A Newly Established Cuproptosis-Related Gene Signature for Predicting Prognosis and Immune Infiltration in Uveal Melanoma
Abstract
1. Introduction
2. Results
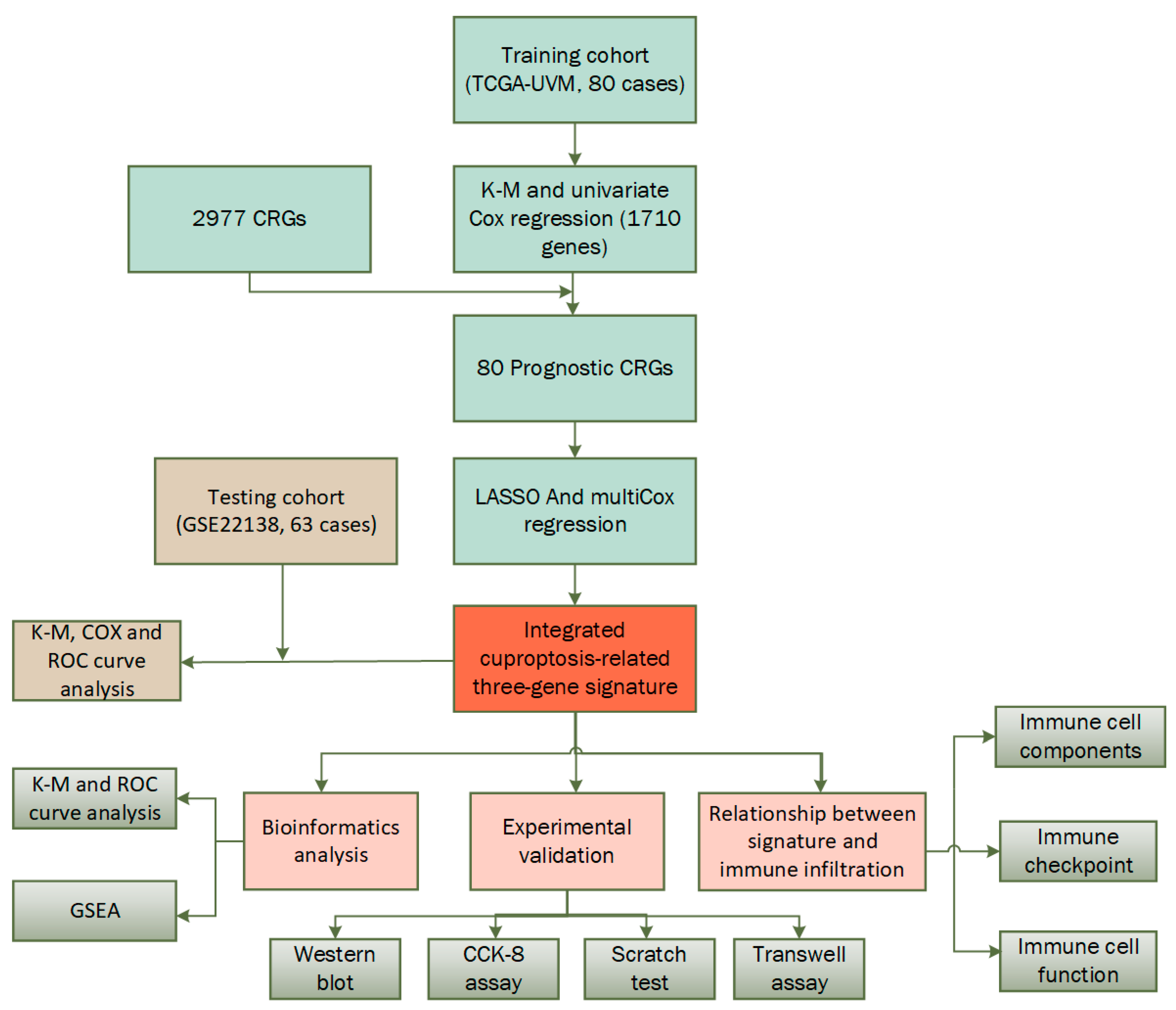
2.1. Construction of a Prognostic CRTG Signature
2.2. Verifying the Prognostic Capacity of the CRTG Signature
2.3. Correlation Analysis of the CRTG Signature and UVM Common Mutations
2.4. Functional Enrichment Analysis
2.5. Correlation Analysis of the CRTG Signature and Immune Cell Infiltration
2.6. Experimental Validation Analysis
3. Discussion
4. Materials and Methods
4.1. Cohorts and CRGs
4.2. Identification and Validation of the Prognostic Cuproptosis-Related Gene Signature
4.3. Correlation between the Gene Signature and UVM Common Mutations
4.4. Gene Set Enrichment Analysis
4.5. Tumor Immune Microenvironment Analysis
4.6. Cell Culture and siRNA Transfection
4.7. Western Blot
4.8. Cell Proliferation
4.9. Scratch Test
4.10. Transwell Assay
4.11. The Effect of Cuproptosis on the Viability of MUM-2B Cells
4.12. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Singh, A.D.; Turell, M.E.; Topham, A.K. Uveal melanoma: Trends in incidence, treatment, and survival. Ophthalmology 2011, 118, 1881–1885. [Google Scholar] [CrossRef] [PubMed]
- Shields, C.L.; Kaliki, S.; Shah, S.U.; Luo, W.; Furuta, M.; Shields, J.A. Iris melanoma: Features and prognosis in 317 children and adults. J. AAPOS 2012, 16, 10–16. [Google Scholar] [CrossRef] [PubMed]
- Kujala, E.; Makitie, T.; Kivela, T. Very long-term prognosis of patients with malignant uveal melanoma. Invest. Ophthalmol. Vis. Sci. 2003, 44, 4651–4659. [Google Scholar] [CrossRef]
- Jager, M.J.; Shields, C.L.; Cebulla, C.M.; Abdel-Rahman, M.H.; Grossniklaus, H.E.; Stern, M.H.; Carvajal, R.D.; Belfort, R.N.; Jia, R.; Shields, J.A.; et al. Uveal melanoma. Nat. Rev. Dis. Primers 2020, 6, 24. [Google Scholar] [CrossRef] [PubMed]
- Damato, E.M.; Damato, B.E. Detection and time to treatment of uveal melanoma in the United Kingdom: An evaluation of 2,384 patients. Ophthalmology 2012, 119, 1582–1589. [Google Scholar] [CrossRef] [PubMed]
- Damato, B. Ocular treatment of choroidal melanoma in relation to the prevention of metastatic death—A personal view. Prog. Retin. Eye Res. 2018, 66, 187–199. [Google Scholar] [CrossRef]
- Zhang, Z.; Zeng, X.; Wu, Y.; Liu, Y.; Zhang, X.; Song, Z. Cuproptosis-Related Risk Score Predicts Prognosis and Characterizes the Tumor Microenvironment in Hepatocellular Carcinoma. Front. Immunol. 2022, 13, 925618. [Google Scholar] [CrossRef]
- Tsvetkov, P.; Coy, S.; Petrova, B.; Dreishpoon, M.; Verma, A.; Abdusamad, M.; Rossen, J.; Joesch-Cohen, L.; Humeidi, R.; Spangler, R.D.; et al. Copper induces cell death by targeting lipoylated TCA cycle proteins. Science 2022, 375, 1254–1261. [Google Scholar] [CrossRef]
- Lv, H.; Liu, X.; Zeng, X.; Liu, Y.; Zhang, C.; Zhang, Q.; Xu, J. Comprehensive Analysis of Cuproptosis-Related Genes in Immune Infiltration and Prognosis in Melanoma. Front. Pharmacol. 2022, 13, 930041. [Google Scholar] [CrossRef]
- Wang, W.; Lu, Z.; Wang, M.; Liu, Z.; Wu, B.; Yang, C.; Huan, H.; Gong, P. The cuproptosis-related signature associated with the tumor environment and prognosis of patients with glioma. Front. Immunol. 2022, 13, 998236. [Google Scholar] [CrossRef]
- Zhang, G.; Sun, J.; Zhang, X. A novel Cuproptosis-related LncRNA signature to predict prognosis in hepatocellular carcinoma. Sci. Rep. 2022, 12, 11325. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Q.; Hu, X.M.; Zhao, W.J.; Ban, X.X.; Li, Y.; Huang, Y.X.; Wan, H.; He, Y.; Liao, L.S.; Shang, L.; et al. Targeting Necroptosis: A Novel Therapeutic Option for Retinal Degenerative Diseases. Int. J. Biol. Sci. 2023, 19, 658–674. [Google Scholar] [CrossRef] [PubMed]
- Yan, W.T.; Zhao, W.J.; Hu, X.M.; Ban, X.X.; Ning, W.Y.; Wan, H.; Zhang, Q.; Xiong, K. PANoptosis-like cell death in ischemia/reperfusion injury of retinal neurons. Neural Regen. Res. 2023, 18, 357–363. [Google Scholar] [CrossRef] [PubMed]
- Hoth, M.; Niemeyer, B.A. The neglected CRAC proteins: Orai2, Orai3, and STIM2. Curr. Top. Membr. 2013, 71, 237–271. [Google Scholar] [CrossRef] [PubMed]
- Wu, S.; Chen, M.; Huang, J.; Zhang, F.; Lv, Z.; Jia, Y.; Cui, Y.Z.; Sun, L.Z.; Wang, Y.; Tang, Y.; et al. ORAI2 Promotes Gastric Cancer Tumorigenicity and Metastasis through PI3K/Akt Signaling and MAPK-Dependent Focal Adhesion Disassembly. Cancer Res. 2021, 81, 986–1000. [Google Scholar] [CrossRef]
- Sanchez-Collado, J.; Lopez, J.J.; Cantonero, C.; Jardin, I.; Regodon, S.; Redondo, P.C.; Gordillo, J.; Smani, T.; Salido, G.M.; Rosado, J.A. Orai2 Modulates Store-Operated Ca2+ Entry and Cell Cycle Progression in Breast Cancer Cells. Cancers 2021, 14, 114. [Google Scholar] [CrossRef]
- Singh, A.K.; Roy, N.K.; Bordoloi, D.; Padmavathi, G.; Banik, K.; Khwairakpam, A.D.; Kunnumakkara, A.B.; Sukumar, P. Orai-1 and Orai-2 regulate oral cancer cell migration and colonisation by suppressing Akt/mTOR/NF-kappaB signalling. Life Sci. 2020, 261, 118372. [Google Scholar] [CrossRef]
- Arden, K.C.; Viars, C.S.; Fu, K.; Rozen, R. Localization of short/branched chain acyl-CoA dehydrogenase (ACADSB) to human chromosome 10. Genomics 1995, 25, 743–745. [Google Scholar] [CrossRef]
- Lu, D.; Yang, Z.; Xia, Q.; Gao, S.; Sun, S.; Luo, X.; Li, Z.; Zhang, X.; Li, X. ACADSB regulates ferroptosis and affects the migration, invasion, and proliferation of colorectal cancer cells. Cell Biol. Int. 2020, 44, 2334–2343. [Google Scholar] [CrossRef]
- Liu, X.; Zhang, W.; Wang, H.; Zhu, L.; Xu, K. Decreased Expression of ACADSB Predicts Poor Prognosis in Clear Cell Renal Cell Carcinoma. Front. Oncol. 2021, 11, 762629. [Google Scholar] [CrossRef]
- Yao, X.; Watkins, N.H.; Brown-Harding, H.; Bierbach, U. A membrane transporter determines the spectrum of activity of a potent platinum-acridine hybrid anticancer agent. Sci. Rep. 2020, 10, 15201. [Google Scholar] [CrossRef] [PubMed]
- Robertson, A.G.; Shih, J.; Yau, C.; Gibb, E.A.; Oba, J.; Mungall, K.L.; Hess, J.M.; Uzunangelov, V.; Walter, V.; Danilova, L.; et al. Integrative Analysis Identifies Four Molecular and Clinical Subsets in Uveal Melanoma. Cancer Cell 2017, 32, 204–220.e15. [Google Scholar] [CrossRef]
- Amaro, A.; Gangemi, R.; Piaggio, F.; Angelini, G.; Barisione, G.; Ferrini, S.; Pfeffer, U. The biology of uveal melanoma. Cancer Metastasis Rev. 2017, 36, 109–140. [Google Scholar] [CrossRef] [PubMed]
- Durante, M.A.; Rodriguez, D.A.; Kurtenbach, S.; Kuznetsov, J.N.; Sanchez, M.I.; Decatur, C.L.; Snyder, H.; Feun, L.G.; Livingstone, A.S.; Harbour, J.W. Single-cell analysis reveals new evolutionary complexity in uveal melanoma. Nat. Commun. 2020, 11, 496. [Google Scholar] [CrossRef]
- Kraehenbuehl, L.; Holland, A.; Armstrong, E.; O’Shea, S.; Mangarin, L.; Chekalil, S.; Johnston, A.; Bomalaski, J.S.; Erinjeri, J.P.; Barker, C.A.; et al. Pilot Trial of Arginine Deprivation Plus Nivolumab and Ipilimumab in Patients with Metastatic Uveal Melanoma. Cancers 2022, 14, 2638. [Google Scholar] [CrossRef] [PubMed]
- Pant, A.; Medikonda, R.; Lim, M. Alternative Checkpoints as Targets for Immunotherapy. Curr. Oncol. Rep. 2020, 22, 126. [Google Scholar] [CrossRef]
- Tocheva, A.S.; Mor, A. Checkpoint Inhibitors: Applications for Autoimmunity. Curr. Allergy Asthma Rep. 2017, 17, 72. [Google Scholar] [CrossRef] [PubMed]
- Karydis, I.; Chan, P.Y.; Wheater, M.; Arriola, E.; Szlosarek, P.W.; Ottensmeier, C.H. Clinical activity and safety of Pembrolizumab in Ipilimumab pre-treated patients with uveal melanoma. Oncoimmunology 2016, 5, e1143997. [Google Scholar] [CrossRef]
- Kottschade, L.A.; McWilliams, R.R.; Markovic, S.N.; Block, M.S.; Villasboas Bisneto, J.; Pham, A.Q.; Esplin, B.L.; Dronca, R.S. The use of pembrolizumab for the treatment of metastatic uveal melanoma. Melanoma Res. 2016, 26, 300–303. [Google Scholar] [CrossRef]
- Zhao, L.; Xia, W.; Zhang, Y.; Zou, P.; Zhu, Q.; Zhang, R. Efficacy and Safety of Immune Checkpoint Blockades in the Treatment of Ocular Melanoma: A Systematic Review and Meta-Analysis. Front. Oncol. 2021, 11, 781162. [Google Scholar] [CrossRef]
- Meyer, K.D.; Jaffrey, S.R. The dynamic epitranscriptome: N6-methyladenosine and gene expression control. Nat. Rev. Mol. Cell Biol. 2014, 15, 313–326. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; He, Q.; Shen, H.; Xia, A.; Tian, W.; Yu, W.; Sun, B. TOX promotes the exhaustion of antitumor CD8+ T cells by preventing PD1 degradation in hepatocellular carcinoma. J. Hepatol. 2019, 71, 731–741. [Google Scholar] [CrossRef] [PubMed]
- Luo, H.; Ma, C. A Novel Ferroptosis-Associated Gene Signature to Predict Prognosis in Patients with Uveal Melanoma. Diagnostics 2021, 11, 219. [Google Scholar] [CrossRef] [PubMed]









| Characteristic | Training Group (n = 80) | Testing Group (n = 63) | Total (n = 143) | p Value |
|---|---|---|---|---|
| Age | 0.36 | |||
| <65 | 36 (45.00%) | 9 (11.25%) | 45 (56.25%) | |
| ≥65 | 24 (30.00%) | 11 (13.75%) | 35 (43.75%) | |
| Gender | 1.00 | |||
| Female | 26 (32.50%) | 9 (11.25%) | 35 (43.75%) | |
| Male | 34 (42.50%) | 11 (13.75%) | 45 (56.25%) | |
| T classification | 0.58 | |||
| T2 | 4 (5.00%) | 1 (1.25%) | 5 (6.25%) | |
| T3 | 25 (31.25%) | 11 (13.75%) | 36 (45.00%) | |
| T4 | 31 (38.75%) | 8 (10.00%) | 39 (48.75%) | |
| N classification | 0.55 | |||
| N0 | 56 (70.00%) | 20 (25.00%) | 76 (95.00%) | |
| NX | 4 (5.00%) | 0 (0.00 + 0%) | 4 (5.00%) | |
| M classification | 0.94 | |||
| M0 | 55 (68.75%) | 18 (22.50%) | 73 (91.25%) | |
| M1 | 2 (2.50%) | 1 (1.25%) | 3 (3.75%) | |
| MX | 3 (3.75%) | 1 (1.25%) | 4 (5.00%) | |
| Tumor stage | 0.49 | |||
| Stage II | 27 (33.75%) | 12 (15.00%) | 39 (48.75%) | |
| Stage III | 30 (37.50%) | 7 (8.75%) | 37 (46.25%) | |
| Stage IV | 3 (3.75%) | 1 (1.25%) | 4 (5.00%) | |
| Tumor thickness, mm | 0.69 | |||
| <10 | 23 (28.75%) | 6 (7.50%) | 29 (36.25%) | |
| ≥10 | 37 (46.25%) | 14 (17.50%) | 51 (63.75%) | |
| Tumor diameter, mm | 0.16 | |||
| <20 | 41 (51.25%) | 18 (22.50%) | 59 (73.75%) | |
| NA | 1 (1.25%) | 0 (0.00 + 0%) | 1 (1.25%) | |
| ≥20 | 18 (22.50%) | 2 (2.50%) | 20 (25.00%) |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Huang, W.; Yang, F.; Zhang, Y.; Fang, Q.; Lai, Y.; Lan, Y. A Newly Established Cuproptosis-Related Gene Signature for Predicting Prognosis and Immune Infiltration in Uveal Melanoma. Int. J. Mol. Sci. 2023, 24, 11358. https://doi.org/10.3390/ijms241411358
Huang W, Yang F, Zhang Y, Fang Q, Lai Y, Lan Y. A Newly Established Cuproptosis-Related Gene Signature for Predicting Prognosis and Immune Infiltration in Uveal Melanoma. International Journal of Molecular Sciences. 2023; 24(14):11358. https://doi.org/10.3390/ijms241411358
Chicago/Turabian StyleHuang, Wei, Fan Yang, Yichi Zhang, Qianqi Fang, Yitao Lai, and Yuqing Lan. 2023. "A Newly Established Cuproptosis-Related Gene Signature for Predicting Prognosis and Immune Infiltration in Uveal Melanoma" International Journal of Molecular Sciences 24, no. 14: 11358. https://doi.org/10.3390/ijms241411358
APA StyleHuang, W., Yang, F., Zhang, Y., Fang, Q., Lai, Y., & Lan, Y. (2023). A Newly Established Cuproptosis-Related Gene Signature for Predicting Prognosis and Immune Infiltration in Uveal Melanoma. International Journal of Molecular Sciences, 24(14), 11358. https://doi.org/10.3390/ijms241411358






