Identification and Characterization of an HtrA Sheddase Produced by Coxiella burnetii
Abstract
:1. Introduction
2. Results
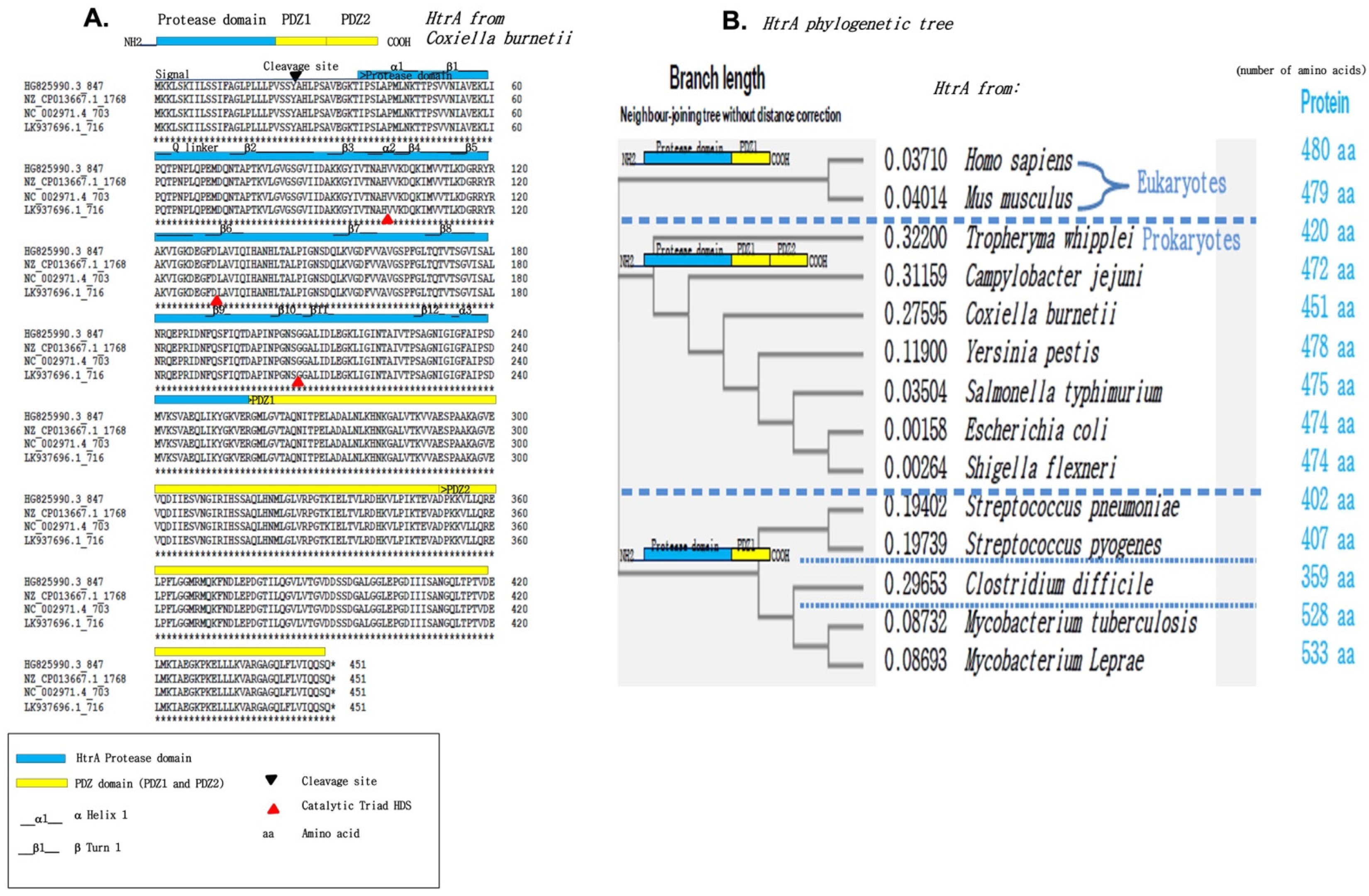
2.1. In Silico Search for a Hypothetical Sheddase in the Genome of C. burnetii
2.2. Identification of an HtrA-like Sheddase Produced by Coxiella Burnetii
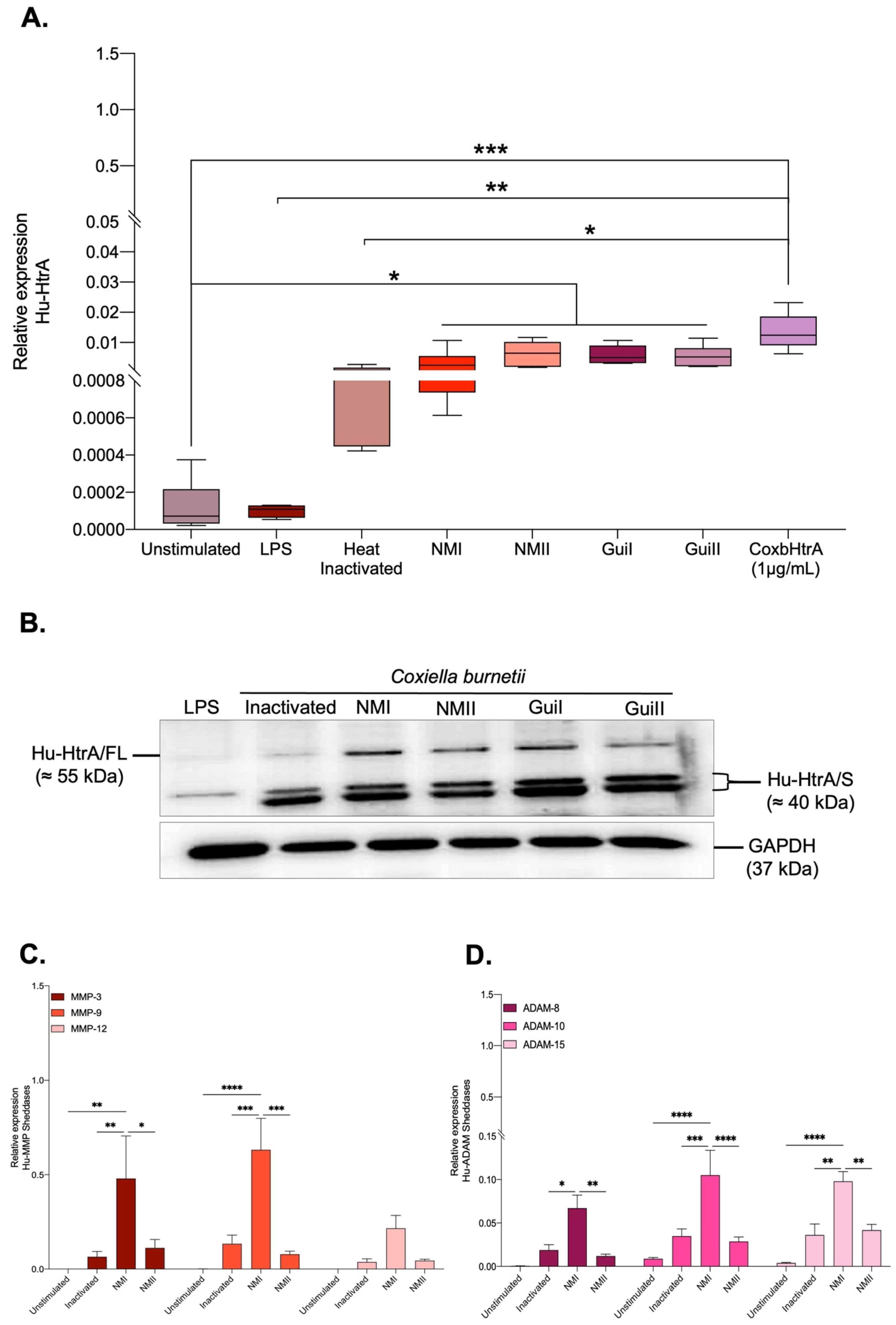
2.3. Transcription of the CbHtrA-like Gene in C. burnetii-Infected Cells
2.4. Effects of CbHtrA from C. burnetii on E-Cadherin Expression in BeWo Cells
2.5. C. burnetii Infection of BeWo Cells Also Modulates the Expression of Human Sheddases
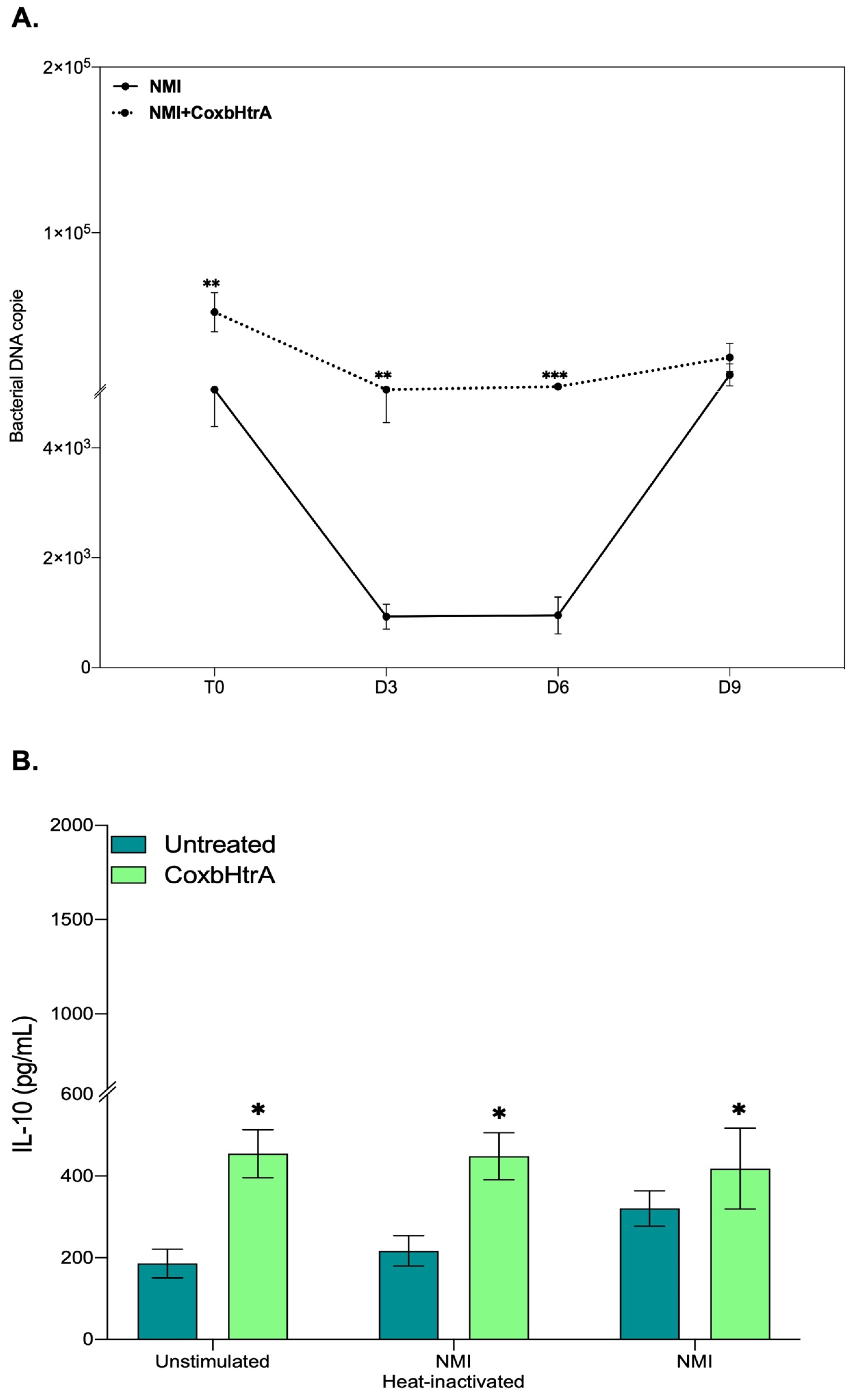
2.6. Evaluation of the Role of CbHtrA in the Physiopathology of C. burnetii Infection
3. Discussion
3.1. The Genome of C. burnetii Contains a Gene Encoding an HtrA Sheddase
3.2. How Could C. burnetii HtrA Sheddase Act to Cleave E-Cad When the Bacterium Is inside a Cell?
3.3. C. burnetii Induces a Complexe Profile of Sheddases Expression during the Virulent and Avirulent Phases of Infection
3.4. What Could Be the Consequences of the Expression of the HtrA Sheddase in the Intracellular Cycle of Replication of C. burnetii and the Pathophysiology of Q Fever?
4. Materials and Methods
4.1. Coxiella Burnetii
4.2. In Silico Sequence Analysis
4.3. Production of Recombinant CbHtrA
4.4. In Vitro Cell Culture Model and Infection
4.5. DNA/RNA Extraction and Reverse Transcription Assay
4.6. Polymerase Chain Reaction (PCR) Standard
4.7. Quantitative Reverse Transcription–Polymerase Chain Reaction (qRT-PCR)
4.8. Evaluation of Coxb-HtrA Functional Activity
4.9. Flow Cytometry Assay
4.10. Confocal Microscopy Analysis
4.11. Antimicrobial Activity of Macrophage-Differentiated THP-1
4.12. Immunoassays
4.13. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Maurin, M.; Raoult, D. Q fever. Clin. Microbiol. Rev. 1999, 12, 518–553. [Google Scholar] [CrossRef] [Green Version]
- Barry, A.O.; Mege, J.-L.; Ghigo, E. Hijacked phagosomes and leukocyte activation: An intimate relationship. J. Leukoc. Biol. 2011, 89, 373–382. [Google Scholar] [CrossRef] [PubMed]
- Pechstein, J.; Schulze-Luehrmann, J.; Bisle, S.; Cantet, F.; Beare, P.A.; Ölke, M.; Bonazzi, M.; Berens, C.; Lührmann, A. The Coxiella burnetii T4SS Effector AnkF Is Important for Intracellular Replication. Front. Cell. Infect. Microbiol. 2020, 10, 559915. [Google Scholar] [CrossRef] [PubMed]
- McClure, E.E.; Chávez, A.S.O.; Shaw, D.K.; Carlyon, J.A.; Ganta, R.R.; Noh, S.M.; Wood, D.O.; Bavoil, P.M.; Brayton, K.A.; Martinez, J.J.; et al. Engineering of obligate intracellular bacteria: Progress, challenges and paradigms. Nat. Rev. Genet. 2017, 15, 544–558. [Google Scholar] [CrossRef] [PubMed]
- Van Schaik, E.J.; Chen, C.; Mertens, K.; Weber, M.M.; Samuel, J.E. Molecular pathogenesis of the obligate intracellular bacterium Coxiella burnetii. Nat. Rev. Microbiol. 2013, 11, 561–573. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Melenotte, C.; Protopopescu, C.; Million, M.; Edouard, S.; Carrieri, M.P.; Eldin, C.; Angelakis, E.; Djossou, F.; Bardin, N.; Fournier, P.-E.; et al. Clinical Features and Complications of Coxiella burnetii Infections From the French National Reference Center for Q Fever. JAMA Netw. Open 2018, 1, e181580. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Melenotte, C.; Million, M.; Audoly, G.; Gorse, A.; Dutronc, H.; Roland, G.; Dekel, M.; Moreno, A.; Cammilleri, S.; Carrieri, M.P.; et al. B-cell non-Hodgkin lymphoma linked to Coxiella burnetii. Blood 2016, 127, 113–121. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Melenotte, C.; Izaaryene, J.-J.; Gomez, C.; Delord, M.; Prudent, E.; Lepidi, H.; Mediannikov, O.; Lacoste, M.; Djossou, F.; Mania, A.; et al. Coxiella burnetii: A Hidden Pathogen in Interstitial Lung Disease? Clin. Infect. Dis. 2018, 67, 1120–1124. [Google Scholar] [CrossRef] [Green Version]
- Melenotte, C.; Caputo, A.; Bechah, Y.; Lepidi, H.; Terras, J.; Kowalczewska, M.; Di Pinto, F.; Nappez, C.; Raoult, D.; Brégeon, F. The hypervirulent Coxiella burnetii Guiana strain compared in silico, in vitro and in vivo to the Nine Mile and the German strain. Clin. Microbiol. Infect. 2019, 25, 1155.e1–1155.e8. [Google Scholar] [CrossRef]
- van Roeden, S.E.; Wever, P.C.; Oosterheert, J.J. “Chronic Q fever-related complications and mortality: Data from a nationwide cohort”—Author’s reply. Clin. Microbiol. Infect. 2019, 25, 1436–1437. [Google Scholar] [CrossRef]
- Weehuizen, J.M.; E van Roeden, S.; Hogewoning, S.J.; van der Hoek, W.; Bonten, M.J.M.; Hoepelman, A.I.M.; Bleeker-Rovers, C.P.; Wever, P.C.; Oosterheert, J.J. No increased risk of mature B-cell non-Hodgkin lymphoma after Q fever detected: Results from a 16-year ecological analysis of the Dutch population incorporating the 2007–2010 Q fever outbreak. Leuk. Res. 2022, 51, 1481–1488. [Google Scholar] [CrossRef] [PubMed]
- Elkington, P.T.G.; O’kane, C.M.; Friedland, J.S. The paradox of matrix metalloproteinases in infectious disease. Clin. Exp. Immunol. 2005, 142, 12–20. [Google Scholar] [CrossRef]
- Vanlaere, I.; Libert, C. Matrix Metalloproteinases as Drug Targets in Infections Caused by Gram-Negative Bacteria and in Septic Shock. Clin. Microbiol. Rev. 2009, 22, 224–239. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ortega, F.E.; Rengarajan, M.; Chavez, N.; Radhakrishnan, P.; Gloerich, M.; Bianchini, J.; Siemers, K.; Luckett, W.S.; Lauer, P.; Nelson, W.J.; et al. Adhesion to the host cell surface is sufficient to mediate Listeria monocytogenes entry into epithelial cells. Mol. Biol. Cell 2017, 28, 2945–2957. [Google Scholar] [CrossRef] [PubMed]
- Kague, E.; Thomazini, C.M.; Pardini, M.I.D.C.M.; de Carvalho, F.; Leite, C.V.; Pinheiro, N.A. Methylation status of CDH1 gene in samples of gastric mucous from brazilian patients with chronic gastritis infected by Helicobacter pylori. Arq. Gastroenterol. 2010, 47, 7–12. [Google Scholar] [CrossRef] [Green Version]
- Barisani-Asenbauer, T.; Inic-Kanada, A.; Grdovic, N.; Stein, E.; Ghasemian, E.; Schuerer, N.; Vidakovic, M. Chlamydia trachomatis infection induces epithelial-mesenchymal transition in conjunctival epithelial cells. Investig. Ophthalmol. Vis. Sci. 2017, 58, 5778. [Google Scholar]
- Jacobs, G.; Hellmig, S.; Huse, K.; Titz, A.; Franke, A.; Kwiatkowski, R.; Ott, S.; Kosmahl, M.; Fischbach, W.; Lucius, R.; et al. Polymorphisms in the 3′-untranslated region of the CDH1 gene are a risk factor for primary gastric diffuse large B-cell lymphoma. Haematologica 2011, 96, 987–995. [Google Scholar] [CrossRef]
- Wu, S.; Lim, K.C.; Huang, J.; Saidi, R.F.; Sears, C.L. Bacteroides fragilis enterotoxin cleaves the zonula adherens protein, E-cadherin. Proc. Natl. Acad. Sci. USA 1998, 95, 14979–14984. [Google Scholar] [CrossRef] [Green Version]
- Chung, L.; Orberg, E.T.; Geis, A.L.; Chan, J.L.; Fu, K.; Shields, C.E.D.; Dejea, C.M.; Fathi, P.; Chen, J.; Finard, B.B.; et al. Bacteroides fragilis Toxin Coordinates a Pro-carcinogenic Inflammatory Cascade via Targeting of Colonic Epithelial Cells. Cell Host Microbe 2018, 23, 203–214.e5. [Google Scholar] [CrossRef] [Green Version]
- Kumar, R.; Herold, J.L.; Schady, D.; Davis, J.; Kopetz, S.; Martinez-Moczygemba, M.; Murray, B.E.; Han, F.; Li, Y.; Callaway, E.; et al. Streptococcus gallolyticus subsp. gallolyticus promotes colorectal tumor development. PLoS Pathog. 2017, 13, e1006440. [Google Scholar] [CrossRef] [Green Version]
- Niessen, C.M.; Leckband, D.; Yap, A.S.; Zhao, B.; Knepper, M.A.; Chou, C.-L.; Pisitkun, T. Tissue Organization by Cadherin Adhesion Molecules: Dynamic Molecular and Cellular Mechanisms of Morphogenetic Regulation. Physiol. Rev. 2011, 91, 691–731. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hyafil, F.; Babinet, C.; Jacob, F. Cell-cell interactions in early embryogenesis: A molecular approach to the role of calcium. Cell 1981, 26, 447–454. [Google Scholar] [CrossRef] [PubMed]
- Shields, B.D.; Koss, B.; Taylor, E.M.; Storey, A.J.; West, K.L.; Byrum, S.D.; Mackintosh, S.G.; Edmondson, R.; Mahmoud, F.; Shalin, S.C.; et al. Loss of E-Cadherin Inhibits CD103 Antitumor Activity and Reduces Checkpoint Blockade Responsiveness in Melanoma. Cancer Res. 2019, 79, 1113–1123. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- McCrea, P.D.; Maher, M.T.; Gottardi, C.J. Nuclear Signaling from Cadherin Adhesion Complexes. Curr. Top. Dev. Biol. 2015, 112, 129–196. [Google Scholar]
- Grabowska, M.M.; Day, M.L. Soluble E-cadherin: More than a symptom of disease. Front. Biosci. 2012, 17, 1948–1964. [Google Scholar] [CrossRef] [Green Version]
- Syrigos, K.N.; Harrington, K.J.; Karayiannakis, A.; Baibas, N.; Katirtzoglou, N.; Roussou, P. Circulating soluble E-cadherin levels are of prognostic significance in patients with multiple myeloma. Anticancer Res. 2004, 24, 2027–2031. [Google Scholar] [PubMed]
- Mezouar, S.; Osman, I.O.; Melenotte, C.; Slimani, C.; Chartier, C.; Raoult, D.; Mege, J.-L.; Devaux, C.A. High Concentrations of Serum Soluble E-Cadherin in Patients With Q Fever. Front. Cell. Infect. Microbiol. 2019, 9, 219. [Google Scholar] [CrossRef] [Green Version]
- Devaux, C.A.; Mezouar, S.; Mege, J.-L. The E-Cadherin Cleavage Associated to Pathogenic Bacteria Infections Can Favor Bacterial Invasion and Transmigration, Dysregulation of the Immune Response and Cancer Induction in Humans. Front. Microbiol. 2019, 10, 2598. [Google Scholar] [CrossRef]
- Krajinović, L.C.; Šoprek, S.; Korva, M.; Dželalija, B.; Rode, O.Đ.; Škerk, V.; Avšič-Županc, T.; Markotić, A. Serum levels of metalloproteinases and their inhibitors during infection with pathogens having integrin receptor-mediated cellular entry. Scand. J. Infect. Dis. 2012, 44, 663–669. [Google Scholar] [CrossRef]
- Jansen, A.; Schoffelen, T.; Textoris, J.; Mege, J.; Bleeker-Rovers, C.; Roest, H.; Wever, P.; Joosten, L.; Netea, M.; van de Vosse, E.; et al. Involvement of matrix metalloproteinases in chronic Q fever. Clin. Microbiol. Infect. 2017, 23, 487.e7–487.e13. [Google Scholar] [CrossRef] [Green Version]
- Osman, I.O.; Mezouar, S.; Brahim-Belhaouari, D.; Mege, J.-L.; Devaux, C.A. Modulation of the E-cadherin in human cells infected in vitro with Coxiella burnetii. PLoS ONE 2023, 18, e0285577. [Google Scholar] [CrossRef] [PubMed]
- Abfalter, C.M.; Schubert, M.; Götz, C.; Schmidt, T.P.; Posselt, G.; Wessler, S. HtrA-mediated E-cadherin cleavage is limited to DegP and DegQ homologs expressed by gram-negative pathogens. Cell Commun. Signal. 2016, 14, 30. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Israeli, M.A.; Elia, U.; Rotem, S.; Cohen, H.; Tidhar, A.; Bercovich-Kinori, A.; Cohen, O.; Chitlaru, T. Distinct Contribution of the HtrA Protease and PDZ Domains to Its Function in Stress Resilience and Virulence of Bacillus anthracis. Front. Microbiol. 2019, 10, 255. [Google Scholar] [CrossRef]
- Kim, D.-Y.; Kim, K.K. Structure and Function of HtrA Family Proteins, the Key Players in Protein Quality Control. BMB Rep. 2005, 38, 266–274. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kim, H.; Wu, K.; Lee, C. Stress-Responsive Periplasmic Chaperones in Bacteria. Front. Mol. Biosci. 2021, 8, 678697. [Google Scholar] [CrossRef]
- Amara, A.B.; Bechah, Y.; Mege, J.L. Immune Response and Coxiella burnetii Invasion. In Coxiella burnetii: Recent Advances and New Perspectives in Research of the Q Fever Bacterium; Toman, R., Heinzen, R.A., Samuel, J.E., Mege, J.-L., Eds.; Advances in Experimental Medicine and Biology; Springer: Dordrecht, The Netherlands, 2012; pp. 287–298. [Google Scholar]
- Capo, C.; Zaffran, Y.; Zugun, F.; Houpikian, P.; Raoult, D.; Mege, J.L. Production of interleukin-10 and transforming growth factor beta by peripheral blood mononuclear cells in Q fever endocarditis. Infect. Immun. 1996, 64, 4143–4147. [Google Scholar] [CrossRef] [Green Version]
- Ghigo, E.; Capo, C.; Raoult, D.; Mege, J.-L. Interleukin-10 Stimulates Coxiella burnetii Replication in Human Monocytes through Tumor Necrosis Factor Down-Modulation: Role in Microbicidal Defect of Q Fever. Infect. Immun. 2001, 69, 2345–2352. [Google Scholar] [CrossRef] [Green Version]
- Maguire, B.A.; Wild, D.G. The roles of proteins L28 and L33 in the assembly and function of Escherichia coli ribosomes in vivo. Mol. Microbiol. 1997, 23, 237–245. [Google Scholar] [CrossRef]
- Gieldon, A.; Zurawa-Janicka, D.; Jarzab, M.; Wenta, T.; Golik, P.; Dubin, G.; Lipinska, B.; Ciarkowski, J. Distinct 3D Architecture and Dynamics of the Human HtrA2(Omi) Protease and Its Mutated Variants. PLoS ONE 2016, 11, e0161526. [Google Scholar] [CrossRef] [Green Version]
- Filloux, A. Filloux, Bacterial protein secretion systems: Game of types. Microbiology 2022, 168, 001193. [Google Scholar] [CrossRef]
- Desvaux, M.; Hébraud, M.; Talon, R.; Henderson, I.R. Secretion and subcellular localizations of bacterial proteins: A semantic awareness issue. Trends Microbiol. 2009, 17, 139–145. [Google Scholar] [CrossRef] [PubMed]
- Crane, J.M.; Randall, L.L. The Sec System: Protein Export in Escherichia coli. EcoSal Plus 2017, 7. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ruiz-Perez, F.; Nataro, J.P. Bacterial serine proteases secreted by the autotransporter pathway: Classification, specificity, and role in virulence. Cell. Mol. Life Sci. 2013, 71, 745–770. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Clarke, K.R.; Hor, L.; Pilapitiya, A.; Luirink, J.; Paxman, J.J.; Heras, B. Phylogenetic Classification and Functional Review of Autotransporters. Front. Immunol. 2022, 13, 921272. [Google Scholar] [CrossRef]
- Hoy, B.; Geppert, T.; Boehm, M.; Reisen, F.; Plattner, P.; Gadermaier, G.; Sewald, N.; Ferreira, F.; Briza, P.; Schneider, G.; et al. Distinct Roles of Secreted HtrA Proteases from Gram-negative Pathogens in Cleaving the Junctional Protein and Tumor Suppressor E-cadherin. J. Biol. Chem. 2012, 287, 10115–10120. [Google Scholar] [CrossRef] [Green Version]
- Boehm, M.; Haenel, I.; Hoy, B.; Brøndsted, L.; Smith, T.G.; Hoover, T.; Wessler, S.; Tegtmeyer, N. Extracellular secretion of protease HtrA from Campylobacter jejuni is highly efficient and independent of its protease activity and flagellum. Eur. J. Microbiol. Immunol. 2013, 3, 163–173. [Google Scholar] [CrossRef] [Green Version]
- Neddermann, M.; Backert, S. Quantification of serine protease HtrA molecules secreted by the foodborne pathogen Campylobacter jejuni. Gut Pathog. 2019, 11, 14. [Google Scholar] [CrossRef]
- Chang, Z. The function of the DegP (HtrA) protein: Protease versus chaperone. IUBMB Life 2016, 68, 904–907. [Google Scholar] [CrossRef] [Green Version]
- Kim, S.; Grant, R.A.; Sauer, R.T. Covalent Linkage of Distinct Substrate Degrons Controls Assembly and Disassembly of DegP Proteolytic Cages. Cell 2011, 145, 67–78. [Google Scholar] [CrossRef] [Green Version]
- Huston, W.M.; Theodoropoulos, C.; Mathews, S.A.; Timms, P. Chlamydia trachomatis responds to heat shock, penicillin induced persistence, and IFN-gamma persistence by altering levels of the extracytoplasmic stress response protease HtrA. BMC Microbiol. 2008, 8, 190. [Google Scholar] [CrossRef] [Green Version]
- Zhong, G. Chlamydia Trachomatis Secretion of Proteases for Manipulating Host Signaling Pathways. Front. Microbiol. 2011, 2, 14. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wu, X.; Lei, L.; Gong, S.; Chen, D.; Flores, R.; Zhong, G. The chlamydial periplasmic stress response serine protease cHtrA is secreted into host cell cytosol. BMC Microbiol. 2011, 11, 87. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hua, K.; Li, Y.; Zhou, H.; Hu, X.; Chen, Y.; He, R.; Luo, R.; Zhou, R.; Bi, D.; Jin, H. Haemophilus parasuis Infection Disrupts Adherens Junctions and Initializes EMT Dependent on Canonical Wnt/β-Catenin Signaling Pathway. Front. Cell. Infect. Microbiol. 2018, 8, 324. [Google Scholar] [CrossRef] [Green Version]
- Overall, C.M.; López-Otín, C. Strategies for MMP inhibition in cancer: Innovations for the post-trial era. Nat. Rev. Cancer 2002, 2, 657–672. [Google Scholar] [CrossRef] [PubMed]
- Takeda, S. ADAM and ADAMTS Family Proteins and Snake Venom Metalloproteinases: A Structural Overview. Toxins 2016, 8, 155. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Okada, Y.; Nakanishi, I. Activation of matrix metalloproteinase 3 (stromelysin) and matrix metalloproteinase 2 (‘gelatinase’) by human neutrophil elastase and cathepsin G. FEBS Lett. 1989, 249, 353–356. [Google Scholar] [CrossRef] [Green Version]
- Garbett, E.A.; Reed, M.; Brown, N.J. Proteolysis in human breast and colorectal cancer. Br. J. Cancer 1999, 81, 287–293. [Google Scholar] [CrossRef] [Green Version]
- Noe, V.; Fingleton, B.; Jacobs, K.; Crawford, H.; Vermeulen, S.; Steelant, W.; Bruyneel, E.; Matrisian, L.; Mareel, M. Release of an invasion promoter E-cadherin fragment by matrilysin and stromelysin-1. J. Cell Sci. 2001, 114, 111–118. [Google Scholar] [CrossRef]
- Ogata, Y.; Enghild, J.; Nagase, H. Matrix metalloproteinase 3 (stromelysin) activates the precursor for the human matrix metalloproteinase 9. J. Biol. Chem. 1992, 267, 3581–3584. [Google Scholar] [CrossRef]
- Nagase, H.; Visse, R.; Murphy, G. Structure and function of matrix metalloproteinases and TIMPs. Cardiovasc. Res. 2006, 69, 562–573. [Google Scholar] [CrossRef] [Green Version]
- Parks, W.C.; Wilson, C.L.; López-Boado, Y.S. Matrix metalloproteinases as modulators of inflammation and innate immunity. Nat. Rev. Immunol. 2004, 4, 617–629. [Google Scholar] [CrossRef] [PubMed]
- Yan, W.; Li, S.X.; Wei, M.; Gao, H. Identification of MMP9 as a novel key gene in mantle cell lymphoma based on bioinformatic analysis and design of cyclic peptides as MMP9 inhibitors based on molecular docking. Oncol. Rep. 2018, 40, 2515–2524. [Google Scholar] [CrossRef] [PubMed]
- Welgus, H.G.; Campbell, E.J.; Cury, J.D.; Eisen, A.Z.; Senior, R.M.; Wilhelm, S.M.; Goldberg, G.I. Neutral metalloproteinases produced by human mononuclear phagocytes. Enzyme profile, regulation, and expression during cellular development. J. Clin. Investig. 1990, 86, 1496–1502. [Google Scholar] [CrossRef] [PubMed]
- Kuefer, R.; Day, K.C.; Kleer, C.G.; Sabel, M.S.; Hofert, M.D.; Varambally, S.; Zorn, C.S.; Chinnaiyan, A.M.; Rubin, M.A.; Day, M.L. ADAM15 Disintegrin Is Associated with Aggressive Prostate and Breast Cancer Disease. Neoplasia 2006, 8, 319–329. [Google Scholar] [CrossRef] [Green Version]
- Najy, A.J.; Day, K.C.; Day, M.L. ADAM15 Supports Prostate Cancer Metastasis by Modulating Tumor Cell–Endothelial Cell Interaction. Cancer Res. 2008, 68, 1092–1099. [Google Scholar] [CrossRef] [Green Version]
- Najy, A.J.; Day, K.C.; Day, M.L. The Ectodomain Shedding of E-cadherin by ADAM15 Supports ErbB Receptor Activation. J. Biol. Chem. 2008, 283, 18393–18401. [Google Scholar] [CrossRef] [Green Version]
- de Waal Malefyt, R.; Abrams, J.; Bennett, B.; Figdor, C.G.; De Vries, J.E. Interleukin 10(IL-10) inhibits cytokine synthesis by human monocytes: An autoregulatory role of IL-10 produced by monocytes. J. Exp. Med. 1991, 174, 1209–1220. [Google Scholar] [CrossRef] [Green Version]
- Toman, R.; Škultéty, L. Structural study on a lipopolysaccharide from Coxiella burnetii strain Nine Mile in avirulent phase II. Carbohydr. Res. 1996, 283, 175–185. [Google Scholar] [CrossRef]
- Hoover, T.A.; Culp, D.W.; Vodkin, M.H.; Williams, J.C.; Thompson, H.A. Chromosomal DNA Deletions Explain Phenotypic Characteristics of Two Antigenic Variants, Phase II and RSA 514 (Crazy), of the Coxiella burnetii Nine Mile Strain. Infect. Immun. 2002, 70, 6726–6733. [Google Scholar] [CrossRef] [Green Version]
- Capo, C.; Lindberg, F.P.; Meconi, S.; Zaffran, Y.; Tardei, G.; Brown, E.J.; Raoult, D.; Mege, J.L. Subversion of monocyte functions by Coxiella burnetii: Impairment of the cross-talk between alphavbeta3 integrin and CR3. J. Immunol. 1999, 163, 6078–6085. [Google Scholar] [CrossRef]
- Seshadri, R.; Paulsen, I.T.; Eisen, J.A.; Read, T.D.; Nelson, K.E.; Nelson, W.C.; Ward, N.L.; Tettelin, H.; Davidsen, T.M.; Beanan, M.J.; et al. Complete genome sequence of the Q-fever pathogen Coxiella burnetii. Proc. Natl. Acad. Sci. USA 2003, 100, 5455–5460. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kuley, R.; Smith, H.E.; Janse, I.; Harders, F.L.; Baas, F.; Schijlen, E.; Nabuurs-Franssen, M.H.; Smits, M.A.; Roest, H.I.J.; Bossers, A. First Complete Genome Sequence of the Dutch Veterinary Coxiella burnetii Strain NL3262, Originating from the Largest Global Q Fever Outbreak, and Draft Genome Sequence of Its Epidemiologically Linked Chronic Human Isolate NLhu3345937. Genome Announc. 2016, 4, e00245-16. [Google Scholar] [CrossRef] [Green Version]
- D’amato, F.; Rouli, L.; Edouard, S.; Tyczka, J.; Million, M.; Robert, C.; Nguyen, T.T.; Raoult, D. The genome of Coxiella burnetii Z3055, a clone linked to the Netherlands Q fever outbreaks, provides evidence for the role of drift in the emergence of epidemic clones. Comp. Immunol. Microbiol. Infect. Dis. 2014, 37, 281–288. [Google Scholar] [CrossRef] [PubMed]
- Hyatt, D.; Chen, G.-L.; Locascio, P.F.; Land, M.L.; Larimer, F.W.; Hauser, L.J. Prodigal: Prokaryotic gene recognition and translation initiation site identification. BMC Bioinform. 2010, 11, 119. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Petersen, T.N.; Brunak, S.; von Heijne, G.; Nielsen, H. SignalP 4.0: Discriminating signal peptides from transmembrane regions. Nat. Methods 2011, 8, 785–786. [Google Scholar] [CrossRef] [PubMed]
- Schwende, H.; Fitzke, E.; Ambs, P.; Dieter, P. Differences in the state of differentiation of THP-1 cells induced by phorbol ester and 1,25-dihydroxyvitamin D3. J. Leukoc. Biol. 1996, 59, 555–561. [Google Scholar] [CrossRef]
- Daigneault, M.; Preston, J.A.; Marriott, H.M.; Whyte, M.K.B.; Dockrell, D.H. The Identification of Markers of Macrophage Differentiation in PMA-Stimulated THP-1 Cells and Monocyte-Derived Macrophages. PLoS ONE 2010, 5, e8668. [Google Scholar] [CrossRef]
- Chanput, W.; Mes, J.J.; Savelkoul, H.F.J.; Wichers, H.J. Characterization of polarized THP-1 macrophages and polarizing ability of LPS and food compounds. Food Funct. 2013, 4, 266–276. [Google Scholar] [CrossRef]
- Winer, J.; Jung, C.K.S.; Shackel, I.; Williams, P.M. Development and validation of real-time quantitative reverse transcriptase-polymerase chain reaction for monitoring gene expression in cardiac myocytes in vitro. Anal. Biochem. 1999, 270, 41–49. [Google Scholar] [CrossRef]
- Mezouar, S.; Benammar, I.; Boumaza, A.; Diallo, A.B.; Chartier, C.; Buffat, C.; Boudjarane, J.; Halfon, P.; Katsogiannou, M.; Mege, J.-L. Full-Term Human Placental Macrophages Eliminate Coxiella burnetii Through an IFN-γ Autocrine Loop. Front. Microbiol. 2019, 10, 2434. [Google Scholar] [CrossRef] [Green Version]

| C. burnetii Strain | Protein | Sheddases | % Identity | e-Value | % Coverage | Signal IP | Genomic Position |
|---|---|---|---|---|---|---|---|
| Cb175 | HG825990.3_1452 | MMP-3 | 28 | 5 × 10−5 | 95 | No | 1.208.002–1.208.280 |
| HG825990.3_2153 | ADAM_15 | 30 | 7 × 10−4 | 53 | No | 1.793.727–1.793.329 | |
| HG825990.3_823 | Subtilase | 31 | 3 × 10−5 | 48 | Yes | 676.754–676.359 | |
| HG825990.3_51 | Rhomboid-1 | 26 | 6 × 10−4 | 61 | Yes | 43.537–42.977 | |
| HG825990 3_847 | HtrA | 42 | 2 × 10−112 | 100 | Yes | 699.374–698.019 | |
| NL3262 | NZ_CP013667.1_1221 | MMP-3 | 30 | 5 × 10−4 | 45 | No | 1.131.415–1.131.011 |
| NZ_CP013667.1_2102 | MMP-9 | 25 | 0.001 | 79 | No | 1.982.404–1.983.099 | |
| NZ_CP013667.1_1792 | Subtilase | 31 | 2 × 10−5 | 48 | Yes | 1.679.995–1.680.382 | |
| NZ_CP013667.1_230 | Rhomboid-1 | 26 | 6 × 10−4 | 61 | Yes | 217.053–217.613 | |
| NZ_CP013667.1_1768 | HtrA | 42 | 2 × 10−112 | 100 | Yes | 1.656.010–1.657.365 | |
| RSA 493 | NC_002971.4_1204 | MMP-3 | 30 | 5 × 10−4 | 45 | No | 1.207.961–1.208.365 |
| NC_002971.4_271 | MMP-9 | 25 | 0.001 | 79 | No | 258.686–258.513 | |
| NC_002971.4_682 | Subtilase | 31 | 3 × 10−5 | 48 | Yes | 676.626–676.228 | |
| NC_002971.4_45 | Rhomboid-1 | 26 | 6 × 10−4 | 61 | Yes | 43.528–42.968 | |
| NC_002971.4_703 | HtrA | 42 | 2 × 10−112 | 100 | Yes | 699.241–697.886 | |
| Z3055 | LK937696.1_1129 | MMP-3 | 30 | 5 × 10−4 | 45 | No | 1.100.606–1.102.408 |
| LK937696.1_272 | MMP-9 | 25 | 0.001 | 79 | No | 258.788–258.549 | |
| LK937696.1_695 | Subtilase | 31 | 2 × 10−5 | 48 | Yes | 676.641–676.243 | |
| LK937696.1_44 | Rhomboid-1 | 26 | 6 × 10−4 | 61 | Yes | 43.525–42.965 | |
| LK937696.1_716 | HtrA | 42 | 2 × 10−112 | 100 | Yes | 699.254–697.899 |
| C. burnetii Strains | Sheddases | Mean e-Value | Mean % Coverage | Identification |
|---|---|---|---|---|
| Cb175 | ADAM_15 | 7 × 10−4 | 53 | Hypothetical C. burnetii protein |
| NL3262 RSA 493 Z3055 | MMP-9 | 0.001 | 79 | 50s ribosomal protein L28 |
| Cb175 NL3262 RSA 493 Z3055 | MMP-3 | 5 × 10−4 to 5 × 10−5 | 45 to 95 | Membrane of the nuclear transport 2 (NFT2) |
| Subtilase | 2 × 10−5 to 3 × 10−5 | 48 | Hypothetical C. burnetii protein with a putative Rif-1 domain | |
| Rhomboid-1 | 6 × 10−4 | 61 | Hypothetical C. burnetii protein with a putative OmpA domain | |
| HtrA | 2 × 10−112 | 100 | C. burnetii HtrA protein |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Osman, I.O.; Caputo, A.; Pinault, L.; Mege, J.-L.; Levasseur, A.; Devaux, C.A. Identification and Characterization of an HtrA Sheddase Produced by Coxiella burnetii. Int. J. Mol. Sci. 2023, 24, 10904. https://doi.org/10.3390/ijms241310904
Osman IO, Caputo A, Pinault L, Mege J-L, Levasseur A, Devaux CA. Identification and Characterization of an HtrA Sheddase Produced by Coxiella burnetii. International Journal of Molecular Sciences. 2023; 24(13):10904. https://doi.org/10.3390/ijms241310904
Chicago/Turabian StyleOsman, Ikram Omar, Aurelia Caputo, Lucile Pinault, Jean-Louis Mege, Anthony Levasseur, and Christian A. Devaux. 2023. "Identification and Characterization of an HtrA Sheddase Produced by Coxiella burnetii" International Journal of Molecular Sciences 24, no. 13: 10904. https://doi.org/10.3390/ijms241310904





