossRef]
- Ding, X.; Richter, T.; Chen, M.; Fujii, H.; Seo, Y.S.; Xie, M.; Zheng, X.; Kanrar, S.; Stevenson, R.A.; Dardick, C.; et al. A rice ki-nase-protein interaction map. Plant Physiol. 2009, 149, 1478–1492. [Google Scholar] [CrossRef]
- Singh, A.; Giri, J.; Kapoor, S.; Tyagi, A.K.; Pandey, G.K. Protein phosphatase complement in rice: Genome-wide identification and transcriptional analysis under abiotic stress conditions and reproductive development. BMC Genom. 2010, 11, 435. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Min, M.K.; Choi, E.H.; Kim, J.A.; Yoon, I.S.; Han, S.; Lee, Y.; Lee, S.; Kim, B.G. Two Clade A Phosphatase 2Cs Expressed in Guard Cells Physically Interact with Abscisic Acid Signaling Components to Induce Stomatal Closure in Rice. Rice 2019, 12, 37. [Google Scholar] [CrossRef] [PubMed]
- Baek, W.; Lim, C.W.; Lee, S.C. Functional analysis of the pepper protein phosphatase, CaAIPP1, and its interacting partner CaAIRF1: Modulation of ABA signalling and the drought stress response. Plant Cell Environ. 2017, 40, 2359–2368. [Google Scholar] [CrossRef]
- Jeong, S.; Lim, C.W.; Lee, S.C. Pepper SnRK2.6-activated MEKK protein CaMEKK23 is directly and indirectly modulated by clade A PP2Cs in response to drought stress. New Phytol. 2023, 238, 237–251. [Google Scholar] [CrossRef] [PubMed]
- Marczak, M.; Cieśla, A.; Janicki, M.; Kasprowicz-Maluśki, A.; Kubiak, P.; Ludwików, A. Protein Phosphatases Type 2C Group A Interact with and Regulate the Stability of ACC Synthase 7 in Arabidopsis. Cells 2020, 9, 978. [Google Scholar] [CrossRef] [PubMed]
- Guo, X.; Hou, X.; Fang, J.; Wei, P.; Xu, B.; Chen, M.; Feng, Y.; Chu, C. The rice GERMINATION DEFECTIVE 1, encoding a B3 domain transcriptional repressor, regulates seed germination and seedling development by integrating GA and carbohydrate metabolism. Plant J. 2013, 75, 403–416. [Google Scholar] [CrossRef]
- Xie, Z.; Zhang, Z.L.; Zou, X.; Yang, G.; Komatsu, S.; Shen, Q.J. Interactions of two abscisic-acid induced WRKY genes in re-pressing gibberellin signaling in aleurone cells. Plant J. 2006, 46, 231–242. [Google Scholar] [CrossRef]
- Liao, Y.; Bai, Q.; Xu, P.; Wu, T.; Guo, D.; Peng, Y.; Zhang, H.; Deng, X.; Chen, X.; Luo, M.; et al. Mutation in Rice Abscisic Acid2 Results in Cell Death, Enhanced Disease-Resistance, Altered Seed Dormancy and Development. Front. Plant Sci. 2018, 9, 405. [Google Scholar] [CrossRef]
- Feng, C.Z.; Chen, Y.; Wang, C.; Kong, Y.H.; Wu, W.H.; Chen, Y.F. Arabidopsis RAV1 transcription factor, phosphorylated by SnRK2 kinases, regulates the expression of ABI3, ABI4, and ABI5 during seed germination and early seedling development. Plant J. 2014, 80, 654–668. [Google Scholar] [CrossRef]
- Shu, K.; Zhou, W.; Yang, W. APETALA 2-domain-containing transcription factors: Focusing on abscisic acid and gibberellins antagonism. New Phytol. 2018, 217, 977–983. [Google Scholar] [CrossRef]
- Lin, Q.; Wu, F.; Sheng, P.; Zhang, Z.; Zhang, X.; Guo, X.; Wang, J.; Cheng, Z.; Wang, J.; Wang, H.; et al. The SnRK2-APC/CTE regulatory module mediates the antagonistic action of gibberellic acid and abscisic acid pathways. Nat. Commun. 2015, 6, 7981. [Google Scholar] [CrossRef]
- Lin, Q.; Zhang, Z.; Wu, F.; Feng, M.; Sun, Y.; Chen, W.; Cheng, Z.; Zhang, X.; Ren, Y.; Lei, C.; et al. The APC/CTE E3 Ubiquitin Ligase Complex Mediates the Antagonistic Regulation of Root Growth and Tillering by ABA and GA. Plant Cell. 2020, 32, 1973–1987. [Google Scholar] [CrossRef] [PubMed]
- Han, S.; Shin, D.; Moon, S.; Jeon, S.; Byun, M.; Kim, B. Optimization of Agrobacterium-mediated Transformation in Japoni-ca-type Rice Oryza sativa L. cv. Dongjin for high Efficiency. Korean J. Breed. Sci. 2012, 44, 221–228. [Google Scholar]
- Karimi, M.; Inze, D.; Depicker, A. GATEWAY vectors for Agrobacterium-mediated plant transformation. Trends Plant Sci. 2002, 7, 193–195. [Google Scholar] [CrossRef] [PubMed]
- Min, M.K.; Kim, R.; Kim, B.G. Reconstitution of the Core ABA Signaling in Protoplasts: Transcriptional Activators. Methods Mol. Biol. 2022, 2462, 31–43. [Google Scholar] [CrossRef] [PubMed]

| Gene ID | WT-Mean * | WT-STDEV * | OX-Mean * | OX-STDEV * | Fold Change of OX/WT | ||
|---|---|---|---|---|---|---|---|
| GA20ox | OsGA20ox1 | Os03g0856700 | 6.387454 | 0.03598653 | 5.953094 | 0.041642482 | −1.35125 |
| OsGA20ox2 (SD1) | Os01g0883800 | 4.000492 | 0.14224135 | 3.644319 | 0.131228565 | −1.28352 | |
| OsGA20ox4 | Os05g0421900 | 5.403216 | 0.28699917 | 7.934473 | 0.125858956 | 5.752747 | |
| GA3ox | OsGA3ox2 (D18) | Os01g0177400 | 6.607055 | 0.07167825 | 6.690712 | 0.095721887 | 1.058955 |
| GA2ox (I) | OsGA2ox3 | Os01g0757200 | 8.866322 | 0.25283603 | 9.214727 | 0.112922287 | 1.273661 |
| OsGA2ox4 | Os05g0514600 | 6.277192 | 0.1829343 | 4.578638 | 0.125073243 | −3.25041 | |
| GA2ox (II) | OsGA2ox1 | Os05g0158600 | 6.667921 | 0.06508819 | 6.4263 | 0.130932063 | −1.18283 |
| GA2ox (III) | OsGA2ox6 | Os04g0522500 | 7.123396 | 0.28365293 | 6.383509 | 0.079399404 | −1.66837 |
| CPS | OsCPS | Os02g0278700 | 7.044363 | 0.06118494 | 6.434432 | 0.158792429 | −1.52498 |
| KS | OsKS | Os04g0611800 | 9.16339 | 0.09306083 | 8.827319 | 0.090157625 | −1.26251 |
| KO | OsKO/CYP701A (D35) | Os06g0570100 | 9.297471 | 0.13797206 | 9.716885 | 0.088412978 | 1.337714 |
| KAO | OsKAO/CYP88A5 | Os06g0110000 | 6.218438 | 0.19921268 | 7.706477 | 0.121483696 | 2.80157 |
| GA16, 17ox | CYP714D1 (EUI) | Os05g0482400 | 6.432289 | 0.30009856 | 6.971563 | 0.031743112 | 1.450763 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Song, J.; Ga, E.; Park, S.; Lee, H.; Yoon, I.S.; Lee, S.B.; Lee, J.-Y.; Kim, B.-G. PROTEIN PHOSPHATASE 2C08, a Negative Regulator of Abscisic Acid Signaling, Promotes Internode Elongation in Rice. Int. J. Mol. Sci. 2023, 24, 10821. https://doi.org/10.3390/ijms241310821
Song J, Ga E, Park S, Lee H, Yoon IS, Lee SB, Lee J-Y, Kim B-G. PROTEIN PHOSPHATASE 2C08, a Negative Regulator of Abscisic Acid Signaling, Promotes Internode Elongation in Rice. International Journal of Molecular Sciences. 2023; 24(13):10821. https://doi.org/10.3390/ijms241310821
Chicago/Turabian StyleSong, Jaeeun, Eunji Ga, Sangkyu Park, Hyo Lee, In Sun Yoon, Saet Buyl Lee, Jong-Yeol Lee, and Beom-Gi Kim. 2023. "PROTEIN PHOSPHATASE 2C08, a Negative Regulator of Abscisic Acid Signaling, Promotes Internode Elongation in Rice" International Journal of Molecular Sciences 24, no. 13: 10821. https://doi.org/10.3390/ijms241310821
APA StyleSong, J., Ga, E., Park, S., Lee, H., Yoon, I. S., Lee, S. B., Lee, J.-Y., & Kim, B.-G. (2023). PROTEIN PHOSPHATASE 2C08, a Negative Regulator of Abscisic Acid Signaling, Promotes Internode Elongation in Rice. International Journal of Molecular Sciences, 24(13), 10821. https://doi.org/10.3390/ijms241310821






