Gut Microbiome and Microbiome-Derived Metabolites in Patients with End-Stage Kidney Disease
Abstract
1. Introduction
2. Results
2.1. Clinical and Biochemical Characteristics
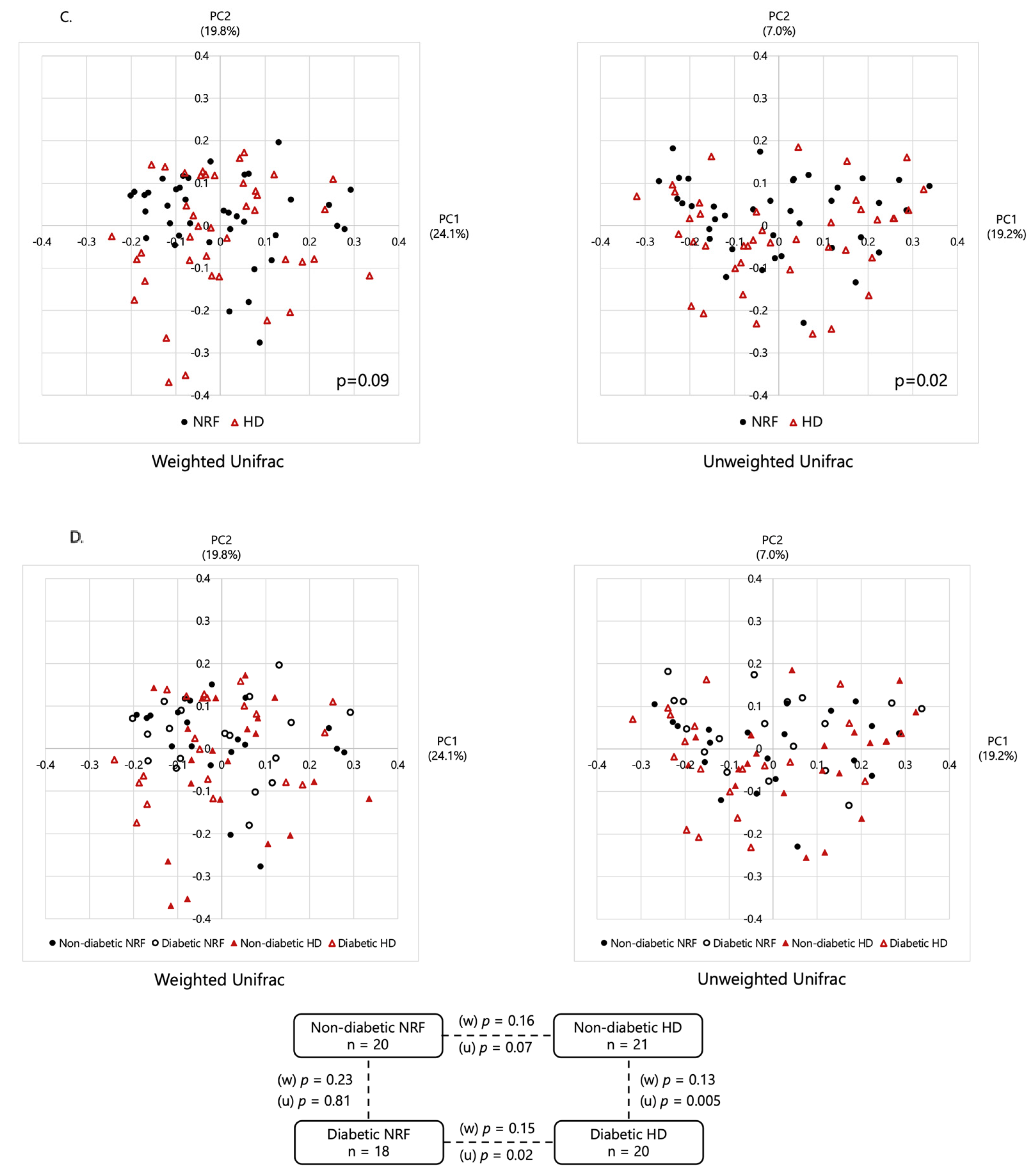
2.2. Microbial Diversity
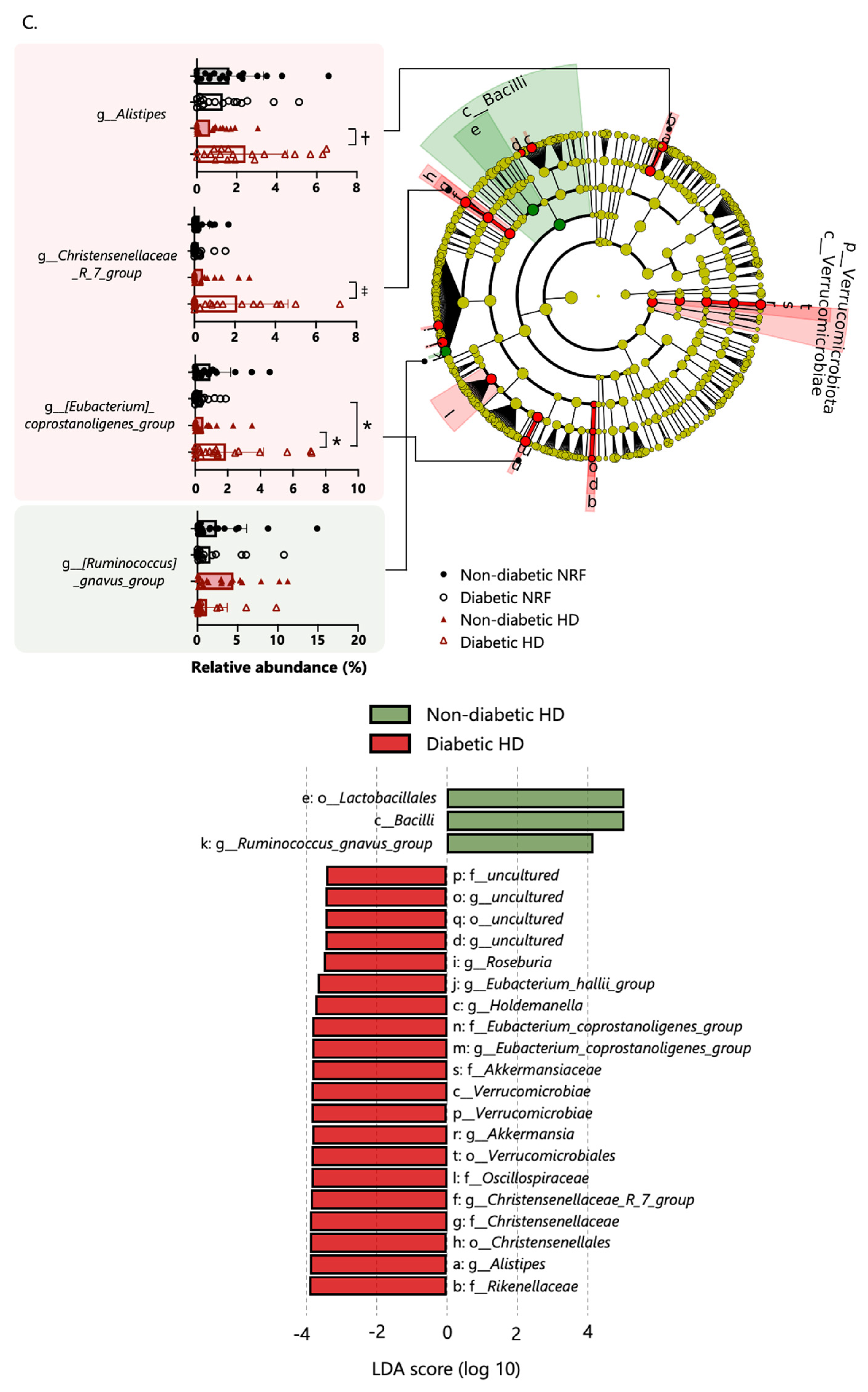
2.3. Bacterial Taxonomic Differences
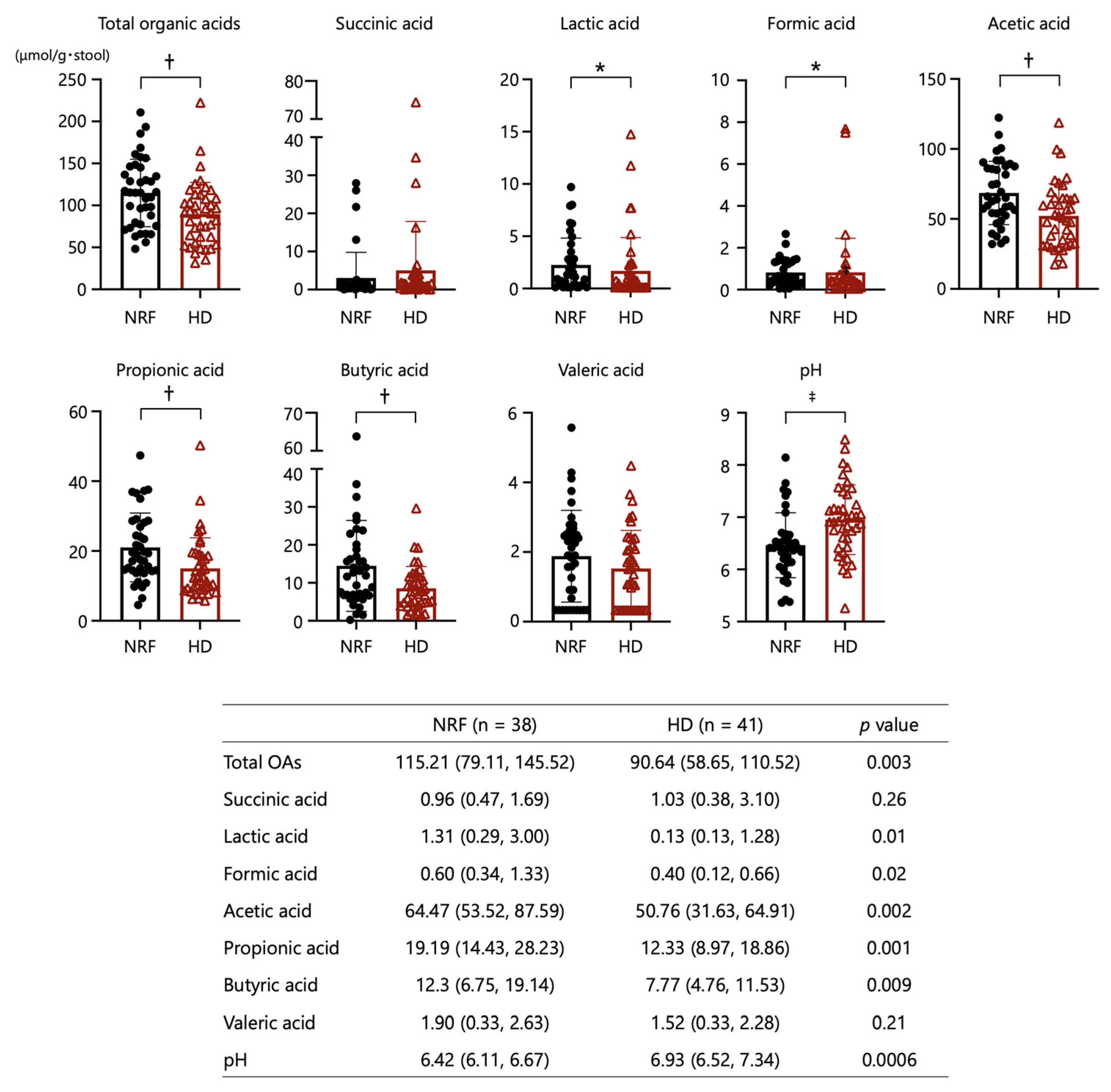
2.4. Organic Acids and pH
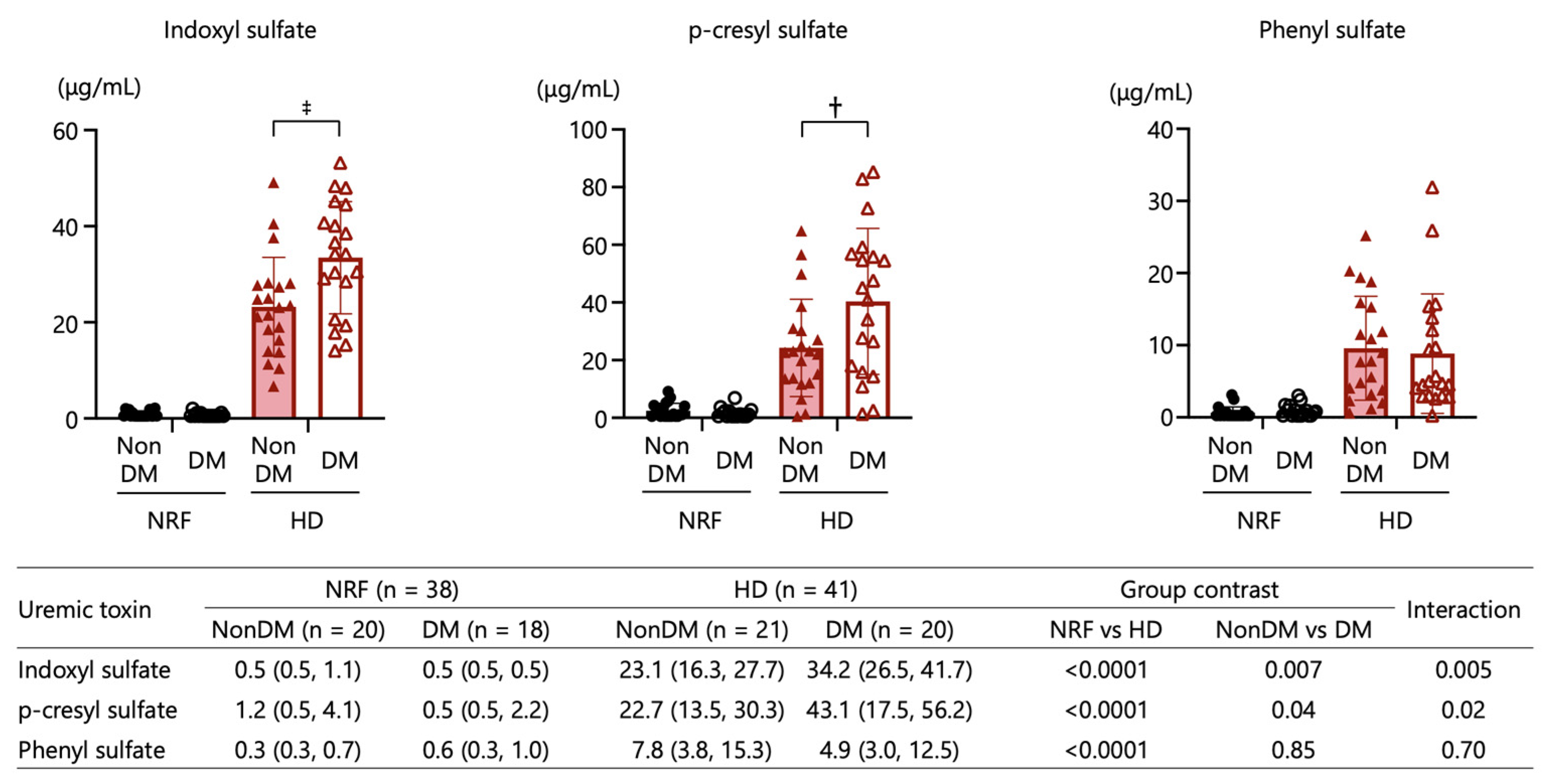
2.5. Uremic Toxins
2.6. Functional Prediction
2.7. Associations among Organic Acids, Inflammatory Markers, and Uremic Toxins
2.8. Microbiomes Associated with ESKD or Uremic Toxins
3. Discussion
4. Materials and Methods
4.1. Research Design
4.2. Collection of Stool Samples and DNA Extraction
4.3. 16S ribosomal RNA Gene Amplicon Sequencing and Analysis
4.4. Measurements of Fecal Organic Acids and pH
4.5. Measurements of Inflammatory Markers and Uremic Toxins
4.6. Functional Prediction of Microbial Communities
4.7. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| AKI | acute kidney injury |
| ANOVA | analysis of variance |
| ASVs | amplicon sequence variants |
| BMI | body mass index |
| BP | blood pressure |
| CKD | chronic kidney disease |
| DKD | diabetic kidney disease |
| DPP-4 | dipeptidyl peptidase-4 |
| eGFR | estimated glomerular filtration rate |
| ELISA | enzyme-linked immunosorbent assay |
| ESKD | end-stage kidney disease |
| GLP-1RA | glucagon-like peptide-1 receptor agonist |
| GPR | G protein-coupled receptor |
| HD | hemodialysis |
| hsCRP | high-sensitive c-reactive protein |
| IL | interleukin |
| IS | indoxyl sulfate |
| KEGG | Kyoto Encyclopedia of Genes and Genomes |
| LBP | lipopolysaccharide-binding protein |
| LC-MS/MS | liquid chromatography coupled to dual mass spectrometry |
| LDA | linear discriminant analysis |
| LEfSe | linear discriminant analysis effect size |
| LPS | lipopolysaccharide |
| NOS | nitric oxide synthase |
| NRF | normal renal function |
| OA | organic acid |
| UT | uremic toxin |
| pCS | p-cresyl sulfate |
| PERMANOVA | permutational multivariate analysis of variance |
| PICRUSt2 | phylogenetic investigation of communities by reconstruction of unobserved states 2 |
| PPI | proton pump inhibitor |
| PS | phenyl sulfate |
| SCFA | short-chain fatty acid |
| SGLT2 | sodium-glucose cotransporter-2 |
| TNF | tumor necrosis factor |
| α-GI | alpha-glucosidase inhibitor |
References
- Nenci, A.; Becker, C.; Wullaert, A.; Gareus, R.; van Loo, G.; Danese, S.; Huth, M.; Nikolaev, A.; Neufert, C.; Madison, B.; et al. Epithelial NEMO links innate immunity to chronic intestinal inflammation. Nature 2007, 446, 557–561. [Google Scholar] [CrossRef]
- Anders, H.J.; Andersen, K.; Stecher, B. The intestinal microbiota, a leaky gut, and abnormal immunity in kidney disease. Kidney Int. 2013, 83, 1010–1016. [Google Scholar] [CrossRef]
- Zhao, L.; Zhang, F.; Ding, X.; Wu, G.; Lam, Y.Y.; Wang, X.; Fu, H.; Xue, X.; Lu, C.; Ma, J.; et al. Gut bacteria selectively promoted by dietary fibers alleviate type 2 diabetes. Science 2018, 359, 1151–1156. [Google Scholar] [CrossRef]
- Vaziri, N.D.; Wong, J.; Pahl, M.; Piceno, Y.M.; Yuan, J.; DeSantis, T.Z.; Ni, Z.; Nguyen, T.-H.; Andersen, G.L. Chronic kidney disease alters intestinal microbial flora. Kidney Int. 2013, 83, 308–315. [Google Scholar] [CrossRef]
- Sato, J.; Kanazawa, A.; Ikeda, F.; Yoshihara, T.; Goto, H.; Abe, H.; Komiya, K.; Kawaguchi, M.; Shimizu, T.; Ogihara, T.; et al. Gut Dysbiosis and Detection of “Live Gut Bacteria” in Blood of Japanese Patients With Type 2 Diabetes. Diabetes Care 2014, 37, 2343–2350. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.J.; Chen, X.; Kwan, T.K.; Loh, Y.W.; Singer, J.; Liu, Y.; Ma, J.; Tan, J.; Macia, L.; Mackay, C.R.; et al. Dietary Fiber Protects against Diabetic Nephropathy through Short-Chain Fatty Acid–Mediated Activation of G Protein–Coupled Receptors GPR43 and GPR109A. J. Am. Soc. Nephrol. 2020, 31, 1267–1281. [Google Scholar] [CrossRef] [PubMed]
- Yang, T.; Richards, E.M.; Pepine, C.J.; Raizada, M.K. The gut microbiota and the brain–gut–kidney axis in hypertension and chronic kidney disease. Nat. Rev. Nephrol. 2018, 14, 442–456. [Google Scholar] [CrossRef] [PubMed]
- Lekawanvijit, S.; Adrahtas, A.; Kelly, D.J.; Kompa, A.R.; Wang, B.H.; Krum, H. Does indoxyl sulfate, a uraemic toxin, have direct effects on cardiac fibroblasts and myocytes? Eur. Heart J. 2010, 31, 1771–1779. [Google Scholar] [CrossRef] [PubMed]
- Lano, G.; Burtey, S.; Sallée, M. Indoxyl Sulfate, a Uremic Endotheliotoxin. Toxins 2020, 12, 229. [Google Scholar] [CrossRef]
- Kikuchi, K.; Saigusa, D.; Kanemitsu, Y.; Matsumoto, Y.; Thanai, P.; Suzuki, N.; Mise, K.; Yamaguchi, H.; Nakamura, T.; Asaji, K.; et al. Gut microbiome-derived phenyl sulfate contributes to albuminuria in diabetic kidney disease. Nat. Commun. 2019, 10, 1835. [Google Scholar] [CrossRef]
- Marzocco, S.; Fazeli, G.; Di Micco, L.; Autore, G.; Adesso, S.; Piaz, F.D.; Heidland, A.; Di Iorio, B. Supplementation of Short-Chain Fatty Acid, Sodium Propionate, in Patients on Maintenance Hemodialysis: Beneficial Effects on Inflammatory Parameters and Gut-Derived Uremic Toxins, A Pilot Study (PLAN Study). J. Clin. Med. 2018, 7, 315. [Google Scholar] [CrossRef] [PubMed]
- Borges, N.A.; Barros, A.F.; Nakao, L.; Dolenga, C.J.; Fouque, D.; Mafra, D. Protein-Bound Uremic Toxins from Gut Microbiota and Inflammatory Markers in Chronic Kidney Disease. J. Ren. Nutr. 2016, 26, 396–400. [Google Scholar] [CrossRef]
- Andrade-Oliveira, V.; Amano, M.T.; Correa-Costa, M.; Castoldi, A.; Felizardo, R.J.; de Almeida, D.C.; Bassi, E.J.; Moraes-Vieira, P.M.; Hiyane, M.I.; Rodas, A.C.; et al. Gut Bacteria Products Prevent AKI Induced by Ischemia-Reperfusion. J. Am. Soc. Nephrol. 2015, 26, 1877–1888. [Google Scholar] [CrossRef]
- Masui, R.; Sasaki, M.; Funaki, Y.; Ogasawara, N.; Mizuno, M.; Iida, A.; Izawa, S.; Kondo, Y.; Ito, Y.; Tamura, Y.; et al. G Protein-Coupled Receptor 43 Moderates Gut Inflammation Through Cytokine Regulation from Mononuclear Cells. Inflamm. Bowel Dis. 2013, 19, 2848–2856. [Google Scholar] [CrossRef] [PubMed]
- Vinolo, M.A.R.; Rodrigues, H.G.; Hatanaka, E.; Sato, F.T.; Sampaio, S.C.; Curi, R. Suppressive effect of short-chain fatty acids on production of proinflammatory mediators by neutrophils. J. Nutr. Biochem. 2011, 22, 849–855. [Google Scholar] [CrossRef]
- Walker, A.W.; Duncan, S.H.; McWilliam Leitch, E.C.; Child, M.W.; Flint, H.J. pH and Peptide Supply Can Radically Alter Bacterial Populations and Short-Chain Fatty Acid Ratios within Microbial Communities from the Human Colon. Appl. Environ. Microbiol. 2005, 71, 3692–3700. [Google Scholar] [CrossRef] [PubMed]
- Sakon, H.; Nagai, F.; Morotomi, M.; Tanaka, R. Sutterella parvirubra sp. nov. and Megamonas funiformis sp. nov., isolated from human faeces. Int. J. Syst. Evol. Microbiol. 2008, 58 Pt 4, 970–975. [Google Scholar] [CrossRef]
- Lun, H.; Yang, W.; Zhao, S.; Jiang, M.; Xu, M.; Liu, F.; Wang, Y. Altered gut microbiota and microbial biomarkers associated with chronic kidney disease. Microbiologyopen 2019, 8, e00678. [Google Scholar] [CrossRef]
- Sommer, F.; Bäckhed, F. The gut microbiota—masters of host development and physiology. Nat. Rev. Microbiol. 2013, 11, 227–238. [Google Scholar] [CrossRef]
- Li, Q.; Zhang, S.; Wu, Q.-J.; Xiao, J.; Wang, Z.-H.; Mu, X.-W.; Zhang, Y.; Wang, X.-N.; You, L.-L.; Wang, S.-N.; et al. Serum total indoxyl sulfate levels and all-cause and cardiovascular mortality in maintenance hemodialysis patients: A prospective cohort study. BMC Nephrol. 2022, 23, 231. [Google Scholar] [CrossRef]
- Itoh, Y.; Ezawa, A.; Kikuchi, K.; Tsuruta, Y.; Niwa, T. Correlation between Serum Levels of Protein-Bound Uremic Toxins in Hemodialysis Patients Measured by LC/MS/MS. Mass Spectrom. 2013, 2, S0017. [Google Scholar] [CrossRef] [PubMed]
- Kaur, H.; Das, C.; Mande, S.S. In Silico Analysis of Putrefaction Pathways in Bacteria and Its Implication in Colorectal Cancer. Front. Microbiol. 2017, 8, 2166. [Google Scholar] [CrossRef] [PubMed]
- Bone, E.; Tamm, A.; Hill, M. The production of urinary phenols by gut bacteria and their possible ce:role in the causation of large bowel cancer. Am. J. Clin. Nutr. 1976, 29, 1448–1454. [Google Scholar] [CrossRef]
- Hansen, S.H.; Andersen, M.L.; Birkedal, H.; Cornett, C.; Wibrand, F. The Important Role of Taurine in Oxidative Metabolism. Adv. Exp. Med. Biol. 2006, 583, 129–135. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Su, X.; Zhang, L.; Liu, Y.; Shi, M.; Lv, C.; Gao, Y.; Xu, D.; Wang, Z. Dysbiosis of the gut microbiome is associated with CKD5 and correlated with clinical indices of the disease: A case–controlled study. J. Transl. Med. 2019, 17, 228. [Google Scholar] [CrossRef] [PubMed]
- Louca, P.; Nogal, A.; Wells, P.M.; Asnicar, F.; Wolf, J.; Steves, C.J.; Spector, T.D.; Segata, N.; Berry, S.E.; Valdes, A.M.; et al. Gut microbiome diversity and composition is associated with hypertension in women. J. Hypertens. 2021, 39, 1810–1816. [Google Scholar] [CrossRef]
- Li, Q.; Chang, Y.; Zhang, K.; Chen, H.; Tao, S.; Zhang, Z. Implication of the gut microbiome composition of type 2 diabetic patients from northern China. Sci. Rep. 2020, 10, 5450. [Google Scholar] [CrossRef]
- Lecamwasam, A.; Nelson, T.M.; Rivera, L.; Ekinci, E.I.; Saffery, R.; Dwyer, K.M. Gut Microbiome Composition Remains Stable in Individuals with Diabetes-Related Early to Late Stage Chronic Kidney Disease. Biomedicines 2020, 9, 19. [Google Scholar] [CrossRef] [PubMed]
- Imhann, F.; Bonder, M.J.; Vila, A.V.; Fu, J.; Mujagic, Z.; Vork, L.; Tigchelaar, E.F.; Jankipersadsing, S.A.; Cenit, M.C.; Harmsen, H.J.M.; et al. Proton pump inhibitors affect the gut microbiome. Gut 2016, 65, 740–748. [Google Scholar] [CrossRef]
- Saadat, Y.R.; Niknafs, B.; Khatibi, S.M.H.; Ardalan, M.; Majdi, H.; Bahmanpoor, Z.; Abediazar, S.; Vahed, S.Z. Gut microbiota; an overlooked effect of phosphate binders. Eur. J. Pharmacol. 2020, 868, 172892. [Google Scholar] [CrossRef]
- Yang, M.; Shi, F.-H.; Liu, W.; Zhang, M.-C.; Feng, R.-L.; Qian, C.; Liu, W.; Ma, J. Dapagliflozin Modulates the Fecal Microbiota in a Type 2 Diabetic Rat Model. Front. Endocrinol. 2020, 11, 635. [Google Scholar] [CrossRef]
- Ryan, P.M.; Patterson, E.; Carafa, I.; Mandal, R.; Wishart, D.S.; Dinan, T.G.; Cryan, J.F.; Tuohy, K.M.; Stanton, C.; Ross, R.P. Metformin and Dipeptidyl Peptidase-4 Inhibitor Differentially Modulate the Intestinal Microbiota and Plasma Metabolome of Metabolically Dysfunctional Mice. Can. J. Diabetes 2020, 44, 146–155.e2. [Google Scholar] [CrossRef]
- Zhang, L.; Song, P.; Zhang, X.; Metea, C.; Schleisman, M.; Karstens, L.; Leung, E.; Zhang, J.; Xu, Q.; Liu, Y.; et al. Alpha-Glucosidase Inhibitors Alter Gut Microbiota and Ameliorate Collagen-Induced Arthritis. Front. Pharmacol. 2020, 10, 1684. [Google Scholar] [CrossRef] [PubMed]
- Kubota, H.; Tsuji, H.; Matsuda, K.; Kurakawa, T.; Asahara, T.; Nomoto, K. Detection of Human Intestinal Catalase-Negative, Gram-Positive Cocci by rRNA-Targeted Reverse Transcription-PCR. Appl. Environ. Microbiol. 2010, 76, 5440–5451. [Google Scholar] [CrossRef] [PubMed]
- Matsuki, T.; Watanabe, K.; Fujimoto, J.; Takada, T.; Tanaka, R. Use of 16S rRNA Gene-Targeted Group-Specific Primers for Real-Time PCR Analysis of Predominant Bacteria in Human Feces. Appl. Environ. Microbiol. 2004, 70, 7220–7228. [Google Scholar] [CrossRef] [PubMed]
- Sugimoto, T.; Shima, T.; Amamoto, R.; Kaga, C.; Kado, Y.; Watanabe, O.; Shiinoki, J.; Iwazaki, K.; Shigemura, H.; Tsuji, H.; et al. Impacts of Habitual Diets Intake on Gut Microbial Counts in Healthy Japanese Adults. Nutrients 2020, 12, 2414. [Google Scholar] [CrossRef] [PubMed]
- Caporaso, J.G.; Lauber, C.L.; Walters, W.A.; Berg-Lyons, D.; Lozupone, C.A.; Turnbaugh, P.J.; Fierer, N.; Knight, R. Global patterns of 16S rRNA diversity at a depth of millions of sequences per sample. Proc. Natl. Acad. Sci. USA 2011, 108 (Suppl. S1), 4516–4522. [Google Scholar] [CrossRef]
- Douglas, G.M.; Maffei, V.J.; Zaneveld, J.R.; Yurgel, S.N.; Brown, J.R.; Taylor, C.M.; Huttenhower, C.; Langille, M.G.I. PICRUSt2 for prediction of metagenome functions. Nat. Biotechnol. 2020, 38, 685–688. [Google Scholar] [CrossRef]
- Brown, C.T.; Davis-Richardson, A.G.; Giongo, A.; Gano, K.A.; Crabb, D.B.; Mukherjee, N.; Casella, G.; Drew, J.C.; Ilonen, J.; Knip, M.; et al. Gut Microbiome Metagenomics Analysis Suggests a Functional Model for the Development of Autoimmunity for Type 1 Diabetes. PLoS ONE 2011, 6, e25792. [Google Scholar] [CrossRef]



| Normal Renal Function (n = 38) | Hemodialysis (n = 41) | p Value | |
|---|---|---|---|
| Clinical characteristics | |||
| eGFR (mL/min/1.73 m2) | 77.1 ± 15.3 | NA | |
| Age (years) | 69.1 ± 5.8 | 71.2 ± 5.0 | 0.08 |
| Male (%) | 65.8% | 58.5% | 0.64 |
| BMI | 24.3 ± 3.2 | 22.8 ± 3.7 | 0.07 |
| Systolic BP (mmHg) | 127 ± 12 | 149 ± 24 | <0.001 |
| Diastolic BP (mmHg) | 75 ± 9 | 77 ± 15 | 0.61 |
| Type 2 diabetes (%) | 47.4% | 48.8% | >0.999 |
| Inflammatory markers | |||
| hsCRP (mg/dL) | 0.058 (0.027, 0.113) | 0.096 (0.032, 0.219) | 0.049 |
| LBP (ng/mL) | 13.1 (11.3, 14.4) | 15.4 (13.4, 20.0) | <0.001 |
| Medication | |||
| Proton pump inhibitor | 7.9% | 63.4% | <0.001 |
| Phosphate binder | 0.0% | 80.5% | <0.001 |
| DPP-4 inhibitor | 36.8% | 24.4% | 0.33 |
| α-GI | 5.3% | 7.3% | >0.999 |
| Glinide | 7.9% | 7.3% | >0.999 |
| GLP-1 receptor agonist | 0.0% | 4.9% | 0.49 |
| SGLT2 inhibitor | 10.5% | 0.0% | 0.049 |
| Metformin | 15.8% | 0.0% | 0.01 |
| Insulin | 5.3% | 0.0% | 0.23 |
| Acetic Acid | Propionic Acid | Butyric Acid | Formic Acid | Lactic Acid | hsCRP | LBP | IS | pCS | PS | |
|---|---|---|---|---|---|---|---|---|---|---|
| Total organic acid | 0.91 § | 0.79 § | 0.73 § | 0.11 | 0.47 § | −0.04 | −0.21 | −0.34 † | −0.33 † | −0.27 * |
| Acetic acid | 0.69 § | 0.66 § | 0.13 | 0.37 ‡ | −0.01 | −0.17 | −0.34 † | −0.32 † | −0.25 * | |
| Propionic acid | 0.57 § | −0.04 | 0.34 † | 0.01 | −0.18 | −0.28 * | −0.35 † | −0.15 | ||
| Butyric acid | 0.01 | 0.26 * | 0.04 | −0.14 | −0.26 * | −0.15 | −0.19 | |||
| Formic acid | 0.42 ‡ | 0.04 | 0.00 | −0.22 | −0.29 * | −0.18 | ||||
| Lactic acid | −0.01 | −0.07 | −0.30 † | −0.42 ‡ | −0.26 * | |||||
| hsCRP | 0.67 § | −0.21 | 0.13 | 0.24 * | ||||||
| LBP | 0.43 § | 0.27 * | 0.44 § | |||||||
| IS | 0.82 § | 0.80 § | ||||||||
| p-CS | 0.60 § |
| Variable | OR (95% CI) | p Value |
|---|---|---|
| Total organic acid | 0.46 (0.27–0.78) | 0.004 |
| Acetic acid | 0.43 (0.25–0.74) | 0.002 |
| Propionic acid | 0.49 (0.29–0.81) | 0.005 |
| Butyric acid | 0.61 (0.37–1.01) | 0.055 |
| Formic acid | 0.62 (0.38–1.00) | 0.051 |
| Lactic acid | 0.54 (0.33–0.87) | 0.01 |
| hsCRP | 1.63 (0.99–2.71) | 0.057 |
| LBP | 3.70 (1.70–8.07) | 0.001 |
| Microbiome | Model 1 | Model 2 |
|---|---|---|
| OR (95% CI) | ||
| c__Negativicutes | 0.37 (0.20, 0.69) † | 0.40 (0.20, 0.80) † |
| o__Veillonellales_Selenomonadales | 0.54 (0.33, 0.88) * | 0.56 (0.32, 0.98) * |
| o__Burkholderiales | 0.41 (0.22, 0.76) † | 0.58 (0.28, 1.22) |
| f__Selenomonadaceae | 0.32 (0.15, 0.68) † | 0.30 (0.11, 0.80) * |
| f__Sutterellaceae | 0.39 (0.22, 0.68) † | 0.48 (0.24, 0.94) * |
| g__Agathobacter | 0.60 (0.38, 0.95) * | 0.74 (0.41, 1.34) |
| g__Fusicatnibacter | 0.55 (0.34, 0.89) * | 0.46 (0.25, 0.86) * |
| g__Megamonas | 0.24 (0.08, 0.71) † | 0.26 (0.08, 0.86) * |
| c__Bacilli | 2.01 (1.21, 3.35) † | 2.25 (1.18, 4.30) * |
| o__Lactobacillales | 1.88 (1.15, 3.07) * | 1.38 (0.75, 2.52) |
| f__Streptococcaceae | 1.62 (1.01, 2.60) * | 1.42 (0.82, 2.46) |
| g__Clostridium_innocuum_group (*1) | 2.14 (1.29, 3.56) * | 2.05 (1.04, 4.05) * |
| g__Christensenellaceae_R_7_group (*2) | 1.49 (0.94, 2.34) | 2.01 (1.06–3.80) * |
| Microbiome | Model 1: IS | Model 2: pCS | Model 3: PS |
|---|---|---|---|
| Standardized Coefficient | |||
| o__Burkholderiales | −0.08 | −0.13 | −0.14 |
| f__Sutterellaceae | −0.13 | −0.11 | −0.26 * |
| g__UBA1819 (*1) | 0.06 | 0.11 | 0.16 |
| o__Clostridia.sp | 0.23 * | 0.41 ‡ | −0.03 |
| o__Christensenellales | 0.26 * | 0.38 ‡ | 0.04 |
| o__Clostridia_vadinBB60_group (*2) | 0.23 * | 0.36 † | −0.004 |
| f__Eggerthellaceae | 0.23 * | 0.24 * | 0.10 |
| c__Clostridia;__;__;__ | 0.23 * | 0.41 ‡ | −0.03 |
| f__Christensenellaceae | 0.26 * | 0.38 † | 0.04 |
| f__Clostridia_vadinBB60_group (*3) | 0.23 * | 0.36 † | −0.004 |
| g__[Clostridium]_innocuum_group (*4) | 0.14 | 0.18 | 0.24 * |
| c__Clostridia;__;__ | 0.23 * | 0.41 ‡ | −0.03 |
| g__Christensenellaceae_R_7_group (*5) | 0.27 † | 0.39 ‡ | 0.06 |
| g__Clostridia_vadinBB60_group (*6) | 0.23 * | 0.35 † | −0.004 |
| g__Family_XIII_AD3011_group (*7) | 0.19 | 0.34 † | 0.06 |
| g__Megamonas | −0.26 * | −0.23 * | −0.13 |
| c__Negativicutes | −0.16 | −0.15 | −0.08 |
| f__Selenomonadaceae | −0.26 † | −0.23 | −0.14 |
| g__Fusicatenibacter | −0.23 * | −0.20 | −0.16 |
| o__Clostridia | 0.13 | 0.34 † | 0.04 |
| f__Hungateiclostridiaceae | 0.13 | 0.34 † | 0.07 |
| f__Oscillospiraceae | 0.06 | 0.18 | −0.05 |
| f__Anaerovoracaceae | 0.17 | 0.30 † | 0.07 |
| g__Ruminiclostridium | 0.13 | 0.33 † | 0.06 |
| g__Intestinimonas | 0.22 | 0.37 † | 0.19 |
| g__NK4A214_group (*8) | 0.20 | 0.49 ‡ | −0.03 |
| f__Ruminococcaceae;__ | 0.10 | 0.30 † | −0.04 |
| g__Anaerofilum | 0.17 | 0.30 † | 0.04 |
| g__Negativibacillus | 0.15 | 0.31 † | −0.01 |
| g__[Eubacterium]_brachy_group (*7) | 0.09 | 0.29 † | 0.03 |
| o__Clostridiales | 0.10 | −0.02 | 0.26 * |
| f__Clostridiaceae | 0.10 | −0.02 | 0.26 * |
| g__Clostridium_sensu_stricto_1 | 0.09 | −0.03 | 0.28 * |
| g__GCA-900066755 (*9) | 0.12 | 0.02 | 0.39 ‡ |
| g__Parasutterella | −0.08 | −0.07 | −0.21 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Koshida, T.; Gohda, T.; Sugimoto, T.; Asahara, T.; Asao, R.; Ohsawa, I.; Gotoh, H.; Murakoshi, M.; Suzuki, Y.; Yamashiro, Y. Gut Microbiome and Microbiome-Derived Metabolites in Patients with End-Stage Kidney Disease. Int. J. Mol. Sci. 2023, 24, 11456. https://doi.org/10.3390/ijms241411456
Koshida T, Gohda T, Sugimoto T, Asahara T, Asao R, Ohsawa I, Gotoh H, Murakoshi M, Suzuki Y, Yamashiro Y. Gut Microbiome and Microbiome-Derived Metabolites in Patients with End-Stage Kidney Disease. International Journal of Molecular Sciences. 2023; 24(14):11456. https://doi.org/10.3390/ijms241411456
Chicago/Turabian StyleKoshida, Takeo, Tomohito Gohda, Takuya Sugimoto, Takashi Asahara, Rin Asao, Isao Ohsawa, Hiromichi Gotoh, Maki Murakoshi, Yusuke Suzuki, and Yuichiro Yamashiro. 2023. "Gut Microbiome and Microbiome-Derived Metabolites in Patients with End-Stage Kidney Disease" International Journal of Molecular Sciences 24, no. 14: 11456. https://doi.org/10.3390/ijms241411456
APA StyleKoshida, T., Gohda, T., Sugimoto, T., Asahara, T., Asao, R., Ohsawa, I., Gotoh, H., Murakoshi, M., Suzuki, Y., & Yamashiro, Y. (2023). Gut Microbiome and Microbiome-Derived Metabolites in Patients with End-Stage Kidney Disease. International Journal of Molecular Sciences, 24(14), 11456. https://doi.org/10.3390/ijms241411456








